Abstract
For the purpose of supervision and academic research on GMO, it was necessary to establish a simple and effective on-site qualitative and quantitative method for GMO detection. In this paper, double-stranded displacement probes were first designed to monitor the quantitative LAMP amplification, and calcein was used effectively in LAMP reaction for qualitative measurement. High specificity of the methods was identified by testing 16 crops. The rice endogenous gene SPS and M12 event-specific gene can both be quantified in a wide dynamic range (200–20,000 copies). The LOD and LOQ of both genes were 20 and 200 copies, respectively. When the methods were used to analyze rice samples, 0.5% (w/w) of the GM rice M12 event could be quantified accurately and 0.1% (w/w) could be detected qualitatively. These results indicated that the developed methods were specific, sensitive and repeatable, and proven to be an efficient alternative to real-time PCR. Moreover, the developed methods were more suitable for the reliable quantification in poor-equipped laboratories or on-site detection.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In recent years, many qualitative and quantitative methods for the detection of nucleic acid ingredients of genetically modified (GM) crops were explored, such as polymerase chain reaction (PCR), real-time PCR and digital PCR [1,2,3,4,5,6]. However, these methods required rapid thermo-cycling to complete the amplification, and precise temperature control equipments were needed. Thus, these methods were more suitable to be used in the well-equipped laboratories rather than in on-site or poor-equipped laboratories. Loop-mediated isothermal amplification (LAMP), depending on auto-cycling strand displacement DNA synthesis performed using the Bst DNA polymerase, has been developed to amplify nucleic acids specifically at a stable temperature by using 4–6 primers. Meanwhile, a simple isothermal condition can make sure the experiment to go smoothly. Thus, the LAMP method may be more suitable to be used in on-site or poor-equipped laboratories. Indeed, the LAMP method has been already applied to qualitatively detect GM crops, such as oilseed rape, soybean, maize, cotton, sugarcane, cattle and rice with cry1Ab gene [7,8,9,10,11,12]. However, compared with real-time PCR, on-site quantitative isothermal amplification techniques have not been developed for rapid and effective quantitation of GMO. So, quantitative LAMP method for on-site testing or for grass-root laboratories was still needed.
In LAMP assay, a set of four to six specially designed primers are used to amplify specific sequences of DNA under isothermal conditions between 60 and 65 °C. Thereby, it obviates the need for a thermal cycler and provides the exceptionally higher specificity of amplification than conventional PCR method [13, 14]. Meanwhile, the results of LAMP reactions can be determined visually by adding the manganese ion indicator calcein: the color of the solution changes to green in the presence of LAMP amplicons; otherwise, it remains orange when there is no amplification in the reaction system [15]. At the beginning of the invention of LAMP technology, it was reported that LAMP technology can be used to quantify nucleic acid by real-time monitoring the turbidity of the reaction system [16, 17]. Subsequently, two other research teams used real-time turbidimetry to study the quantitative method of LAMP technology [7, 18]. However, the real-time turbidimetry depended on the white precipitates of magnesium pyrophosphate, which cannot form a homogeneous system. Thus, the baseline would vary in the amplification process resulting in poor repeatability and a standard curve with an unsatisfactory linear relationship. Meanwhile, it was reported that the LAMP method can be used to quantify nucleic acid by adding fluorescent dyes [19]. However, there was no detailed information on how to use the real-time LAMP method for quantification. In real-time PCR, the problem of baseline instability and non-specificity amplification could be solved by the application of fluorescence labeled probes [4, 20]. Therefore, it might be feasible to apply fluorescent labeled probes into LAMP to establish a real-time isothermal amplification method. However, the strand-displacing Bst DNA polymerase without hydrolytic activity was used in the LAMP reaction, instead of Taq polymerase [14]. Therefore, the conventional hydrolytic probes were not suitable for the real-time fluorescent LAMP reaction because it cannot release the fluorescent signals during the positive LAMP amplification. If a pair of appropriate reverse complementary and replacement probes were used in the LAMP reaction, the problem might be solved.
The GM rice (Oryza sativa L.) M12 event, showed strong potential for resistance to bacterial leaf blight in a field trial [21], was one of the potentially commercialized GM crops. Wherein, rapid and accurate methods for the screening and quantification of GM rice M12 event was still needed. Here, we assessed real-time quantitative LAMP method with double-stranded displacement probes to quantify the concentration of GM rice M12 event, and the results were also compared with those from real-time PCR and digital PCR method. Meanwhile, in order to establish a visual on-site screening method for GM rice M12 event, calcein was used to monitor the color change in the reactions.
Materials and methods
Materials
The 100% (w/w) GM rice M12 event was a gift from the Chinese Academy of Agricultural Sciences; GM rice event TT51-1, KF6, KMD, conventional wild-type rice cultivar “minghui 63” and GM soybean GTS-40-3-2 and GM corn NK603 were stored in our own laboratory at – 20 °C. Wheat (Triticum aestivum), barley (Hordeum vulgare), sugarcane (Saccharum officinarum), buckwheat (Fagopyrum esculentum), Soybean (Glycine max), corn (Zea mays), rapeseed (Brassica napus), tomato (Lycopersicon esculentum), potato (Solanum tuberosum) were collected from the local market and then stored in our own laboratory at – 20 °C.
DNA extraction
The extraction of genomic DNA was performed according to the CTAB protocol [22]. All the purified DNA were measured and evaluated using the Qubit nucleic acid and protein analyzer (Qubit™ 3 Fluorometer, ThermoFisher, USA).
Oligonucleotide primers and probes for LAMP and digital PCR
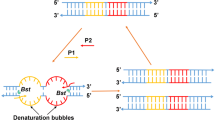
Two sets of LAMP primers, containing two inner primers FIP (F1c-F2) and BIP (B1c-B2), two outer primers (F3 and B3) and two loop primers (FLP and BLP), were designed corresponding to the sequence of rice endogenous gene SPS (sucrose phosphate synthase) and the left border flanking sequence of GM rice M12 event by online software PrimerExplorer V5 (Fig. 1). A pair of reverse complementary probes for quantitative LAMP was designed to target the template between the F1 and B1 regions of the amplicons and were labeled with FAM and BHQ1 at their 5′ and 3′ ends, respectively. Following the optimum design principle of the LAMP primer14, all of the primers and probes were designed and checked for homology with software BLASTNr within the GenBank databases to increase the specificity and sensitivity of the method. The same sequence of SPS and M12 event-specific gene were used to design the primers and probes for digital PCR by the Primer Premier 5.0 software program (PREMIER Biosoft International, Palo Alto, USA). The details of the primers and probes used in this study are given in Table 1 and synthesized by Invitrogen Co. Ltd. (Shanghai, China).
The operating procedure of LAMP
All of the LAMP assays were performed using 5 μL of template DNA (10 ng/μL) in a final volume of 20 μL of the LAMP reaction mixture composed of 200 mM of each dNTP (Takara, Japan), 0.8 M betaine (Sigma, USA), 20 mM Tris–HCl (pH 8.8), 10 mM KCl, 10 mM (NH4)2SO4, 2 mM MgSO4, 0.1% Tween 20 and 6.4 U of Bst DNA polymerase large fragment (New England Bio Labs, USA). The final concentrations of the six primers were as follows: 0.2 mM each of the outer primers F3 and B3, 1.6 mM of the inner primers FIP and BIP, and 0.4 mM each of the loop primers FLP and BLP. The LAMP reaction was performed with 80 min at 65 °C. For visual LAMP reaction, 0.1 mM calcein was added into the reaction mixture. After the reaction, the derived products were removed from the heat block (Eppendorf, Germany) and directly observed by the naked eye. In real-time quantitative LAMP reaction, instead of the calcein, 0.1 mM of reverse complementary probes labeled with the dyes FAM and BHQ1, respectively, were added into the reaction mixture to hybridize with the amplicons during amplification. The signals were measured using FAM channel (excitation, 460–490 nm; emission, 520–550 nm) of fluorometric thermal cycler (LC480; Roche, USA).
Digital PCR procedure and data analysis
Different from LAMP reaction, the probes used in digital PCR were hydrolysis probes as there were Taq DNA polymerase with hydrolytic activity in the digital PCR reaction. The digital PCR mixture was prepared as follows: 10 μL of 2 × droplet digital PCR Master Mix (Bio-Rad, Pleasanton, USA), 1 μL of each primer (final concentration of 300 nM) and probe (final concentration of 180 nM), 5 μL of template DNA with the concentration of 5 pg/μL, 50 pg/μL, 0.5 ng/μL, 2 ng/μL and 2 μL of nuclease and protease-free water were mixed to complete a 20-μL reaction volume, respectively. A QX200™ droplet generator (Bio-Rad, Pleasanton, CA) was used to generate the droplets. The reactions were performed on the GeneAmp® PCR System 9700 (ABI, USA) with the following step: 52 °C for 5 min, 95 °C for 5 min (1 °C/s); 60 cycles of 94 °C for 15 s and 60 °C for 1 min (0.5 °C/s), 98 °C for 10 min (1 °C/s). After amplification, the plate was loaded onto the QX200 system. The fluorescence was monitored using FAM and HEX channel, and the analysis of the droplet fluorescence was done with QuantaSoft Software version 1.6.6.0320 (Bio-Rad, Pleasanton, USA). All of the thresholds were set up manually to allow the distinction between positive and negative droplets, and the concentrations of DNA were calculated automatically by the instrument according to the Poisson distribution principle. Only the reactions with more than 10,000 accepted droplets were used for analysis.
Specificity of the assay
The specificity of SPS and event-specific gene of GM rice M12 event using in LAMP method was duplex confirmed using double-stranded displacement probes and calcein analyses mentioned above. The genomic DNA used for the specificity analysis of rice endogenous gene SPS were wild-type rice “minghui 63” (positive control), 4 GM rice events (i.e. M12, TT51-1, KF6 and KMD) and 9 gramineae closely related to rice in taxonomy and common crops (wheat, barley, sugarcane, buckwheat, soybean, corn, rapeseed, tomato, potato). The specificity of M12 event-specific gene was verified using genomic DNA from non-GM wild-type rice “minghui 63” (negative control), GM rice M12 event (positive control), GM rice events TT51-1, KF6 and KMD, GM soybean GTS-40–3-2 and GM corn NK603.
Calibration curve and the accuracy of the quantitative LAMP for GM rice event M12
The DNA concentration of the GM rice M12 event was determined by Qubit™ 3 fluorometer, then diluted with tenfold serial into five gradients (from 10 ng/μL to 1 pg/μL) and 5 μL of each template DNA, theoretically containing 100,000 copies, 10,000 copies, 1000 copies, 100 copies, and 10 copies of SPS and M12 event-specific genes, respectively, was added into a final volume of 20 μL of the LAMP reaction mixture. Since the rice single-copy genomic mass is about 0.5 pg these two target genes were all single-copy genes [21, 23, 24]. Then, the LAMP test was carried out with three repeats for each reaction and the POI (point of inflection), which was defined as the time corresponding to the maximum slope in the real-time fluorescence curve [19], was adopted for the LAMP quantification. The calibration curve was made using the POI value of quantitative LAMP (X axis) and the logarithmic theoretical concentration of target genes (Y axis). Furthermore, the SPS and M12 event-specific genes with theoretical content of 20,000 copies, 5000 copies, 500 copies and 50 copies, which were determined by Qubit™ 3 Fluorometer, were all absolute quantified in triplicate using both digital PCR and LAMP, respectively. The accuracy was determined by evaluating the bias between theoretical value and experimental value, and the precision was determined by the standard deviations (SD) and the relative repeatability standard deviation (RSDr). The quantitative results of the LAMP method were analyzed and compared with the results of digital PCR for the same sample in this experiment and other results of real-time PCR in literature.
LOD and LOQ of the quantitative LAMP for GM rice M12 event
To determinate the LOD and LOQ of the quantitative method, four levels of serial dilution samples with the concentration of 200, 100, 10 and 1 pg of each template DNA, theoretically containing 400, 200, 20 and 2 copies of SPS and M12 event-specific genes, were added into a final volume of 20 μL of the LAMP reaction mixture and quantitatively analyzed using the calibration curve. Each concentration of the samples was performed in triplicate. Meanwhile, calcein was added into the LAMP reaction of the above gradients of template DNA to get the visual results.
Quantification of the contents of GM rice M12 event in rice samples
To verify the practical application of this quantitative LAMP method, different contents of GM rice M12 event samples were used for quantitative analysis. The samples with different mass ratios of GM rice M12 event mixed with “wild type indica rice cultivar “minghui 63”, which were 100%, 5%, 0.5% and 0.1% (w/w), were prepared as follows: a mixture content 5% (w/w) M12 was made by mixing 0.5 g of rice seed of homozygous M12 event with 9.5 g of “minghui 63” rice, and then the mixture was homogenized into a powder; then a mixture content 0.5% (w/w) M12 was made by mixing 1 g 5% (w/w) M12 content with 9 g of “minghui 63” rice, followed by homogenization into a powder. The mixture content 0.1% (w/w) was prepared in the similar way. The qualitative and quantitative LAMP assays of rice endogenous genes SPS and M12 event-specific gene mentioned above were used to test all these samples in three parallels. None-GM rice “minghui 63” was used as the negative control. The absolute copy numbers of the two target genes were calculated by quantitative LAMP and the GM content (the ratio of exogenous and endogenous genes) was compared with the mass concentration of the samples.
Results
Specificity of the qualitative and quantitative LAMP for GM rice M12 event
First, the specificity of SPS gene was evaluated using the double-stranded displacement probes and calcein. The results showed that no unspecific amplification was observed with any of the tested nine crops (i.e. wheat, barley, sugarcane, buckwheat, soybean, corn, rapeseed, tomato and potato), while non-GM rice (the wild-type rice “minghui 63”, positive control) and 4 GM rice events (i.e. M12, TT51-1, KF6 and KMD) can be detected specifically (Fig. 2a, b). It indicated that the LAMP detection system for rice endogenous gene SPS was highly specific whether using calcein or double-stranded displacement probes. Thus, the methods could be used to distinguish rice from other crops. Furthermore, the specificity of the methods using the double-stranded displacement probes and calcein for M12 event was further identified using seven samples including “minghui 63” rice, 4 GM rice events (i.e. M12, TT51-1, KF6 and KMD1), GM soybean GTS-40-3-2 and GM corn NK603. It was found that positive results were only obtained for GM rice M12 event in both the double-stranded displacement probes and calcein analyses, while negative results were seen for other six crops, since other threr GM rices (i.e. TT51-1, KF6 and KMD rice), GM soybean GTS-40-3-2 and GM corn NK603 contained no target genes (Fig. 2c, d). Therefore, these results showed that the visible and real-time quantitative LAMP assays in this study were all specific for the identification of the target genes.
The specificity of SPS and M12 event specific gene analyses using double-stranded displacement probes and calcein. a Amplification curve of LAMP using double-stranded displacement probes for the SPS gene. From left to right above the baseline were positive control (the wild-type rice “minghui 63”), M12, KF6, TT51-1 and KMD, the wheat, barley, sugarcane, buckwheat, soybean, corn, rapeseed, tomato, potato and no template control (NTC) (ddH2O) were below the baseline. b Visible products of SPS gene amplified by LAMP with calcein. The meaning of the labeling was as follows: 1–2, positive control (the wild-type rice “minghui 63”); 3–4, M12; 5–6, TT51-1; 7–8, KF6; 9–10, KMD; 11–12, wheat; 13–14, barley; 15–16, sugarcane; 17–18, buckwheat; 19–20, soybean; 21–22, corn; 23–24, rapeseed; 25–26, tomato; 27–28, potato; 29, NTC (ddH2O). c Amplification curve of LAMP using displacement probes for the M12 event specific gene. The positive control (the GM rice event M12) was above the baseline, and the GM rice event TT51-1, KF6, KMD, GM soybean GTS-40-3-2, GM corn NK603, negative control (the wild-type rice “minghui 63”) and NTC (ddH2O) were below the baseline. d Visible products of M12 event specific gene amplified by LAMP with calcein. The meaning of the labeling was as follows: 1–2, GM rice event M12; 3–4, GM rice event Bt 63; 5–6, GM rice event KF6; 7–8, GM rice event KMD; 9–10, GM soybean GTS-40-3-2; 11–12, GM corn NK603; 13–14, negative control; 15, NTC (ddH2O). Notes: 1 cycle = 1 min
Accuracy of the quantitative LAMP
For easy operation, two standard curves of the target genes (i.e. SPS and M12 event specific gene) were drawn using the POI (point of inflection) value of quantitative LAMP (X axis) and the logarithmic theoretical concentration of target genes (Y axis). The R2 of the standard curve for SPS and M12 event-specific gene from the quantitative LAMP methods were 0.998 and 0.997, respectively (Fig. 3). Thus, the POI values were linearly related to the amount of target genes (log copy numbers of target gene), indicating that the standard curves can be used for the quantitation. Furthermore, the methods were used to amplify the target genes with theoretical content of 20,000, 5000, 500 and 50 copies, so as to obtain the corresponding POI value. Then the established standard curves were used to calculate the concentration of target genes according to the corresponding POI values. The results showed that the negative control samples for both genes were 0 copy, while the average concentrations of these four samples were 68.29 ± 17.66 copies to 19,231.48 ± 2679.27 copies for SPS gene and 56.63 ± 17.09 copies to 18,029.01 ± 843.10 copies for M12 event-specific gene in the reaction, respectively (Table 2), wherein, for the sample with a theoretical content of 50 copies, the RSDr of the two target genes among three replicates exceeded 25%, which were inconsistent with the current international quantitative method required [25, 26]. Therefore, the accuracy of the quantitative result of the sample with theoretical content of 50 copies was not very well. However, the RSDr values of the other three replicates of the sample with a theoretical content of 20,000, 5000 and 500 copies were all within 25%. And the bias ranged from -3.84 to 12.88%, meeting the quantitative requirements (i.e. within ± 25%) [25, 26]. Therefore, the quantification of the sample with a theoretical content of 20,000, 5000 and 500 copies was considered to be accurate and reliable.
The standard curves of the LAMP assay with double-stranded displacement probes. The standard curves were established using the POI (point of inflection) value (X axis) and the logarithmic concentration of target gene (Y axis). The theoretical concentrations of the target gene used for constructing the standard curves were 100,000, 10,000, 1000, 100 and 10 copies in each reaction. a The standard curve of rice endogenous gene SPS. b The standard curve of M12 event specific gene
In order to further ensure the accuracy of the developed quantitative LAMP methods, digital PCR was also used to quantify the SPS and M12 event-specific gene in the homozygous GM rice M12 event with the theoretical contents of 50, 500, 5000 and 20,000 copies. The results of digital PCR showed that the quantitative concentration of SPS were from 48.13 ± 2.88 to 21,414.05 ± 275.02 copies, and the concentration of M12 event-specific gene was from 51.33 ± 6.43 to 22,980.00 ± 367.15 copies. Moreover, compared withthe results from real-time LAMP and digital PCR (Table 2), the quantitative results were consistent for the samples with theoretical concentration of 500 copies and higher. Therefore, when the theoretical content of the two target genes was equal or greater than 500 copies, it was reliable to use the developed real-time LAMP method for rapid on-site quantification [26].
The LOD and LOQ of the quantitative LAMP for GM rice M12
Since the theoretical content of the two target genes was 50 copies for SPS or M12 event-specific gene, the result from the developed LAMP methods was not reliable. Thus, it was needed to identify the LOD and LOQ of the real-time and visible LAMP methods. The samples with a theoretical level of 400, 200, 20 and 2 copies were analyzed using the calibration curves established by the above methods. The results showed that only two of the three repetitions of the sample with two copies of SPS gene had positive fluorescent signal, while the M12 event-specific gene had only one positive signal (Table 3). Meanwhile, it was also impossible for the naked eye to observe the color changes in all three repetitions of 2 copies of target genes in the LAMP reaction with the calcein (Fig. 4). However, for the samples with 20 copies of SPS and M12 event-specific gene, although positive signals could be both observed in all the LAMP reaction with calcein or double-stranded displacement probes, the quantitative RSDr value or bias was more than 25%, which did not meet the quantitative requirements at this concentration [25, 26]. Thus, the quantitative result was also not reliable when the concentration of target gene was low to 20 copies. For the samples with 200 and 400 copies of SPS and M12 event-specific gene, the quantitative results were 175.36 ± 31.66 and 432.51 ± 29.72 for SPS gene, and 162.78 ± 35.46 and 365.34 ± 51.89 for M12 event-specific gene (Table 3), respectively. The bias was within acceptable range (< 25%), and the RSDr values of all the three replicates were not more than 25%, which indicated the quantitative results were accurate for the samples with 200 and 400 copies of the two target genes. Therefore, for the samples with equal or greater than 20 copies of SPS and M12 event-specific gene, it could be qualitatively detected by the developed LAMP methods, either with calcein or probes. And for the samples with equal or greater than 200 copies of SPS and M12 event-specific gene, it could be quantified accurately and reliably by the developed quantitative LAMP method with double-stranded displacement probes. Thus, the LOD and LOQ of the LAMP method used in this study were 20 and 200 copies for the two target genes, respectively.
Visible color change of the LAMP assays for quantify different concentration of template DNA. a The visible LAMP reaction of SPS gene. b The visible LAMP reaction of M12 event specific gene. The meaning of the labeling was as follows: 1–3, 400 copies of GM rice M12 event; 4–6, 200 copies of GM rice M12 event; 7–9, 20 copies of GM rice M12 event; 10–12, 2 copies of GM rice M12 event; 13, negative control; 14, NTC (ddH2O)
Quantification of the contents of GM rice M12 event in rice samples
Furthermore, in order to verify the developed LAMP method for quantifying different contents of GM rice M12 event in rice sample, the initial concentration of the two target genes in the 0.1–100% (w/w) GM rice M12 event were calculated by the developed real-time LAMP. It was found that no amplification signal of M12 event specific gene was observed in the non-GM rice “minghui 63”, while other assays showed positive results (Fig. 5, Table 4). For the sample containing 0.1% GM rice M12 event, the calculating GM concentration varied from 0.2 to 1.3% for three parallels with a RSDr of 91.2%, though the positive results were both obtained with the calcein and double-stranded displacement probes. Taking into account the performance requirements of European Network of GMO Laboratories (ENGL) [26], these data were assumed to be unacceptable at this concentration. However, for the higher concentration levels 0.5%, 5% and 100%, they were determined as 0.46% ± 0.07%, 5.28% ± 0.53% and 92.84% ± 3.94%, respectively. Thus, the accuracy and precision were acceptable since the bias was from 5.57 to 8.64% and the RSDr was from 4.2 to 14.2%. These data showed that when the content of M12 was equal or greater than 0.5% in rice samples, the results from the developed quantitative LAMP methods were accurate and reliable. In addition, since the word’s lowest detection limit 0.9% was established in European Union [27], the developed quantitative LAMP methods were effective and reliable and can be used widely.
Detection of rice sample with different GM contents by visible LAMP amplification with calcein. a SPS gene, b M12 event specific gene. The meaning of the labeling was as follows: 1–3, 100% GM rice event M12; 4–6, 5%GM rice event M12; 7–9, 0.5%GM rice event M12; 10–12, 0.1%GM rice event M12; 13–15, wild-type rice “minghui 63” 16, NTC (ddH2O)
Discussion
Currently, real-time PCR and digital PCR were widely used in quantitative detection of GMO, as they had relatively high levels of precision and accuracy [4, 28,29,30,31,32,33,34]. However, they required precise laboratory instrument, which restricted their use in on-site or poor-equipped laboratories. Compared with real-time PCR and digital PCR, LAMP showed comparable or even better results with simple experimental condition. In LAMP, its specificity was not worse or even better than that in real-time PCR theoretically [15], since it contained 4–6 distinct primers targeting 6–8 sequences. In another study, LAMP was even shown to be less susceptible to background DNA than real-time PCR [14]. Usually, the LAMP method was used to qualitatively detect GM crops. However, in this study, LAMP was developed as an efficient quantitative method by using the new designed double-stranded displacement probes, and it was proved to be an alternative to real-time quantitative PCR analyses, especially in on-site or poor-equipped laboratories.
For real-time PCR and digital PCR, the quantitative limits ranged from 5 to 300 copies approximately [3,4,5, 35,36,37,38,39], and it was reported that the LOD and LOQ were 10 and 100 copies, respectively, when the real-time PCR was used to analyse the GM rice M12 event [40]. In this study, the SPS and M12 event-specific genes with a content of less than 50 copies were quantified by digital PCR, and the accuracy and precision meet the general quantitative requirements. While using the developed quantitative LAMP method to analyze the same GM sample in this study, the LOD and LOQ were 20 and 200 copies respectively. Thus, the sensitivity of the quantitative LAMP using the double-stranded displacement probes in this study was slightly lower compared with the real-time PCR and digital PCR. Moreover, when the copy numbers of the target genes were more than 200 or the GM content was more than 0.5% (w/w), it could be quantified accurately by the quantitative LAMP with double-stranded displacement probes. However, for the quantification of GM material using real-time PCR or digital PCR, as low as 0.1% of the GM content was reliably quantified [29, 41]. Though the sensitivity of the quantitative LAMP using the double-stranded displacement probes in this study was slightly lower compared with real-time PCR and digital PCR method, the developed LAMP method still meets the international minimum labeling requirements (i.e. 0.9% in European Union, 5% in Japan, 1% in Brazil, etc.) [27]. Meanwhile, the LAMP assay developed in this study can provide a simple approach for quantifying GMO content in the basic laboratory or on-site quantitative testing which cannot be completed by the other two methods.
Although quantitative LAMP methods with turbidity had been proved to be feasible theoretically [7, 16,17,18], there were few follow-up studies in the past few years. In the real-time LAMP, there were insufficient data to prove the accuracy and accuracy of the quantification [17, 18]. It may be due to the instability of turbidity in LAMP amplification or the inaccuracy of portable instruments. However, when the double-stranded displacement probes were used in this study, the positive signals were more stable and the precision of the experiment turned to be better, and the linear correlation between POI value and log(template concentration) was high enough to enable the quantitative analysis. Therefore, the novel real-time LAMP assays with double-stranded displacement probes designed in this study were considered to be an improvement of the LAMP assay. Moreover, the developed LAMP method would be a good supplementary method for DNA-based quantitative techniques.
Conclusions
In this study, it was already proved that the developed LAMP was an efficient alternative to screen and quantify the GM rice M12 event. It had ideal specificity on the detection of SPS and M12 event-specific gene. Meanwhile, the methods had the good performance to quantify GM rice M12 event ingredients in rice sample. Moreover, the LAMP reaction does not require expensive equipment (particular thermal cycle instrument or Gel electrophoresis equipment), and the reaction can be conducted quantitatively using a simple and portable device (a thermo-stable block). Furthermore, when the GM content was equal or greater than 0.5% (w/w), it could be quantified accurately by the quantitative LAMP with double-stranded displacement probes. Thus, the LAMP assay developed in this study can provide a simple method aiming to quantify GM ingredients in imported and domestic foods, and it can be used as an effective alternative to quantify GMO content in the basic laboratory or on-site quantitative testing.
Abbreviations
- LAMP:
-
Loop-mediated isothermal amplification
- GM:
-
Genetically modified
- PCR:
-
Polymerase chain reaction
- LOD:
-
Limit of detection
- LOQ:
-
Limit of quantification
References
Dörries HH, Remus I, Grönewald A, Grönewald C, Berghof-Jäger K (2010) Development of a qualitative, multiplex real-time PCR kit for screening of genetically modified organisms (GMOs). Anal Bioanal Chem 396:2043–2054
Wang X, Teng D, Yang Y, Tian F, Guan Q, Wang J (2011) Construction of a reference plasmid molecule containing eight targets for the detection of genetically modified crops. Appl Microb Biotech 90:721–731
Xu X, Chen X, Lai Y, Zhang Q, Wu Y, Li J, Peng C, Wang X, Miao Q, Liu H, Xu J (2019) Event-specific qualitative and quantitative detection of genetically modified rice G6H1. Food Anal Method 12:440–447
Grohmann L, Reiting R, Mäde D, Uhlig S, Simon K, Frost K, Randhawa GJ, Zur K (2015) Collaborative trial validation of cry1Ab/Ac and Pubi-cry TaqMan-based real-time PCR assays for detection of DNA derived from genetically modified Bt plant products. Accred Qual Assur 20:85–96
Barbau-Piednoir E, Stragier P, Roosens N, Mazzara M, Savini C, Van den Eede G, Van den Bulcke M (2014) Inter-laboratory testing of GMO detection by combinatory SYBR®Green PCR screening (CoSYPS). Food Anal Method 7:1719–1728
Yang L, Yang Y, Jin W, Zhang X, Li X, Wu Y, Li J, Li L (2018) Development and interlaboratories validation of event-specific quantitative real-time PCR method for genetically modified rice G6H1 event. J Agr Food Chem 66:8179–8186
Huang X, Chen L, Xu J, Ji HF, Zhu S, Chen H (2014) Rapid visual detection of phytase gene in genetically modified maize using loop-mediated isothermal amplification method. Food Chem 156:184–189
Wang C, Li R, Quan S, Shen P, Zhang D, Shi J, Yang L (2015) GMO detection in food and feed through screening by visual loop-mediated isothermal amplification assays. Anal Bioanal Chem 407:4829–4834
Zhou D, Guo J, Xu L, Gao S, Lin Q, Wu Q, Wu L, Que Y (2014) Establishment and application of a loop-mediated isothermal amplification (LAMP) system for detection of cry1Ac transgenic sugarcane. Sci Rep UK 4:4912
Takabatake R, Kagiya Y, Minegishi Y, Yeasmin S, Futo S, Noguchi A, Kondo K, Mano J, Kitta K (2018) Development and evaluation of rapid screening detection methods for genetically modified crops using loop-mediated isothermal amplification. Food Chem 252:390–396
Li Q, Fang J, Liu X, Xi X, Li M, Gong Y, Zhang M (2013) Loop-mediated isothermal amplification (LAMP) method for rapid detection of cry1Ab gene in transgenic rice (Oryza sativa L.). Eur Food Res Technol 236:589–598
Chen X, Wang X, Jin N, Zhou Y, Huang S, Miao Q, Zhu Q, Xu J (2012) Endpoint visual detection of three genetically modified rice events by loop-mediated isothermal amplification. Int J Mol Sci 13:14421–14433
Norihiro T, Yasuyoshi M, Hidetoshi K, Tsugunori N (2008) Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat Protoc 3:877
Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T (2000) Loop-mediated isothermal amplification of DNA. Nucleic Acids Res 28:E63
Nagamine K, Hase T, Notomi T (2002) Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol Cell Probe 16:223–229
Mori Y, Kitao M, Tomita N, Notomi T (2004) Real-time turbidimetry of LAMP reaction for quantifying template DNA. J Biochem Biophys Method 59:145–157
Parida M, Sannarangaiah S, Dash PK, Rao PVL, Morita K (2008) Loop mediated isothermal amplification (LAMP): a new generation of innovative gene amplification technique; perspectives in clinical diagnosis of infectious diseases. Rev Med Virol 18:407–421
Soleimani M, Shams S, Majidzadeh-A K (2013) Developing a real-time quantitative loop-mediated isothermal amplification assay as a rapid and accurate method for detection of Brucellosis. J Appl Microbiol 115:828–834
Li C, Li Z, Jia H, Yan J (2011) One-step ultrasensitive detection of microRNAs with loop-mediated isothermal amplification (LAMP). Chem Commun 47:2595–2597
Navarro E, Serrano-Heras G, Castaño MJ, Solera J (2015) Real-time PCR detection chemistry. Clin Chim Acta 439:231–250
Yang J, Ni D, Wu J, Xu C, Jia S, Tang Y, Fauquet C, Zhang S (2006) Breeding and food safety evaluation of transgenic hybrid rice harboring Xa21 gene. Mol Plant Breeding 4:174–180
Wang W, Han J, Wu Y, Yuan F, Chen Y, Ge Y (2011) Simultaneous detection of eight food allergens using optical thin-film biosensor chips. J Agr Food Chem 59:6889–6894
Yu J, Hu S, Wang J, Wong GKS, Li S, Liu B, Deng Y, Dai L et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296:79–92
Ding J, Jia J, Yang L, Wen H, Zhang C, Liu W, Zhang D (2004) Validation of a rice specific gene, sucrose phosphate synthase, used as the endogenous reference gene for qualitative and real-time quantitative PCR detection of transgenes. J Agr Food Chem 52:3372–3377
International Standard Orgarnization (1994) Accuracy (trueness and precision) of measurement methods and results. International Organization for Standardization, ISO) 5725. In: Geneva: International Standard Orgarnization, pp 5–6
European Network of GMO Laboratories (ENGL) (2015) Definition of minimum preformance requirements for analytical methods of GMO testing. EURL-Scientific and Technical Research Reports JRC95544. https://gmo-crl.jrc.ec.europa.eu/doc/MPR%20Report%20Application%2020_10_2015.pdf. Accessed 18 Aug 2017
Xu L, Liu P, Li W, Sun Z, Song G (2018) Analysis of the recent trends of international labeling policies for genetically modified products and the enlightenment to China's labeling management. China Biotechnol 38:94–98
Meti-Buh G, Torstein T, La Jose Luis P, Holst-Jensen A, Maria P, Teresa E, Jana Ž, Kristina G (2010) Comparison of nine different real-time PCR chemistries for qualitative and quantitative applications in GMO detection. Anal Bioanal Chem 396:2023–2029
Fu W, Zhu P, Wang C, Huang K, Du Z, Tian W, Wang Q, Wang H, Xu W, Zhu S (2015) A highly sensitive and specific method for the screening detection of genetically modified organisms based on digital PCR without pretreatment. Sci Rep UK 5:12715
Zhang L, Cao Y, Liu X, Wu G, Wu Y, Lu C (2012) In-depth analysis of the endogenous reference genes used in the quantitative PCR detection systems for rice. Eur Food Res Technol 234:981–993
Sina-Elisabeth BA, Zita Erika M, Rupert H, David Q, Bernhard P, Haslberger AG, Christian B (2014) Mutation scanning in a single and a stacked genetically modified (GM) event by real-time PCR and high resolution melting (HRM) analysis. Int J Mol Sci 15:19898–19923
Zhu P, Wang C, Huang K, Luo Y, Xu W (2016) A novel pretreatment-free duplex chamber digital PCR detection system for the absolute quantitation of GMO samples. Int J Mol Sci 17:402. https://doi.org/10.3390/ijms17030402
Fan C, Ma J, Guo Q, Li X, Wang H, Lu M (2013) Selection of reference genes for quantitative real-time PCR in bamboo (Phyllostachys edulis). PLoS ONE 8:e56573
Ling H, Wu Q, Guo J, Xu L, Que Y (2014) Comprehensive selection of reference genes for gene expression normalization in sugarcane by real time quantitative RT-PCR. PLoS ONE 9:e97469
Kim JH, Park SB, Roh HJ, Shin MK, Moon GI, Hong JH, Kim HY (2017) Event-specific qualitative and quantitative detection of five genetically modified rice events using a single standard reference molecule. Food Chem 226:187–192
Wu G, Wu Y, Nie S, Zhang L, Xiao L, Cao Y, Lu C (2010) Real-time PCR method for detection of the transgenic rice event TT51-1. Food Chem 119:417–422
Hernández M, Esteve T, Prat S, Pla M (2004) Development of real-time PCR systems based on SYBR® Green I, Amplifluor™ and TaqMan® technologies for specific quantitative detection of the transgenic maize event GA21. J Cereal Sci 39:99–107
Taverniers I, Windels P, Vaïtilingom M, Milcamps A, Van Bockstaele E, Van den Eede G, De Loose M (2005) Event-specific plasmid standards and real-time PCR methods for transgenic Bt11, Bt176, and GA21 maize and transgenic GT73 canola. J Agr Food Chem 53:3041–3052
Liu E, Lu L, Lv Y, Jiang X, Zhang W, Tang J, Zheng G, Lin X, Fu Q (2015) Event-specific qualitative PCR detection method of genetically modified alfalfa events J101. J Food Saf Qual 6:1936–1940
Li P, Zhang L, Ye J, He S, Jia J, Pan A, Tang X (2018) A qualitative and quantitative PCR detection method for disease-resistant genetically modified rice M12 and its derivates. Acta Agron Sin 44:949–955
Wan J, Li S, Wu Y, Brzoska P, Keys D, Chen C, Valliyodan B, Shannon JG, Nguyen HT (2016) Application of digital PCR in the analysis of transgenic soybean plants. Adv Biosci Biotechnol 7:403–417
Acknowledgements
This work was supported by National GMO Cultivation Major Project of New Varieties (Grant no. 2018ZX08012001).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Compliance with ethics requirements
This article does not contain any studies with human or animal subjects.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Deng, T., Huang, W., Xing, R. et al. Establishment and application of a loop-mediated isothermal amplification method with double-stranded displacement probes to quantify the genetically modified rice M12 event. Eur Food Res Technol 246, 631–641 (2020). https://doi.org/10.1007/s00217-020-03430-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00217-020-03430-1