Abstract
A standard plasmid containing eight targets was developed for quantitative detection of genetically modified (GM) soybeans and cotton. These eight targets were joined in tandem to form the pTLE8 plasmid with a length of 3,680 bp. This plasmid contains part of the endogenous soybean Lec1 gene, the Cauliflower mosaic virus (CaMV) 35S promoter, the Agrobacterium tumefaciens nopaline synthase (NOS) terminator, the PAT gene of the soybean line A2704-12, the event-specific 5′-junction region of Roundup-Ready Soya (RRS, 35SG), the Cry1A(c) gene from Bacillus thuringiensis (Bt), the endogenous cotton Sad1 gene, and a part of RRS EPSPS gene. The PCR efficiencies with pTLE8 as a calibrator ranged from 99.4% to 100.2% for the standard curves of the RRS EPSPS gene and the taxon-specific Lec1 gene (R 2 ≥ 0.996). The limits of detection and quantification were nine and 15 copies, respectively. The standard deviation (SD) and relative standard deviation (RSD) values of repeatability were from 0.09 to 0.52 and from 0.28% to 2.11%, and those for reproducibility were from 0.12 to 1.15 and 0.42% to 3.85%, respectively. The average conversion factor (Cf) for the CRMs RRS quantification was 0.91. The RSD of the mean values for known samples ranged from 3.09% to 18.53%, and the biases were from 0.5% to 40%. These results show that our method using the pTLE8 plasmid as a reference material (RM) is reliable and feasible in the identification of GM soybeans, thus paving the way for the establishment of identification management systems for various products containing GMO components.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The production of approved GM crops and derived products continues to increase in more and more countries. In an attempt to regulate the increasing consumption of genetically modified organism (GMO)-derived products, a uniform traceability system for the documentation of the flow of GMO-derived commodities has been established in the European Community (EC 50/European Commission Regulation EC 2000; EC 1830/European Commission Regulation EC 2003). Regulation (EC) number 1829/European Commission Regulation EC 2003 on GM food and feed regulates the placement on the market of food and feed products containing GMOs and mandates the labeling of such products.
To enforce the GMO labeling regulations, the application of reference molecules as calibrators is becoming essential for practical identification of GMOs (Huang and Pan 2005). Because of the advantages of plasmid standards in terms of easy preparation, low cost, universal applicability, and long-term stability, plasmid DNA calibrators are preferred over genomic DNA calibrators from certified reference materials (CRMs). Validated plasmid calibrators are widely used for GMO quantification. At present, single- or multiple-target plasmids, containing either one GM-specific sequence or endogenous sequence or several specific GM sequences from one species, have been developed as RMs (Kuribara et al. 2002; Yang et al. 2005; Burns et al. 2006; Toyota et al. 2006; Sivaganesan et al. 2008; Wang et al. 2009; Köppel et al. 2010; Lievens et al. 2010; Scholtens et al. 2010).
In this study, a new multiplex calibrator was constructed by integrating eight fragments [1, the endogenous soybean gene (Lec1); 2, the 35S promoter; 3, the NOS terminator; 4, the PAT gene of the soybean line A2704-12; 5, the RRS event-specific 5′-junction (35SG); 6, the Cry1A(c) gene modified from Bt; 7, the endogenous cotton reference control gene (Sad1); and 8, the RRS EPSPS gene] into the same plasmid. The above eight DNA fragments were spliced together and subsequently cloned into an appropriate vector for GM detection based on previous protocols (Kuribara et al. 2002; Yu et al. 2004; Lu 2005; Hong et al. 2006; Charels et al. 2007; Pang et al. 2007; Sivaganesan et al. 2008). We also developed a quantitative PCR assay for GM soybean detection using the above plasmid as a reference molecule in place of CRMs, and we further evaluated the established method, including determination of the Cf value calculated by the copy number ratios of exogenous to endogenous DNA sequences of genuine RRS (Arumuganathan and Earle 1991).
Materials and methods
Soybean samples
Non-GM soybeans (ERM-BF410a) and CRM RRS (ERM-BF410e, containing 20 g/kg GMO) were purchased from the EU Joint Research Centre, IRMM (Institute for Reference Materials and Measurements, Belgium). The RRS flours were initially mixed with non-GM soybean flour to obtain samples containing 0.25, 0.5, 1, and 2 g/kg RRS [0.25%, 0.5%, 1%, and 2% RRS (w/w), respectively]. Genomic DNA was isolated from powdered CRMs according to a SDS-based method (Ogunkanmi et al. 2008). The DNA pellets were dissolved in 100 μl nuclease-free water. The samples were further diluted to give 50 ng/μl working stock solutions with sterile distilled water. The gDNA was stored in aliquots at −80 °C.
Fusion PCR
Primers
Primers were designed for the endogenous soybean Lec1 gene and for segments of the sequences of 35S, NOS, PAT, 35SG, Cry1A(c), Sad1, and EPSPS from GenBank (Table 1). The two primers in the overlapping primer pairs of Lec1-R/35S-F, 35S-R/NOS-F, NOS-R/Pat-F, 35G-R/Cry-F, Cry-R/Sad-F, and Sad-R/Eps-F were complementary to each other. Four restriction enzyme sites, NotI, SalI, EcoRI, and XbaI, were introduced at the ends of the primers for the Lec1, Pat, 35SG, and EPSPS genes. All primers were synthesized and purified by Shanghai Sangon Biotech.
First-round PCR
Eight parallel PCRs were performed to amplify eight individual DNA fragments in a reaction volume of 50 μl containing 2.5 U of Taq DNA polymerase and 1× buffer in the presence of 200 μM dNTP, 0.8 mmol of corresponding primers (Table 1), and 10 ng of genomic DNA from GM soybean or cotton. The optimized PCR program for Lec1 was as follows: 94 °C for 5 min; then 30 cycles of 94°C for 30 s for denaturation, 58.6°C for 30 s, and 72°C for 1 min; followed by 72°C for 10 min. The reaction conditions for the other genes were similar except for the annealing temperature and the extension time. The annealing temperatures used for fragments 2–8 were 56.8 °C, 58.8 °C, 59.5 °C, 55.1 °C, 56.5 °C, 58.6 °C, and 58.6 °C, respectively, and the extension times used for fragments 2–8 were 50 s, 30 s, 30 s, 30 s, 40 s, 30 s, and 60 s, respectively. The amplified products were separated on a 1.5% agarose electrophoresis gel and purified using the TIANgel Midi Gel Extraction Kit (Tiangen Biotech, Beijing, China).
Second-round PCR
The 3,680-bp full-length fragment containing the eight genes was assembled step-by-step (Fig. 1). Firstly, equimolar aliquots of every two or three adjacent DNA fragments (70 ng for each) were mixed in the presence of Taq DNA polymerase (2.5 U), 1× buffer, and 200 μmol of each dNTP in a final volume of 50 μl. The reaction program consisted of 15 cycles of PCR without primers with denaturation at 94 °C for 20 s and annealing and extension conditions as follows: 59.6 °C for 1 min 25 s for the Lec1 and 35S fragments; 58.8 °C for 1 min 25 s for the NOS and PAT fragments; 59.5 °C for 1 min 25 s for the 35SG and Cry fragments; and 60 °C for 1 min 20 s for the 35SG, Cry, Sad1, and EPSPS fragments. Then, 20 μl each of two or three products from the above step were mixed together and 20 cycles of 94 °C for 20 s and 72 °C for 30 s run without primers. Finally, both outer primers (40 pmol for each) were added into the reaction system, and PCR was conducted using the following cycles: 94 °C for 30 s followed by 28 cycles of 94 °C for 30 s, 58.6 °C for 1 min, and 72 °C for 3 min. Then, the 3,680-bp DNA band was excised and recovered from an agarose gel with the TIANgel Mini Purification Kit (Tiangen Biotech).
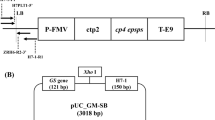
Schematic illustration of fragments of eight genes fused by overlap PCR and the construction of the pTLE8 plasmid. a First-round PCR: synthesis of DNA fragments by PCR; b double-mixing and pre-extension in second-round PCR: every two adjacent fragments were mixed to implement overlap PCR without primers to obtain full-length reassembled DNA; c synthesis of the entire DNA of interest by PCR with the outermost primers and template DNA from step 1 in second-round PCR; d post-extension in second-round PCR: 28 cycles of PCR with an extension temperature of 72 °C were implemented; e schematic diagrams of pTLE8: the integrated fragment was inserted into a vector
Construction of a multiple-target plasmid DNA reference molecule
The 3,680-bp full-length fragment containing the amplified segments of the eight genes was cloned into the pMDTM19-T Simple Vector (Takara, China) and transformed to Escherichia coli DH5α. Three positive clones were selected and sequenced. Plasmid DNA was extracted using a Beijing TIANprep Mini Plasmid Kit and was digested with NotI and XbaI. Purified plasmids were then linearized with EcoRI for 3 h at 37 °C and purified with the TIANquick Midi Purification Kit.
Real-time PCR
PCR primers and probes specific for Lec1 and RRS EPSPS were designed using Primer V 5.0 software (Table 2) and synthesized by Takara Co. The probe contained a reporter dye (TET/FAM) at the 5′ end and a quencher dye (BHQ/Eclipse) at the 3′ end.
TaqMan Universal PCRs were performed in a 15 μl final volume containing 7.5 μl of Premix Ex TaqTM Mix (Takara), 0.3 μl of ROX Dye II (50×), 200 nM of each primer, 250 nM of the probe for Lec1 or EPSPS, and 1.5 μl DNA extracts (50 ng/μl). The PCRs were all run on an ABI PRISM 7500 Sequence Detection System (Applied Biosystems, USA) using the following program: 50 °C for 2 min, 95 °C for 10 min, and 45 cycles of 95 °C for 15 s and 60 °C for 1 min. The fluorescence signals were measured once during each cycle at the extension step, and the data were then analyzed using the Detection System V2.03 (Applied Biosystems). Each reaction was repeated three times in triplicate.
Standard curves, repeatability, and sensitivity
Five serially diluted plasmid DNA samples (54.9, 549, 5,490, 54,900, and 549,000 copies per reaction in 1.5 μl) were used as a calibrators for the construction of a standard curve. The standard curve was constructed by plotting the Ct values against the logarithm of the DNA copy number.
To test the repeatability and reproducibility, three independent real-time PCR runs completed at different times over a period of 30 days with three replicates per run were performed for each sample dilution. The sensitivity of the method was also assessed by determining the limit of detection (LOD) and the limit of quantification (LOQ) with the lower dilution of 6 copies/μl of pTLE8 DNA.
Determination of conversion factors (Cf) for the reference molecule
A gradient of 25, 50, 75, 100, and 150 ng DNA from 2% (w/w) RRS CRM per reaction was prepared to determine the Cf value. The Cf value was calculated by Eq. 1:
The copy numbers were estimated by standard curves based on the reference molecule pTLE8. The final Cf value was the average of five calculated Cfs.
Real-time quantitative PCR of sample unknowns
To validate the real-time PCR systems based on pTLE8, four samples containing 0.25%, 0.5%, 1.0%, and 2.0% (w/w) RRS were treated as unknown genomic samples. Quantification was performed by linear interpolation using a standard regression curve of cycle threshold (Ct) values generated from DNA samples of known concentrations. The GM contents of the samples were calculated according to Eq. 2:
Results
Fusion PCR
The fusion of the eight targets is illustrated in Fig. 1. First, eight individual target segments were PCR-amplified (Fig. 2). The lengths of the amplified fragments were in agreement with the predicted lengths of 530 bp, 311 bp, 253 bp, 371 bp, 369 bp, 312 bp, 335 bp, and 1,199 bp corresponding to sequences of Lec1, 35S, NOS, PAT, 35SG, Cry, Sad1, and EPSPS, respectively. These products were purified and assembled step-by-step to form four fusion fragments: 35SG-Cry (681 bp), NOS-PAT (624 bp), Lec1-35 S (841 bp), and 35SG-Cry-Sad1-EPSPS (2,215 bp) (Fig. 2). Next, the Lec1-35S and NOS-PAT segments was fused into a four-target segment, Lec1-35S-NOS-PAT (data not shown), which was then merged with another four-target segment, 35SG-Cry-Sad1-EPSPS, to form a 3,680-bp full-length fragment (Fig. 2). As estimated by agarose gel electrophoresis, the final PCR product reached a level of 566.7 ng/50 μl.
Agarose (1.5%) gel electrophoresis of the eight-gene fusion constructed by overlap PCR. M1 DNA marker II, M2 DNA marker III, lane 1 Lec1 gene (530 bp), lane 2 35S promoter (311 bp), lane 3 NOS terminator (253 bp), lane 4 PAT gene (371 bp), lane 5 35SG fragment (369 bp), lane 6 Cry gene (312 bp), lane 7 Sad1 gene (335 bp), lane 8 EPSPS gene (1,199 bp), lane 10 35SG + Cry (681 bp), lane 11 NOS + PAT (624 bp), lane 12 Lec1 + 35S (841 bp), lane 13 35SG + Cry + Sad1 + EPSPS (2,215 bp), lanes 9 and 14 Lec1 + 35S + NOS + PAT + 35SG + Cry + Sad1 + EPSPS (3,680 bp)
Construction of a new plasmid as a reference molecule
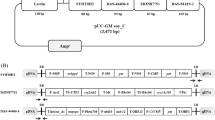
The 3,680-bp full-length fragment was ligated into the pMDTM19-T Simple Vector and transformed into E. coli DH5α. The inserted DNA and the correct structure of the 3,680-bp recombinant product were confirmed by DNA sequencing and restriction enzyme digestion. The sequence of the pTLE8 plasmid contained the DNA sequences of a part of the soybean taxon-specific Lec1 gene, the 35S promoter, the NOS terminator, the PAT gene of the event A2702-12, the RRS event-specific 5′-junction (35SG), a part of the Cry1A(c) gene modified from Bt, a part of the endogenous cotton reference control gene (Sad1), and a part of the RRS EPSPS gene (Fig. 3). The sequence of this 3,680-bp recombinant was consistent with the designed sequencing as determined by Blast via NCBI.
Sequences of the integrated fragments in the standard pTLE8 plasmid. Bold italic letters indicate the sequences of the overlapping primers of two genes, and boxes indicate the sequences of primers and TaqMan probes. Restriction endonuclease enzyme (NotI, SalI, EcoRI, and XbaI) sites are underlined and were inserted into the eight genes in the pTLE8 plasmid reference molecule
The double-stranded plasmid DNA template was subjected to restriction endonuclease digestion, and the DNA fragments were analyzed by agarose gel electrophoresis. The pTLE8 plasmid DNA digested with NotI and XbaI was resolved into two visible bands corresponding to the expected 2,692-bp vector and the 3,680-bp DNA insert fragment, while only one 6,372-bp visible band containing vector and insert DNA was observed when the pTLE8 plasmid DNA was digested with EcoRI (data not shown), showing that the three constructs contained the 3,680-bp region of interest.
Based on the known length of the plasmid vector (2,692 bp), the inserted fragment (3,680 bp), and the molecular mass of single-stranded DNA, the concentration (ng/μl) was used to calculate the number of target copies per PCR reaction (1.5 μl DNA template). For each plasmid DNA marker, 10-fold dilution series were then made from 100,000 to 10 copies. Linearized plasmid solutions containing 36.6, 366, 3,660, 36,600, and 366,000 copies (absolute amounts in 1.0 μl) were prepared as one set.
Calibration curves and sensitivity of the test
For the generation of standard curves, purified plasmid DNA was used as a calibrator. Figure 4 shows two typical standard curves in one of three real-time PCR runs. The regression correlation coefficient (R 2) values of the standard curves were from 0.996 to 0.999 in three PCR runs, indicating excellent linearity between the DNA copy numbers in the template and the fluorescence values (Ct). The PCR reaction efficiencies were from 99.4% to 100.2% for the standard curves of the RRS EPSPS fragment and the taxon-specific Lec1 gene, indicating highly efficient reactions.
Calibration curves for the soybean Lec1 and EPSPS genes. The dilutions contained approximately 6.83 × 105, 6.83 × 104, 6.83 × 103, 6.83 × 102, and 6.83 × 10 initial template copies per PCR. a Construction of the calibration curve and estimation of the amplification efficiency for the Lec1 gene; b construction of the calibration curve and estimation of the amplification efficiency for the EPSPS gene
In this study, the LOD was determined using four serially diluted pTLE8 DNA samples as calibrators. These solutions included 9, 15, 30, and 60 copies per reaction and were analyzed three times, each time with three parallel replicates. The LOD and LOQ of the qualitative PCR were nine and 15 copies of pTLE8 DNA, respectively (Table 3). SD values were in the acceptable range, from 0.13 to 0.34. Based on the lengths of the pTLE8 plasmid (6,392 bp) and the soybean genome (Lievens et al. 2010), we deduced that copy numbers between 15 and 549,000 for pTLE8 would be sufficient to quantify GM soybean DNA representing 0.01% to 100% of a 75 ng genomic DNA sample. Moreover, this range should be sufficient to meet the requirements of the GMO labeling regulations.
Repeatability and reproducibility of the method
The repeatability and reproducibility of the reactions for the real-time PCR system were calculated using data for triplicate reactions obtained from the five concentration levels of pTLE8. The Lec1 and EPSPS genes of the pTLE8 plasmid were repeatedly amplified three times a day. Table 4 shows the results of this reproducibility study of the pTLE8 plasmid for the same day and for three different days using the established method. The values of SD and RSD were all in the acceptable range according to ENGL (ENGL Definition of minimum performance requirements for analytical methods for GMO testing; European Network of GMO Laboratories ENGL 2008) and were from 0.09 to 0.52 and from 0.28% to 2.11% for 1 day, respectively. The SD and RSD values obtained were in the range of 0.12 to 1.15 and 0.42% to 3.85% for the three different days, respectively.
Measurement of Cf
Five different levels of Matrix-CRM DNA for GM soybeans (RRS) were analyzed three times, each time in triplicate, to validate the Cf values. As shown in Table 5, the average Cf value for the RRS quantification was 0.91. The relative standard deviation (RSD) value of the Cf was 11.91%. Theoretically, the transgenic EPSPS sequence and the endogenous Lec1 gene in a haploid RRS genome are both only present as a single copy (Holst-Jensen et al. 2003); therefore, the theoretical Cf of soybeans, including the single copy of the transgene per genome, should be 1.0 for each real-time PCR assay. The discrepancy between the experimental and theoretical Cf values might be the result of deviations in the DNA copy number, variations of the mass-based GM content, and differences in the PCR amplification efficiencies that resulted from the amounts of the non-target sequences in the plasmid and genomic DNA, although no considerable homology to the sequences used for detection by using the DNA database of target crops was found (Holst-Jensen et al. 2003; Huang and Pan 2005; Yang et al. 2007a, b; Wang et al. 2009).
Quantitative analysis of the GM soybean samples using pTLE8 DNA as a calibrator
To investigate the suitability of our calibrator for relative quantification, a series of four samples with well-known GMO contents (in %) were analyzed as unknowns; these samples were 0.25%, 0.5%, 1%, and 2% RRS. Table 6 shows the quantitative data obtained after all runs, and mean relative values of 0.35, 0.66, 1.18, and 2.01 were obtained with the pTLE8 calibrator. The SD values were reasonable, ranging from 0.03 to 0.22, and the RSD values were between 3.09% and 18.53%. The biases for the 2%, 1%, 0.5%, and 0.25% RRS samples were 0.5%, 18%, 32%, and 40%, respectively.
Discussion
A novel reference molecule, pTLE8, containing screen-, event-, and taxon-specific sequences for two major GM crops (soybean and cotton) was constructed in this work. Eight target genes were fused together by overlap extension PCR and then inserted into a destination vector.
Overlap extension PCR was described initially for site-directed mutagenesis by Higuchi et al. (1988) and was then used to splice two short DNA fragments (Yolov and Shabarova 1990). The novelty of the method proposed in this work includes the following. (1) An overlapping sequence between 18 to 25 bp in length was introduced into the adjacent ends of eight target DNA fragments. The desired PCR results were only obtained when these overlapping sequences were included, as reported previously (Zheng et al. 2004; An et al. 2005; Peng et al. 2006), but no amplification products were observed when using overlapping sequences longer than 25 bp (data not shown). (2) In contrast to previous observations (Shevchuk et al. 2004; Taverniers et al. 2004; Simionatto et al. 2009), the fusion of the eight fragments via this overlap extension PCR was more easily performed by Taq DNA polymerase than Pfu DNA polymerase; a higher yield of PCR product and less smearing were observed for Taq (data not shown). (3) Fewer cycles of PCR, 28 instead of 30–35 as used previously (Higuchi et al. 1988; Kuwayama et al. 2002; Yu et al. 2004; An et al. 2005), were sufficient to obtain a high yield of target DNA without non-target bands, and an additional three to five cycles (31–33 total) would have resulted in heavier non-target bands (data not shown).
Quantitative PCR systems based on the pTLE8 plasmid RM described above were developed, and our results showed that the amplification efficiency of 99.4% to 100.2% was within the range of the ENGL criterion of 89.6% to 110.2%, and the values of the R 2 coefficient for all reactions were at least 0.996. The efficiencies of the other quantification systems using the plasmid DNA as a calibrator ranged from 77.0 to 110.0%, and their R 2 coefficients ranged from 0.95 to 0.999 (Taverniers et al. 2004; Huang and Pan 2005; Burns et al. 2006; Toyota et al. 2006; Zhang et al. 2008; Köppel et al. 2010; Lievens et al. 2010). High PCR efficiencies and the linear relationship between copy number and Ct value proved that the real-time PCR assay based on standard calibration plasmids is well suited for quantitative measurements.
In addition, the standard deviations of repeatability and reproducibility (Table 4) were far below the ENGL criterion of 25% and 35%. Values of SD and RSD with similar ranges have been reported for other quantitative detection assays using various plasmid constructs as calibrators (Toyota et al. 2006; Yang et al. 2007a, b; Zhang et al. 2008). The above results indicate that this real-time PCR assay can be used for quantitative nucleic acid analysis.
In a previous study, the RSD and the bias of the quantitative results of known samples were 0.83% to 26.2% and 0.60% to 28%, respectively (Burns et al. 2006; Yang et al. 2007a, b; Zhang et al. 2008). In this study, the RSD ranged from 3.09% to 18.53% for different GM soybean percentages. A bias between 0.5% and 40% was found, and it is possible that mixing affected the bias. The calculated contents of these samples were very close to the expected theoretical values and were reasonable. The results indicate that the method of quantification for practical samples based on the pTLE8 plasmid has sufficient levels of accuracy and precision.
The reference molecule pTLE8 is particularly suitable for the rapid identification of GMO components in unknown samples and can reduce cross contamination (Kuribara et al. 2002; Yang et al. 2007a, b; Wang et al. 2009). CRMs of different species are not needed when the CRMs are used for the quantification of mixed samples. Furthermore, cross contamination resulting from multiple-species genomic DNA of different CRMs can be avoided in the detection of different GM crops in mixed food/feed samples with our protocol based on a plasmid containing multiple genes. Therefore, this plasmid can be used as a calibrator for the identification and quantification of GM soybean or derivates. At the same time, the quantitative assay established in this work can also be applied in management of the labeling of GMO products with a high accuracy and precision. This work provides the essential technical basis for developing this method into an official standard method.
References
An YF, Ji JF, Wu WF, Lv AG, Huang RB, Wei YT (2005) A rapid and efficient method for multiple-site mutagenesis with a modified overlap extension PCR. Appl Microbiol Biotechnol 68:774–778
Arumuganathan K, Earle ED (1991) Nuclear DNA content of some important plant species. Plant Mol Biol Rep 9:208–218
Burns M, Corbisier P, Wiseman G, Valdivia H, McDonald P, Bowler P, Ohara K, Schimmel H, Charels D, Damant A, Harris N (2006) Comparison of plasmid and genomic DNA calibrants for the quantification of genetically modified ingredients. Eur Food Res Technol 224:249–258
Charels D, Broeders S, Corbisier P, Trapmann S, Schimmel H, Emons H (2007) Toward metrological traceability for DNA fragment ratios in GM quantification. 2. Systematic study of parameters influencing the quantitative determination of MON 810 corn by real-time PCR. J Agric Food Chem 55:3258–3267
European Commission Regulation (EC) No. 50/2000 (2000) Off J Eur Commun L6:15–17
European Commission Regulation (EC) No. 1829/2003 and 1830/2003 (2003) Off J Eur Commun L268:1–28
European Network of GMO Laboratories (ENGL) (2008) Definition of minimum performance requirements for analytical methods of GMO testing (13 October 2008). Available at http://gmo-crl.jrc.ec.europa.eu/doc/Min_Perf_Requirements_Analytical_methods.pdf
Higuchi R, Krummel B, Saiki RK (1988) A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res 16:7351–7367
Holst-Jensen A, Rønning SB, Løvseth A, Berdal KG (2003) PCR technology for screening and quantification. Anal Bioanal Chem 375:985–993
Hong SY, Lee JS, Cho KM, Math RK, Kim YH, Hong SJ, Cho YU, Kim H, Yun HD (2006) Assembling a novel bifunctional cellulose–xylanase from Thermotoga maritima by end-to-end fusion. Biotechnol Lett 28:1857–1862
Huang CC, Pan TM (2005) The event-specific real-time detection and quantification of genetically modified soybean Roundup Ready®. J Agric Food Chem 53:3833–3839
Köppel R, Zimmerli F, Breitenmoser A (2010) Multiplex real-time PCR for the simultaneous detection and quantification of DNA from three transgenic rice species and construction and application of an artificial oligonucleotide as reference molecule. Eur Food Res Technol 230:731–736
Kuribara H, Shindo Y, Matsuoka T, Takubo K, Futo S, Aoki N, Hirao T, Akiyama H, Goda Y, Toyoda M, Hino A (2002) Novel reference molecules for quantitation of genetically modified maize and soybean. J AOAC Int 85:1077–1189
Kuwayama H, Obara S, Morio T, Katoh M, Urushihara H, Tanaka Y (2002) PCR-mediated generation of a gene disruption construct without the use of DNA ligase and plasmid vectors. Nucleic Acids Res 30:e2
Lievens A, Bellocchi G, De Bernardi D, Moens W, Savini C, Mazzara M, Van den Eede G, Van den Bulcke M (2010) Use of pJANUS™-02-001 as a calibrator plasmid for Roundup Ready soybean event GTS-40-3-2 detection: an interlaboratory trial assessment. Anal Bioanal Chem 396:2165–2173
Lu Q (2005) Seamless cloning and gene fusion. Trends Biotechnol 23:199–207
Ogunkanmi AL, Oboh B, Onifade B, Ogunjobi AA, Taiwo IA, Ogundipe OT (2008) An improved method of extracting genomic DNA from preserved tissues of Capsicum annuum for PCR amplification. EurAsia J BioSci 2:115–119
Pang S, Qureshi F, Shanahan D, Harris N (2007) Investigation of the use of rolling circle amplification for the detection of GM food. Eur Food Res Technol 225:59–66
Peng RH, Xiong AS, Yao QH (2006) A direct and efficient PAGE-mediated overlap extension PCR method for gene multiple-site mutagenesis. Appl Microbiol Biotechnol 73:234–240
Scholtens IMJ, Kok EJ, Hougs L, Molenaar B, Thissen JTNM, Van der Voet H (2010) Increased efficacy for in-house validation of real-time PCR GMO detection methods. Anal Bioanal Chem 396:2213–2227
Shevchuk NA, Bryksin AV, Nusinovich YA, Cabello FC, Sutherland M, Ladisch S (2004) Construction of long DNA molecules using long PCR-based fusion of several fragments simultaneously. Nucleic Acids Res 32:e19
Simionatto S, Marchioroa SB, Gallia V, Luercea TD, Hartwiga DD, Moreiraa ÂN, Dellagostin OA (2009) Efficient site-directed mutagenesis using an overlap extension-PCR method for expressing Mycoplasma hyopneumoniae genes in Escherichia coli. J Microbiol Methods 79:101–105
Sivaganesan M, Seifring S, Varma M, Haugland RA, Shanks OC (2008) A Bayesian method for calculating real-time quantitative PCR calibration curves using absolute plasmid DNA standards. BMC Bioinform 9:120–131
Taverniers I, Van Bockstaele E, De Loose M (2004) Cloned plasmid DNA fragments as calibrators for controlling GMOs: different real-time duplex quantitative PCR methods. Anal Bioanal Chem 378:1198–1207
Toyota A, Akiyama H, Sugimura M, Watanabe T, Sakata K, Shiramasa Y, Kitta K, Hino A, Esaka M, Maitani T (2006) Rapid quantification methods for genetically modified maize contents using genomic DNAs pretreated by sonication and restriction endonuclease digestion for a capillary-type real-time PCR system with a plasmid reference standard. Biosci Biotechnol Biochem 70:2965–2973
Wang S, Li X, Yang LT, Shen KL, Zhang DB (2009) Development and in-house validation of a reference molecule pMIR604 for simplex and duplex event-specific identification and quantification of GM maize MIR604. Eur Food Res Technol 230:239–248
Yang L, Pan A, Zhang K, Yin C, Qian B, Chen JX, Huang C, Zhang DB (2005) Qualitative and quantitative PCR methods for event-specific detection of genetically modified cotton Mon1445 and Mort531. Transgenic Res 14:817–831
Yang LT, Guo JC, Pan AH, Zhang HB, Zhang KW, Wang ZM, Zhang DB (2007a) Event-specific quantitative detection of nine genetically modified maizes using one novel standard reference molecule. J Agric Food Chem 55:15–24
Yang LT, Liang WQ, Jiang LG, Li WQ, Cao W, Wilson ZA, Zhang DB (2007b) A novel universal real-time PCR system using the attached universal duplex probes for quantitative analysis of nucleic acids. BMC Mol Biol 9:54–66
Yolov AA, Shabarova ZA (1990) Constructing DNA by polymerase recombination. Nucleic Acids Res 18:3983–3986
Yu JH, Hamari Z, Han KH, Seo JA, Reyes-Dominguez Y, Scazzocchio C (2004) Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet Biol 41:973–981
Zhang HB, Yang LT, Guo JC, Li Xiang, Jiang LG, Zhang DB (2008) Development of one novel multiple-target plasmid for duplex quantitative PCR analysis of Roundup Ready Soybean. J Agric Food Chem 56:5514–5520
Zheng L, Baumann U, Reymond JL (2004) An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res 32:e115
Acknowledgments
This study was supported by the National Major Project of Breeding for Genetically Modified Organism in China (no. 2009ZX08012-012B).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wang, X., Teng, D., Yang, Y. et al. Construction of a reference plasmid molecule containing eight targets for the detection of genetically modified crops. Appl Microbiol Biotechnol 90, 721–731 (2011). https://doi.org/10.1007/s00253-011-3159-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-011-3159-9