Abstract
Anaerobic digestion is an effective method for reducing the by-product of waste-activated sludge (WAS) from wastewater treatment plants and for producing bioenergy from WAS. However, only a limited number of studies have attempted to improve anaerobic digestion by targeting the microbial interactions in WAS. In this study, we examined whether different antibiotics positively, negatively, or neutrally influence methane fermentation by evaluating changes in the microbial community and functions in WAS. Addition of azithromycin promoted the microbial communities related to the acidogenic and acetogenic stages, and a high concentration of soluble proteins and a high activity of methanogens were detected. Chloramphenicol inhibited methane production but did not affect the bacteria that contribute to the hydrolysis, acidogenesis, and acetogenesis digestion stages. The addition of kanamycin, which exhibits the same methane productivity as a control (antibiotic-free WAS), did not affect all of the microbial communities during anaerobic digestion. This study demonstrates the simultaneous functions and interactions of diverse bacteria and methanogenic Archaea in different stages of the anaerobic digestion of WAS. The ratio of Caldilinea, Methanosarcina, and Clostridium may correspond closely to the trend of methane production in each antibiotic. The changes in microbial activities and function by antibiotics facilitate a better understanding of bioenergy production.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Wastewater treatment plants (WWTP) systematically treat the wastewater collected from various sources such as households, industrial sites, and hospitals. The secondary treatment in this system creates an abundant by-product called waste-activated sludge (WAS). The Hiagari Wastewater Treatment Plant in Kitakyushu, Japan, produces approximately 7530 m3 of WAS daily (Maeda et al. 2011). Normally, further treatment of the generated WAS accounts for about 60 % of the total WWTP operating costs (Guo et al. 2015). Several attempts have been made to treat and reduce the amount of WAS that gets generated in WWTPs. An ultrasonic treatment was utilized to solubilize WAS (Bougrier et al. 2005; Dewil et al. 2006), and other treatments, such as thermal and chemical treatments (Bougrier et al. 2006; Valo et al. 2004), have been conducted to utilize WAS in bioenergy production.
Anaerobic digestion is another powerful approach that uses a biological reaction for the treatment of WAS. It has been widely applied as an efficient solution for environmental pollution and energy draining. Methane is considered to be the most promising bioenergy generated, because organic matter from WAS can be converted into 60–70 % volume of methane to meet both of the requirements of an efficient solution (Appels et al. 2008; Nguyen et al. 2014). Four anaerobic digestion stages are involved in the biological production of methane, namely hydrolysis, acidogenesis, acetogenesis, and methanogenesis (Bareither et al. 2013; Guo et al. 2015; Mohd Yasin et al. 2015; Nguyen et al. 2014). All stages in this process are driven by various types of microorganisms. WAS acts as a substrate in the process of anaerobic digestion for methane production. In the first hydrolysis stage, large WAS molecules, including proteins or carbohydrates, are degraded and converted into smaller molecules such as amino acids or monosaccharides. The fermentation of these molecules into carboxylic acids (e.g., butyrate or propionate), hydrogen, and carbon dioxide during the acidogenesis stage is combined with the acetogenesis stage, which is driven by acetogenic bacteria to produce acetate (Sanz et al. 1996). Finally, methane producers, Archaea or methanogens, utilize acetate to produce methane during the methanogenesis stage.
Thus, complex microbial communities in WAS are indeed essential for the anaerobic digestion process; however, the best conditions for methane fermentation from WAS have still not been found due to the poor investigations regarding microbial interactions and regulatory systems. Our previous study demonstrated the effect of azithromycin (AZM), a macrolide-type antibiotic for enhanced methane production from WAS (Nguyen et al. 2014). The results of a brief microbial community analysis by denaturing gradient gel electrophoresis (DGGE) determined the activation of Clostridium species. AZM functions to inhibit the biological protein synthesis through the interaction with the 50S subunit of the bacterial ribosome (Shimada et al. 2008). Chloramphenicol (CM) and kanamycin (KAN) antibiotics have the same inhibitory action as AZM; however, they may have a different antibacterial spectrum. The antibacterial spectrum for AZM is primarily gram-positive bacteria, including streptococci and staphylococci, Enterobacteriaceae, and other rapidly growing pyogenic bacteria (Papich 2010; Williams 1991). Besides, CM is well known as having a broad range of antibacterial spectra, including gram-positive, gram-negative, aerobic, and anaerobic bacteria (Schwarz et al. 2004). In addition, KAN antibacterial spectra activity includes gram-negative bacteria and some gram-positive bacteria, but it has a weak activity toward anaerobic bacteria (Papich 2010). Sanz et al. (1996) reported that CM inhibited anaerobic digestion systems even at a low concentration, whereas KAN showed a negligible effect. The different microbial communities under these antibiotics may provide valuable information toward our understanding of microbial interactions and regulatory systems during the anaerobic digestion process of WAS, because quite different patterns of microbial activity should be seen in the WAS with or without each antibiotic due to the different antibacterial spectra. Currently, the composition of a microbial community may be easily evaluated using molecular techniques. In fact, different methodologies have already been developed to determine the microbial community structure of various biological systems such as a clone library, next generation sequencing, fingerprinting techniques, real time polymerase chain reaction (PCR), and so on. Several investigative studies have been done to understand how microbial interactions are altered by antibiotics inside a complicated microbial consortium such as WAS, using next generation sequencing technology (Ng et al. 2015; Su et al. 2015; Zhang et al. 2011). To date, many studies have been carried out using less-advanced methods such as PCR-DGGE (Akyol et al. 2016; Aydin et al. 2015; Lins et al. 2015), terminal restriction fragment length polymorphism (Meng et al. 2015), or fluorescence in situ hybridization (Shimada et al. 2011). The DGGE application has great value for evaluating environmental microbiology but it does have some limitations. For example, the assumption that bands at the same position in the DGGE gel are equal to one genome is invalid. This is because bands that have a genetic similarity may not necessarily have similar migration patterns in the DGGE gel (Jackson et al. 2000). In addition, these conventional methods are unable to detect most of the low-abundance microbes that are present in WAS compared to next generation sequencing technology (Wang et al. 2012).
Therefore, in the situation of methane fermentation (anaerobic digestion) using WAS, the effect of several antibiotics (AZM, CM, and KAN) was investigated in this study to understand and detect the important functions for the efficient anaerobic digestion of WAS. In particular, a microbial community analysis using high-throughput sequencing technology (MiSeq Illumina platform) was performed by using the ribonucleic acid (RNA) extracted from WAS in the presence or absence of each antibiotic to evaluate the microbial dynamics of really active microbes during the fermentation from WAS.
Materials and methods
Activated sludge
Sewage sludge was taken from the Hiagari Wastewater Treatment Plant in Kitakyushu City, Japan. The fresh raw sludge was initially centrifuged at 8000×g for 10 min at 4 °C and the remaining pellet was resuspended with distilled water by shaking thoroughly. Washing steps were done three times before adjusting the final sludge concentration to 10 % (wet sludge weight/w) with distilled water. The sludge was used as the WAS in this study.
Antibiotics
AZM was purchased from LKT Laboratories Inc. (MN, USA) and CM and KAN were purchased from Wako Pure Chemical Industries, Ltd. (Japan). Antibiotics were dissolved in ethanol for AZM and CM stock concentrations of 6 and 30 mg/mL, respectively, and into distilled water for KAN (50 mg/mL) as a stock solution. The stock solutions were used to adjust the final concentrations to 15 μg/mL for each antibiotic into the WAS.
Methane production
A total volume of 30-mL WAS with or without 15 μg/mL of each antibiotic was poured in 66-mL vials. The vials were tightly sealed with butyl rubber stoppers, crimped, and sparged with nitrogen gas for 2 min to provide anaerobic conditions. The vials were then incubated at 37 °C for 120 rpm over 10 days. Each experiment was conducted at least in triplicate. Methane was measured by injecting 50 μL of the headspace gas in the vials into a GC-3200 gas chromatograph (GC) (GL Science, Japan) equipped with a thermal conductive detector and with a molecular sieve 13 × 60/80 mesh column and a Steel Use Stainless (SUS) 2 m × 3 mm inner diameter (GL Science, Japan). Helium gas was used as a carrier gas with a flow rate of 40 mL/min. The GC conditions were set as follows: current of 100 mA, oven, injector, and detector temperatures of 40, 50, and 65 °C, respectively.
Analytical methods
WAS samples during fermentation were used for the following analyses: organic acids, pH, protein concentration, and protease activity. Initially, the fermented WAS samples were centrifuged at 13,000 rpm for 7 min to collect the supernatants which were later filtered through a 0.2-μm membrane syringe filter. The organic acids in each supernatant were analyzed using high-performance liquid chromatography (Shimadzu LC-10 AD), as described previously (Mohd Yusoff et al. 2012). The pH was measured using a compact pH meter (AS ONE, AS-211, Japan). The soluble protein concentration was analyzed by the Lowry method using bovine serum albumin as a standard protein (Lowry et al. 1951). The protease activity was measured as described previously (Maeda et al. 2011). One unit of protease activity was calculated as the quantity of tyrosine (μmol) produced from casein by 1 mg of enzyme per minute.
RNA extraction and cDNA synthesis
RNA extraction was conducted using sludge pellets from raw sludge and each fermented sludge. Prior to the RNA extraction, 3 mL of the RNA later solution (Ambion, Cat#AM7020) was mixed with 4 mL of the sludge sample using a RNA-free falcon tube to avoid RNA degradation through the extraction process, and the mixture was then centrifuged at 15,000 rpm for 2 min at 4 °C to harvest the sludge pellet. The supernatant was discarded and the pellet was immediately resuspended with 1 mL of the RNA later solution in a screw-cap tube and finally centrifuged at 13,000 rpm for 30 s. After removing the supernatant, the remaining sludge pellet was quickly frozen in dry ice with ethanol and stored at −70 °C until the process of RNA extraction. The total RNA was extracted using the RNeasy kit (Qiagen Inc., Valencia, CA), as described in our previous paper (Mohd Yusoff et al. 2012), and the total RNA concentration was measured by using a NanoDrop ND-1000 spectrophotometer (SCRUM Inc., Japan). The cDNA was synthesized from RNA using the PrimeScript RT Reagent kit Perfect Real Time (TAKARA Bio Inc., Shiga, Japan). The synthesis was done using 5 μg of the total RNA in a mixture of 2 μL of 5×PrimeScript buffer, 0.5 μL reverse transcriptase enzyme, 0.5 μL oligo dT primer, and 2 μL random oligomers for a total volume of 10 μL along with the RNA. The mixture was incubated at 25 °C for 10 min, then at 37 °C for 30 min, and finally the enzyme was deactivated at 85 °C for 5 s. Electrophoresis using a 1.4 % agarose gel was performed to determine the quality and quantity of the RNA extracted and the synthesized complementary DNA (cDNA). The cDNA was used later for quantitative real time polymerase chain reaction (qRT-PCR) to determine the bacterial activity, Archaeal activity, and the microbial communities of the bacteria and Archaeal populations.
RT-PCR quantification
The qRT-PCR was performed to quantify the 16S rRNA gene of total bacteria and Archaea using a TaqMan system. The StepOne Real Time PCR System (Applied Biosystems) was used for the amplification and detection of fluorescence by using the primers and probes listed in Table 1. The total 20 μL of the real time PCR (RT-PCR) mixture was prepared as follows: 10 μL of 2×Taqman Fast Advance master mix, 0.72 μL of each primer (25 μM), 0.34 μL of Taqman probe (11.8 μM), and 2 μL of cDNA (100 ng). The quantification for each cDNA template was analyzed in triplicate. The cycling condition was as follows: uracil N-glycosylase (UNG) incubation at 50 °C for 2 min, denaturation and polymerase activation at 95 °C for 20 min, and 40 cycles of annealing and extension at 95 °C for 1 s and 60 °C for 20 s, respectively. The standard curve for universal bacteria and Archaea was calculated as described in our previous paper (Mohd Yasin et al. 2015). Escherichia coli, a rapidly growing aerobic bacteria, was commonly used as the standard for determining the total bacterial numbers because its DNA is consistently detected by RT-PCR (Nadkarni et al. 2002). Archaeaglobus fulgidus was used as a representative for Archaeal activities as Archaeaglobus spp. are closely related to methanogenic Archaea or methanogens in comparative genomic studies (Gao and Gupta 2007). The calculation of copy numbers based on the amount of DNA was performed as described by Lee et al. (Lee et al. 2008).
High-throughput 16S rRNA sequencing
The V4-V5 region of the 16S rRNA gene was used as a target of PCR amplification for the high-throughput 16S rRNA sequencing. The primers used were forward primer 515F (5′-GTGCCAGCMGCCGCGG-3′) and reverse primer 907R (5′-CCGTCAATTCMTTTRAGTTT-3′). The PCR mixture (25 μL) consisted of 10×Taq buffer and Taq polymerase (BioLabs), 20 μM of each primer, 2 mM each dNTPs mix, 25 mM of MgSO4 (Toyobo), and approximately 50 ng of the cDNA template. PCR was carried out under the following conditions: initial denaturation at 94 °C for 3 min followed by 94 °C for 45 s, 50 °C for 60 s, and 72 °C for 90 s of 35 cycles, and a final extension at 72 °C for 10 min. The amplified PCR products were purified using the QIAquick Gel Extraction Kit (Qiagen, Valencia, CA, USA). Each concentration of the purified PCR products was then determined using a Qubit dsDNA HS Assay Kit (Life Technologies, Oregon, USA).
The PCR products were processed using the Nextera XT DNA Library Preparation Kit according to the Illumina manufacturer’s protocol. Briefly, the input DNA was treated for simultaneous fragmentation and tagmentation using the Nextera transposome, as well as being treated for the addition of a dual adapter index with a unique bar code sequence through the PCR amplification to differentiate each sample. Then, the purification was performed by AMPure XP beads and normalized to ensure that an equal library was presented in pooled samples. Pooled samples were diluted in a hybridization buffer and denatured by heat prior to sequencing. A 500-cycle V2 MiSeq reagent cartridge (Illumina) was thawed in a water bath and inverted ten times to mix the reagents inside it prior to use. Finally, the pooled samples were loaded into the cartridge and then onto the MiSeq instrument along with the cleaned flow cell. Sequencing was performed for 251, 8, 8, and 251 cycles for forward, index 1, index 2, and reverse reads, respectively. The data obtained were de-multiplexed and the reads were then classified to different taxonomic levels. The raw sequence data were deposited in the National Center for Biotechnology Information (NCBI) short reads archive database (accession number: SRP072534).
Statistical analysis
The different antibiotics were compared with control WAS using means from at least triplicate data (n = 3). Comparison was performed using means and standard deviations by the Student’s t test (GraphPad software) at a significance level of p < 0.05.
Results
Effect of each antibiotic on methane fermentation from WAS
This study investigated how methane fermentation of WAS can be influenced by three types of antibiotics, AZM, CM, and KAN. They have a certain potential to change the total microbial activity (dynamics) due to the different antibacterial spectra of each antibiotic against the endogenous microbes present in WAS. First, using WAS with 15 μg/mL of AZM, CM, or KAN or antibiotic-free (control), anaerobic fermentation was performed for 10 days to determine the differences of methane production in the presence or absence of each antibiotic. As shown in Fig. 1, the addition of AZM showed a 2× higher methane production than the antibiotic-free WAS. The accumulation of methane significantly increased after the third day of fermentation in the WAS in the presence of AZM (p < 0.05). By contrast, CM had a significant (p < 0.05) inhibitory effect on methane production with the highest production rate of only 684 ± 100 μmol/g VS, and the accumulation of methane was observed after the fifth day of fermentation; there was no methane on the second day. In addition, methane production from WAS in the presence of KAN was significantly similar to the antibiotic-free WAS. Thus, despite the addition of antibiotics which should change the microbial activity due to their bactericidal effects, three patterns of methane production were observed: (1) a positive impact where methane production was improved in the presence of AZM, (2) a negative impact where CM inhibited methane production, and (3) none or less of an impact with or without KAN. Understanding of the different mechanisms between the three patterns of methane production may represent an outstanding finding which should be important for the comprehensive understanding of methane fermentation from WAS, in particular regarding microbial interactions (for example, inhibition, repression, acceleration, and dependence).
Bacterial activity in the presence of each antibiotic at the hydrolysis stage
To determine the differences in the results observed for AZM, CM, and KAN, their bacterial activity at the first hydrolysis step was compared. Since large molecules of WAS should be converted into small molecules, degrading products and enzymes are great targets to evaluate the hydrolysis process. Based on the idea that protein is one of the major components in WAS (Maeda et al. 2009), the soluble protein concentration and protease activity were measured and compared in the three antibiotics (Fig. 2a, b). The concentration of soluble proteins from WAS during anaerobic digestion in the presence of AZM or CM was higher than that in KAN and the control throughout the incubation period. In the WAS with KAN, the protein concentration had the same trend as the control at an early period (2 days); however, it was slightly higher than the control after 7 days. The corroborating evidence was obtained from the result that protease activity in the WAS samples with AZM and CM was higher than that in the control at 2 days (Fig. 2b). Finally, there was a clear relationship between protein concentration and protease activity. These results demonstrate that the hydrolysis process was not inhibited by the addition of AZM, CM, or KAN.
Soluble protein concentration (a) accumulated during anaerobic digestion using waste-activated sludge with AZM (triangle), CM (square), KAN (asterisk), and without any antibiotic as a control (circle). Protease activity (b) detected in the sample with AZM ( ), CM (
), CM ( ), or KAN (
), or KAN ( ), and without any antibiotic as a control (
), and without any antibiotic as a control ( ) during anaerobic digestion. Error bars indicate standard errors (n = 3). *Indicates a significant difference by antibiotic addition
) during anaerobic digestion. Error bars indicate standard errors (n = 3). *Indicates a significant difference by antibiotic addition
Bacterial activity in the presence of each antibiotic at the acidogenesis and acetogenesis stages
Figure 3 shows the profile of organic acids on days 1, 3, 5, and 10 under anaerobic incubation conditions of WAS with or without the addition of an antibiotic (AZM, CM, or KAN) to evaluate the differences of bacterial activity at the acidogenesis and acetogenesis stages. Acetate and butyrate concentrations were significantly higher (p < 0.05) in the presence of AZM compared with those in antibiotic-free WAS. Acetate also accumulated remarkably by day 3 whereas the butyrate concentration was drastically increased by day 5 (Fig. 3a). The addition of CM in WAS had slightly lower intermediates than AZM, especially the butyrate concentration, but still significantly higher than antibiotic-free WAS (p < 0.05) at days 5 and 10 (Fig. 3b). In addition, the production of isobutyrate and propionate was low in both samples with AZM and CM (Fig. 3a, b), unlike the WAS samples with or without KAN where isobutyrate and propionate were accumulated throughout the fermentation (Fig. 3c, d). In particular, only limited concentrations of isobutyrate and propionate were detected in the presence of AZM (Fig. 3a). Interestingly, despite the presence of CM, which inhibited methane fermentation, the production of organic acids (acetate and butyrate) was observed. Taken together, the addition of each antibiotic to WAS did not negatively influence the bacterial activity at the acidogenesis/acetogenesis stage, although different patterns of organic acids were observed for each antibiotic.
Profile of organic acids, acetic acid ( ), propionic acid (
), propionic acid ( ), isobutyric acid (
), isobutyric acid ( ), and butyric acid (
), and butyric acid ( ) during anaerobic digestion of waste-activated sludge with AZM (a), CM (b), KAN (c), and without any antibiotic as a control (d). Error bars indicate standard errors (n = 3). *Indicates a significant difference by antibiotic addition
) during anaerobic digestion of waste-activated sludge with AZM (a), CM (b), KAN (c), and without any antibiotic as a control (d). Error bars indicate standard errors (n = 3). *Indicates a significant difference by antibiotic addition
Bacterial and archaeal activity in the presence of each antibiotic
As shown in Fig. 1, the addition of each antibiotic (AZM, CM, or KAN) showed different patterns of methane production although there was less of an impact in the hydrolysis, acidogenesis, and acetogenesis steps in the presence of antibiotics. Therefore, bacterial and Archaeal activities were determined with qRT-PCR using RNA as a template to evaluate the number of really active bacteria and methanogens in the presence or absence of antibiotics. Figure 4 shows the quantitative activity of bacteria and Archaea in WAS with or without any antibiotic (AZM, CM, KAN, or antibiotic free). As a result, the activity of active bacteria did not show any significant change with or without antibiotics, in agreement with the results that bacterial activity at the hydrolysis and acidogenesis/acetogenesis stages was not negatively influenced in the presence of antibiotics. On the other hand, Archaeal activity in the presence of AZM was significantly higher (p < 0.05) than that without any antibiotic, as supported by a result that methane production was enhanced under AZM (Fig. 1). As expected, the addition of CM in WAS showed a low Archaeal activity relative to the low methane production in the presence of CM. Also, bacterial and Archaeal activities in KAN were similar to those in antibiotic-free samples.
Effect of antibiotics on the dynamics of active microbial communities
The composition of active microbial communities in WAS with or without antibiotics was evaluated using the MiSeq Illumina analyzer. Figure 5 shows the relative abundance of the microbial communities present in each sample at day 2 and day 7 in the presence of AZM, CM, or KAN, as well as in the absence of any antibiotic. The dynamics of microbial communities occurring during anaerobic digestion with or without the addition of antibiotic were evaluated by comparing the communities at day 2 and day 7 within each sample. Based on the taxonomic level (Fig. 5a), Betaproteobacteria was the most abundant class in all samples, although it was slightly decreased by the end of anaerobic digestion. The Proteobacteria, Deltaproteobacteria, Gammaproteobacteria, and Alphaproteobacteria were present in all samples, whereas a very low abundance of Epsilonproteobacteria was detected. The proportion of Clostridia in WAS with AZM was higher than that in other samples. In addition, a slightly higher percentage of Methanomicrobia and Methanobacteria was present in the WAS with AZM, compared to other antibiotics. WAS with KAN or without any antibiotic showed a similar microbial community composition, except for a sample with KAN at day 2 that had a high population of Betaproteobacteria. Nitrospira was detected at a higher percentage during the initial incubation in all samples. The other dominant classes that were present in WAS with or without antibiotics were the Sphingobacteria, Flavobacteria, Anaerolineae, Actinobacteria, and Chloroflexi. The microbial communities in those samples were also evaluated at the genus level (Fig. 5b). Haliangium bacteria were present at a higher percentage and were similar for all samples regardless of the addition of antibiotics. Similar to the class level, Clostridium was present in a high percentage in AZM, particularly at day 7 of fermentation, and CM samples had the lowest percentage of Clostridium. Over 7 days of anaerobic digestion, other bacteria that were present at a higher relative abundance from the AZM sample, compared to the other samples, were Caldilinea and Methanosarcina. Flavobacterium was present at a higher abundance in WAS with the addition of AZM and CM compared to KAN and antibiotic-free WAS. On the other hand, Chondromyces was present at a higher abundance in WAS with the addition of CM, compared to other samples.
Relative abundance of the dominant microbial communities categorized at the class level (a) and genus level (b) in waste-activated sludge in the presence of AZM, CM, KAN, and in the absence of any antibiotic (CTRL) at 2 days and 7 days. The results are derived from high-throughput 16S rRNA sequencing. Minor classes (<1 %) were summed up in the group “others”
Discussion
The effect of different antibiotics was evaluated in this study and three patterns of methane production were observed: (1) a positive effect by AZM, (2) a negative effect by CM, and (3) none or less of an effect by KAN. The different effects on methane fermentation should be due to changes in the microbial activity in WAS at the hydrolysis, acidogenesis/acetogenesis, and methanogenesis stages. In the hydrolysis stage, complex polymers such as proteins, carbohydrates, and lipids are degraded, resulting in the production of readily available organic materials such as amino acids, sugars, and long-chain fatty acids, respectively (Appels et al. 2008; Manyi-Loh et al. 2013). This process was primarily promoted by hydrolytic bacteria that secreted extracellular enzymes such as protease, amylase, or lipase to produce substrates for acidogenic microorganisms. The addition of CM and other antibiotics in WAS did not affect the hydrolytic bacteria that release protease enzymes, as the secretion of those enzymes practically showed the same trend in WAS with or an antibiotic (Fig. 1b).
The profile of soluble metabolites such as acetic, butyric, propionic, and isobutyric fluctuated during the incubation of WAS (Fig. 2), demonstrating that they were intermittently produced and consumed by the microbial community in WAS during the acidogenesis and acetogenesis stages. Clostridia and Bacteroidaceae are the main fermentative bacteria (Guo et al. 2015) that convert dissolved materials into volatile fatty acids, CO2, and H2. Later, acetogenic bacteria, known as hydrogen-producing acetogens or homoacetogens, convert these products to acetate. In all WAS in the presence of antibiotics, these bacteria might not be inhibited by antibiotics as the dissolved materials produced in the hydrolysis stage were utilized to produce intermediates. Thus, these intermediates are very important precursors for methanogens to be utilized and converted to methane at the methanogenesis step. Precisely, specific intermediates were used by specific groups of syntrophic bacteria as substrates for methane production. A higher butyric acid accumulation in WAS with AZM might be due to the inhibition of bacteria responsible for the utilization of butyric acid by AZM. This was supported in a previous report by Amin et al. (2006), where butyric acid utilization was inhibited by a macrolide antibiotic at lower concentrations. Meanwhile, the lower concentrations of propionate and isobutyrate were presumably due to the inhibition of these acid-producing bacteria. A former study by Mellon et al. (2000) found that the application of a macrolide antibiotic suppressed the propionate production by affecting some acidogenic anaerobic bacteria. Besides, pH was also measured for all samples at the end of incubation process, and the accumulation of organic acids in WAS in the presence of AZM and CM may be responsible for the low pH values (pH 5.1 and 5.3, respectively). On the other hand, addition of KAN did not show a low pH (pH 6.2), and it was similar to the control WAS.
In this study, AZM promoted the growth of microbes that contributed to improve methane production. However, other types of macrolides used in anaerobic digestion mostly provide a contradictory impact, unlike the results in this study. For example, a high concentration of tylosin, a 16-membered ring macrolide, showed a certain inhibitory effect on biogas production due to the accumulation of intermediates and a drop in pH (Shimada et al. 2008). Erythromycin, a parenteral antibiotic of AZM, showed only a slight effect on methanogenic activity but inhibited the conversion of some volatile fatty acids (Amin et al. 2006), while Du et al. (2015) obtained opposite results. In their study, CM inhibited methane production almost threefold when compared to the control. Also, it may take longer for some methanogenic Archaea to adapt and acclimatize in this condition since they only started to produce methane after 4 days. Therefore, CM did not affect the microbial community in the initial stages of anaerobic digestion as the protease activity and the soluble intermediates (proteins and organic acids) were detected at a high activity and a high concentration in those samples. Additionally, the presence of CM in WAS suppressed methane production which might be due to the inhibition of microbes only at the methanogenesis stage (Jin et al. 2011; Lallai et al. 2002; Sanz et al. 1996) without inhibiting other pathways. However, Lins et al. (2015) showed that the addition of 100 μg/mL CM showed no difference in gas production compared with the control, unlike our results where a low concentration (15 μg/mL) of CM was used. The Archaeal population plays an important role in the methanogenesis stage that should be responsible for the production of methane from acetate and CO2/H2. A low Archaea activity was detected in the WAS with CM (Fig. 4), indicating that methanogens were unable to convert metabolites such as acetate into methane, even though there were enough available substrates for methane production. Furthermore, the similarity of methane production in the WAS with or without KAN was supported by the amount of active bacteria and Archaea detected in both samples. Despite the addition of KAN, which has a certain antibacterial spectrum, methane production with KAN was the same pattern as the control. This indicates that bacteria sensitive to KAN may not be essential in the anaerobic digestion process. However, methane production was slightly inhibited by the addition of 100 μg/mL KAN due to the suppression of the intermediates degradation pathway (Lins et al. 2015). Thus, a high concentration of antibiotics can provide a different result by changing the microbial community due to stronger antibacterial activity.
Understanding the changes of the microbial communities in the presence of each antibiotic allows us to analyze what microorganisms are important in methane fermentation from WAS. Proteobacteria was an important phylum in this system because it includes the bacteria responsible for glucose, propionate, butyrate, and acetate utilization (Guo et al. 2015). The total percentages of all Proteobacteria classes in WAS with AZM, CM, KAN, and antibiotic-free WAS were 44, 61, 58, and 56 %, respectively. This shows that even though the addition of CM in WAS inhibited methane production, it did not affect the important microbes related to the degradative activity from the hydrolysis to the acetogenesis stages. However, Flavobacterium was detected at a relatively high percentage in AZM and CM samples during fermentation (Fig. 5b), in agreement with a result that showed a similar pattern of organic acid production, particularly acetic and butyric acids. Flavobacterium is one of the acid-phase bacteria that are involved in the anaerobic digestion process (Ali Shah et al. 2014). On the other hand, WAS with AZM contained a high proportion of Clostridium and Caldilinea, which may be important and explain the high methane production. The predominance of Clostridia contributed to a high rate of hydrolysis and degradation of the volatile fatty acid in anaerobic digestion. Caldilinea belongs to the phylum Chloroflexi, which is one of the syntrophic bacteria found ubiquitously in anaerobic digestion systems (Narihiro et al. 2015). Since it is a gram-negative species, AZM might not target this bacterium so it can be highly abundant and contribute to improved methane production. In addition, WAS with AZM was the only sample that contained Methanomicrobia and Methanobacteria which belong to the Archaea. This is also supported by the genus level analysis that showed the presence of Methanosarcina. These methanogens are very well known for their contribution to methane production, and the quantitative data from qRT-PCR also showed a higher population of Archaea compared to those from other samples. This situation was presumably due to the resistance of methanogens in WAS toward macrolide antibiotics such as AZM. Amin et al. (2006) also showed that the presence of erythromycin, a macrolide-type antibiotic at a high concentration, did not affect the methanogens. Conversely, in the WAS with CM that showed lower methane production, the presence of Clostridia was much lower than with AZM but similar to the control (at 7 days), and was still able to produce methane. Thus, this proves that the low methane production was not only due to the low Clostridia population in the sample, but also due to the methanogens as other studies concluded that CM was a powerful inhibitor toward methanogenic Archaea (Lallai et al. 2002; Sanz et al. 1996). In addition, a high percentage of Chondromyces was also present in the WAS with CM. While studies on this bacterium are very scarce, it might not contribute to methane production but may be involved in the production of organic acid such as acetic acid or butyric acid. Chondromyces is an anaerobic myxobacterium (order Myxococcales). According to Veldkamp (1961), anaerobic myxobacteria were able to produce acetic, propionic, and succinic acids from sugar fermentation. Thus, Chondromyces may carry out a similar function for organic acid production. The addition of KAN to WAS had little effect on the dynamics of microbial communities during anaerobic digestion, but it initially supported the growth of Betaproteobacteria. Later, no changes occurred, as was shown by the similar microbial communities in WAS. This might be due to KAN which did not affect the anaerobes involved in methane production. During anaerobic digestion, antibiotic-free WAS showed very slight changes in its microbial community at the initial and final incubation day. This is more evidence that the use of antibiotics altered the complex microbial communities in WAS which are responsible for improving or inhibiting methane production. In conclusion, the application of different antibiotics showed variable effects on the overall anaerobic digestion process through the change of microbial activities. These are useful data that will facilitate the improvement of bioenergy production from WAS.
References
Akyol Ç, Turker G, Ince O, Ertekin E, Üstüner O, Ince B (2016) Performance and microbial community variations in thermophilic anaerobic digesters treating OTC medicated cow manure under different operational conditions. Bioresour Technol 205:191–198
Ali Shah F, Mahmood Q, Maroof Shah M, Pervez A, Ahmad Asad S (2014) Microbial ecology of anaerobic digesters: the key players of anaerobiosis. Sci World J 2014:183752
Amin MM, Zilles JL, Greiner J, Charbonneau S, Raskin L, Morgenroth E (2006) Influence of the antibiotic erythromycin on anaerobic treatment of a pharmaceutical wastewater. Environ Sci Technol 40:3971–3977
Appels L, Baeyens J, Degrève J, Dewil R (2008) Principles and potential of the anaerobic digestion of waste-activated sludge. Prog Energy Combust Sci 34:755–781
Aydin S, Shahi A, Ozbayram EG, Ince B, Ince O (2015) Use of PCR-DGGE based molecular methods to assessment of microbial diversity during anaerobic treatment of antibiotic combinations. Bioresour Technol 192:735–740
Bareither CA, Wolfe GL, McMahon KD, Benson CH (2013) Microbial diversity and dynamics during methane production from municipal solid waste. Waste Manag 33:1982–1992
Bougrier C, Carrère H, Delgenès JP (2005) Solubilisation of waste-activated sludge by ultrasonic treatment. Chem Eng J 106:163–169
Bougrier C, Albasi C, Delgenès JP, Carrère H (2006) Effect of ultrasonic, thermal and ozone pre-treatments on waste activated sludge solubilisation and anaerobic biodegradability. Chem Eng Process 45:711–718
Dewil R, Baeyens J, Goutvrind R (2006) Ultrasonic treatment of waste activated sludge. Environ Prog 25:121–128
Du J, Hu Y, Qi W, Zhang Y, Jing Z, Norton M, Li Y-Y (2015) Influence of four antimicrobials on methane-producing archaea and sulfate-reducing bacteria in anaerobic granular sludge. Chemosphere 140:184–190
Gao B, Gupta RS (2007) Phylogenomic analysis of proteins that are distinctive of Archaea and its main subgroups and the origin of methanogenesis. BMC Genomics 8:1–23
Guo J, Peng Y, Ni B-J, Han X, Fan L, Yuan Z (2015) Dissecting microbial community structure and methane-producing pathways of a full-scale anaerobic reactor digesting activated sludge from wastewater treatment by metagenomic sequencing. Microb Cell Factories 14:1–11
Jackson CR, Roden EE, Churchill PF (2000) Denaturing gradient gel electrophoresis can fail to separate 16S rDNA fragments with multiple base differences. Mol Biol Today 1:49–51
Jin W, Cheng Y-F, Mao S-Y, Zhu W-Y (2011) Isolation of natural cultures of anaerobic fungi and indigenously associated methanogens from herbivores and their bioconversion of lignocellulosic materials to methane. Bioresour Technol 102:7925–7931
Lallai A, Mura G, Onnis N (2002) The effects of certain antibiotics on biogas production in the anaerobic digestion of pig waste slurry. Bioresour Technol 82:205–208
Lee C, Kim J, Shin SG, Hwang S (2008) Monitoring bacterial and archaeal community shifts in a mesophilic anaerobic batch reactor treating a high-strength organic wastewater. FEMS Microbiol Ecol 65:544–554
Lins P, Reitschuler C, Illmer P (2015) Impact of several antibiotics and 2-bromoethanesulfonate on the volatile fatty acid degradation, methanogenesis and community structure during thermophilic anaerobic digestion. Bioresour Technol 190:148–158
Lowry OH, Rosebrough NJ, Farr AL, Randall RJ (1951) Protein measurement with the folin phenol reagent. J Biol Chem 193:265–275
Maeda T, Yoshimura T, Shimazu T, Shirai Y, Ogawa HI (2009) Enhanced production of lactic acid with reducing excess sludge by lactate fermentation. J Hazard Mater 168:656–663
Maeda T, Yoshimura T, García-Contreras R, Ogawa HI (2011) Purification and characterization of a serine protease secreted by Brevibacillus sp. KH3 for reducing waste activated sludge and biofilm formation. Bioresour Technol 102:10650–10656
Manyi-Loh CE, Mamphweli SN, Meyer EL, Okoh AI, Makaka G, Simon M (2013) Microbial anaerobic digestion (bio-digesters) as an approach to the decontamination of animal wastes in pollution control and the generation of renewable energy. Int J Environ Res Publ Health 10:4390–4417
Mellon A, Deshpande S, Mathers J, Bartlett K (2000) Effect of oral antibiotics on intestinal production of propionic acid. Arch Dis Child 82:169–172
Meng L-W, Li X-k, Wang K, Ma K-L, Zhang J (2015) Influence of the amoxicillin concentration on organics removal and microbial community structure in an anaerobic EGSB reactor treating with antibiotic wastewater. Chem Eng J 274:94–101
Mohd Yasin NH, Maeda T, Hu A, Yu C-P, Wood TK (2015) CO2 sequestration by methanogens in activated sludge for methane production. Appl Energy 142:426–434
Mohd Yusoff MZ, Maeda T, Sanchez-Torres V, Ogawa HI, Shirai Y, Hassan MA, Wood TK (2012) Uncharacterized Escherichia coli proteins YdjA and YhjY are related to biohydrogen production. Int J Hydrog Energy 37:17778–17787
Nadkarni MA, Martin FE, Jacques NA, Hunter N (2002) Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiol 148:257–266
Narihiro T, Nobu MK, Kim N-K, Kamagata Y, Liu W-T (2015) The nexus of syntrophy-associated microbiota in anaerobic digestion revealed by long-term enrichment and community survey. Environ Microbiol 17:1707–1720
Ng KK, Shi X, Ng HY (2015) Evaluation of system performance and microbial communities of a bioaugmented anaerobic membrane bioreactor treating pharmaceutical wastewater. Water Res 81:311–324
Nguyen M, Maeda T, Mohd Yusoff M, Ogawa H (2014) Effect of azithromycin on enhancement of methane production from waste activated sludge. J Ind Microbiol Biotechnol 41:1051–1059
Papich MG (2010) Saunders handbook of veterinary drugs: small and large animal. Elsevier Health Sciences
Sanz J, Rodriguez N, Amils áR (1996) The action of antibiotics on the anaerobic digestion process. Appl Microbiol Biotechnol 46:587–592
Schwarz S, Kehrenberg C, Doublet B, Cloeckaert A (2004) Molecular basis of bacterial resistance to chloramphenicol and florfenicol. FEMS Microbiol Rev 28:519–542
Shimada T, Zilles JL, Morgenroth E, Raskin L (2008) Inhibitory effects of the macrolide antimicrobial tylosin on anaerobic treatment. Biotechnol Bioeng 101:73–82
Shimada T, Li X, Zilles JL, Morgenroth E, Raskin L (2011) Effects of the antimicrobial tylosin on the microbial community structure of an anaerobic sequencing batch reactor. Biotechnol Bioeng 108:296–305
Su J-Q, Wei B, Ou-Yang W-Y, Huang F-Y, Zhao Y, Xu H-J, Zhu Y-G (2015) Antibiotic resistome and its association with bacterial communities during sewage sludge composting. Environ Sci Technol 49:7356–7363
Valo A, Carrère H, Delgenès JP (2004) Thermal, chemical and thermo-chemical pre-treatment of waste activated sludge for anaerobic digestion. J Chem Technol Biotechnol 79:1197–1203
Veldkamp H (1961) A study of two marine agar-decomposing, facultatively anaerobic myxobacteria. Microbiol 26:331–342
Wang X, Hu M, Xia Y, Wen X, Ding K (2012) Pyrosequencing analysis of bacterial diversity in 14 wastewater treatment systems in China. Appl Environ Microbiol 78:7042–7047
Williams J (1991) Spectrum of activity of azithromycin. Eur J Clin Microbiol Infect Dis 10:813–820
Zhang T, Zhang X-X, Ye L (2011) Plasmid metagenome reveals high levels of antibiotic resistance genes and mobile genetic elements in activated sludge. PLoS One 6:e26041
Acknowledgments
The authors wish to thank the Japanese Government Scholarship (MEXT) and Science and Technology Research Partnership for Sustainable Development Program (SATREPS) for providing full support to this study. A part of this study was supported by Kitakyushu City.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical approval
This article does not contain any studies performed with human participants or with animals by any of the authors.
Conflict of interest
The authors declare that there are no conflicts of interest.
Rights and permissions
About this article
Cite this article
Mustapha, N.A., Sakai, K., Shirai, Y. et al. Impact of different antibiotics on methane production using waste-activated sludge: mechanisms and microbial community dynamics. Appl Microbiol Biotechnol 100, 9355–9364 (2016). https://doi.org/10.1007/s00253-016-7767-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-016-7767-2


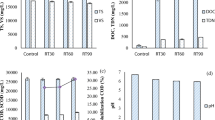




 ) and Archaeal (
) and Archaeal ( ) populations in waste-activated sludge in the presence of AZM, CM, KAN, and without any antibiotic (CTRL). Error bars indicate standard errors (n = 3). *Indicates a significant difference by antibiotic addition
) populations in waste-activated sludge in the presence of AZM, CM, KAN, and without any antibiotic (CTRL). Error bars indicate standard errors (n = 3). *Indicates a significant difference by antibiotic addition