Abstract
The lactic acid bacteria (LAB) communities from must and through alcoholic (AF) and malolactic fermentations (MLF) of Tempranillo red wines were studied in ten wineries from the Designation of Origin Rioja during three consecutive vintages. A statistical study with data from both methods, PCR-DGGE and plating, was performed. Results showed that the LAB community in the D.O. Rioja was highly determined by the type of fermentation and also by the different stages within the winemaking, while other factors such as year, winery, or sampling subzones had not significant effect on the LAB species distribution. Three microbial families, seven genera, and 25 species were described in this research, and Lactobacillus was the most commonly detected genus before MLF. Curiously, genera and species not frequently detected in wines as Weissella, Fructobacillus, and Oenococcus kitaharae were identified during AF, and no-Oenococcus oeni species were described in some MLF by both methods. For the first time, two new O. oeni allelic groups were determined by 16S rDNA/DGGE being randomly adapted to the wine environment. Further studies targeted to understand the implication of the novel species, and O. oeni allelic groups in Rioja wine fermentations could be really interesting.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The wine vinification is a complex fermentative process that starts with grapes from grapevines. Traditionally, those grapes are harvested and conducted to the winery where they are steamed and crushed to become must. Generally, yeasts perform the alcoholic fermentation (AF) converting sugars from must to ethanol and other secondary compounds as CO2. During a second fermentative stage, the lactic acid bacteria (LAB) develop the malolactic fermentation (MLF), which is a bioconversion of l-malic acid from wine into l-lactic acid, CO2, and several intermediate compounds that reduce the total acidity of wines [1]. Both fermentative stages can be depleted sequentially or can be developed simultaneously; moreover, both fermentations can be spontaneous or can be carried out by commercialized starter cultures of yeasts and LAB.
In any case, carrying out the MLF in red wines is advisable for ensuring the wine microbiological stability while the wine sensorial quality results improved [2]. Therefore, monitoring the LAB species taking part in the winemaking environment from must to final MLF could be relevant for elaborating quality wines [3]. Until now, no more than 20 LAB species have been described for must and wine, respectively [4, 5] being Oenococcus oeni the predominant one during MLF. This species is really resistant to low pH and high ethanol content of wines, for these reason is the most frequently detected species that fermentative stage [6].
In a previous study, the O. oeni clonal diversity was analyzed only by culture-dependent methods to develop a future clonal selection [7]. In that previous research, the genera Oenococcus (O.), Lactobacillus (L.) and Pediococcus (P.) were identified, but the influence of different factors as vintages, fermentation types, isolation stages, wineries, and even subzones in microbial population were not statistically assessed. Thus, no such a profound study had been performed about the LAB communities of Tempranillo Rioja red wines. In the current work, ten wineries from the three subzones of the Designation of Origin (D.O.) Rioja were studied during three consecutive years, in must, AF and MLF during different stages of both fermentations and with a more statistical point of view.
Nowadays, the LAB ecology studies are usually supported only by culture-dependent but also by culture-independent methods [8, 9]. Polymerase chain reaction (PCR) and denaturing gradient gel electrophoresis (DGGE) are usually employed with the purpose of analyzing both the species detected by plating and the viable but not cultivable species [10, 11], respectively. Different regions of the genes 16S rDNA and rpoB were used to perform PCR-DGGE because of their conservative sequence and the presence of a unique copy in the bacterial genome, respectively [12, 13]. The combination of results from culture-dependent and culture-independent methods targeted to different genes is considered one of the most adequate strategies to fulfill ecological studies [14].
Therefore, the main aims of the present study were firstly to draw the scenery of the winemaking environment of the D.O. Rioja in relation with the LAB communities and then to describe factors likely to influence in their distribution. Dealing with this study would significantly improve the current oenological knowledge what would be useful for facing probable problems during the elaboration.
Materials and methods
Sampling
One fermentation tank of ten wineries (named with letters from A to J) located in the three subzones of the D.O. Rioja (Rioja Alta, Rioja Baja and Rioja Alavesa) were monitored during three years (I: 2006, II: 2007 and III: 2008) [7]. None of the surveyed wineries had ever used LAB commercial starter cultures. The sampling moments were established in the following stages: must (stage 1), middle AF (stage 2, density near 1025), final AF (stage 3, <2 g/L glucose and fructose), initial MLF (stage 4, LAB population >106 Colony Forming Units/mL), middle MLF (stage 5, 60 % initial malic acid consummated), and final MLF (stage 6, L-malic acid concentration <0.5 g/L) (Table 1). The wines underwent spontaneous MLF with the endogenous microbiota in all cases. Every sample proceeded from Tempranillo red wines and did not present any sensorial deviation. Samples were registered with a number indicating the stage of isolation (from 1 to 6), a capital letter meaning the winery sampled (from A to J) and finally a Roman number in parenthesis that was relative to the isolation year (I, II and III).
Lactic acid bacteria identification
With regards to culture-dependent methods, the enumeration and isolation of LAB able to grow—Colonies Forming Units (CFU)—on modified MRS agar [MRS supplemented with 10 % v/v of tomato juice, 6 g/L of fructose, 0.5 g/L of cysteine-HCl, 5 g/L of d, l-malic acid, 30 g/L of agar and 50 mg/L of pymaricine for inhibiting yeast growth] was carried out. Serial decimal dilutions were plated and incubated under anaerobic conditions at 28 °C during at least 10 days.
After this incubation time, at least 15 colonies were randomly isolated from plates with counts between 10 and 100 CFU—from each sampling stage. Each colony was grown on modified MRS agar and after 48 h the DNA extraction was performed. The pure colony culture was suspended in 1 ml of sterile saline solution (NaCl 0.9 %) and then centrifuged (13,000 rpm, 5 min). After that, 250 μL of the 10 mM buffer lyses (β-mercaptoethanol 100 mM and TRIS pH8 500 mM) was added to the pellet and mixed. After being kept for 10 min at room temperature, it was introduced to boiling water for 10 min and then centrifuged (13,000 rpm, 3 min). The DNA that was in the supernatant after the centrifugation was amplified by the PCR the 16S rRNA genes with WLAB1 and WLAB2 as previously described López et al. [15]. Amplicons were then sequenced by Macrogen Inc. (Seoul, South Korea) to achieve the most suitable identification in the GenBank database [16].
Regarding culture-independent methods, the DNA was directly extracted from red wine using zirconium hydroxide (7 g/L) to facilitate pelleting of the bacteria in wine as Lucore et al. described for milk but with some modifications [17]. A volume of 10 mL of each sample was centrifuged (20 min, 4000×g, 4 °C). The supernatant was discarded and 1.2 mL of saline solution (NaCl 0.9 %) and 2.4 mL of zirconium hydroxide (7 g/L) were added. After 10 min of horizontal shaking at room temperature, the suspension was again centrifuged (10 min, 500×g, 7 °C). The DNA was subsequently extracted and purified from the cell pellet using a PowerSoil® DNA isolation kit (MO BIO Laboratories, Inc., Carlsbad, CA USA) and Fast Prep™ (FP120, BIO 101, Thermo Electron Corporation, USA) bead beater instrument (twice for 45 s at speed setting of 6) as per the manufacturer’s instructions.
PCR was performed using an Applied Biosystem, GeneAmp® PCR System 2700 thermocycler at a final volume of 50 µL. To amplify the region V4 to V5 of 16S rDNA gene, primers WLAB1 and WLAB2GC were used as López et al. described [15]. Moreover, the rpoB1, rpoB1o, and rpoB2GC primers were employed to amplify the region of the rpoB gene as Renouf et al. described [13] with the following modifications: 0.5 μM of each primer, 1 mM dNTP mix and 0.5 μL of PfuUltra II Fusion HS DNA Polymerase (Stratagene).
The separation of the PCR products was performed with the D-CODE™ universal mutation detection system (Bio-Rad, Hercules, Calif.). They were run on 8 % (wt/vol) polyacrylamide gels in TAE buffer (2 M Tris, 1 M glacial acetic acid and 50 mM EDTA pH 8) at a constant temperature of 60 °C. WLAB1—WLAB2GC amplicons were separated with gels containing 35–55 % urea-formamide gradient, and electrophoresis was performed first 10 min at 20 V and then 18 h at 80 V. rpoB amplicons were separated with gels containing 32–50 % urea-formamide gradient, and the electrophoresis was performed for 10 min at 20 V and 15 h at 60 V. Gels were stained in ethidium bromide after the electrophoresis and then were visualized with UV trans-illumination (GelDoc, Bio-Rad). Blocks of the polyacrylamide gels which contained the DGGE bands were excised and subsequently incubated overnight in 20 μL of sterile and pure water at 4 °C to make DNA bands diffuse to the liquid. One microliter of this elution was re-amplified using the PCR conditions described above to DNA sequencing. PCR products were sequenced by Macrogen Inc. (Seoul, South Korea). The 16S rDNA and rpoB sequences were deposited and exposed to the GenBank nucleotide database with the Basic Local Alignment Search Tool (BLAST) [18] under the accession numbers KF753339-KF753505 and KF753506-KF753578, respectively.
Statistical analysis of lactic acid bacteria community distribution
PCR products identified as O. oeni species were aligned with MAFFT multiple sequences alignment software version 7 [19] and then submitted to Modeltest 3.7 [20] in order to know the most adequate phylogenetic model regarding the sequences. Then, the maximum likelihood (ML) that was the most suitable phylogenetic model for data was assessed by PhyML 3.0 interface [21]. The most likely tree obtained was finally edited by MEGA version 4.0.1 [22] to constitute the definitive phylogram. The digitalized images of DGGE gels were entered into FPQuest™ software version 5.1 (Bio-Rad, Hercules, CA, USA) database, and the conversion, normalization, and further processing were made. Due to the complexity of the work, the necessary adjustments were done in order to complete all the information about bands and delete artifacts recognized by the software.
With the aim of including the results of LAB species identified with culture-dependent methods, identification data were codified and also entered into the FPQuest software.
Three experiments were individually designed for results of PCR-16S rDNA-DGGE, PCR-rpoB DGGE and PCR of isolated CFU. Each one of the experiments was statistically analyzed in order to get a dendrogram and a similarity matrix using the Dice coefficient. After this, the Composite Data tool made possible the combination and comparison of the results proceeding from the three approaches with average of each experiment, by UPGMA and setting the weighty of the three experiments at the same level [23]. This created a consensus dendrogram gathering the information about culture-dependent and culture-independent techniques (including the two targeted genes).
The 80 PCR samples were assigned to five groups regarding their isolation. The assigned groups were the subzone, year, winery, fermentation type, and winemaking stage. The FPQuest™ software version 5.1 was then employed in the discriminant analysis of LAB community banding pattern assigned to each group by the Jack-knife method. This statistical method is really useful for evaluating the stability of the clustering and the integrity of banding patterns to defined groups. As other authors have described, the Jack-knife method calculates the Estimated Rate of Correct Classification (ERCC) [24, 25] that is the percentage of correctly pre-assigned observations for each group. The random ERCC was conservatively calculated as James et al. described [25]. Both percentages were statistically compared with the confident limits established for the correctly classified percentages obtained from specific statistical tables [25].
Results
LAB species detected
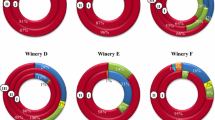
The LAB counts (Fig. 1) widely varied during winemaking. In effect, the AF was characterized by LAB populations between 101 and 102 CFU/mL while during MLF counts ranged from to 105 to 108 CFU/mL. Significant differences were established between the counts of AF and MLF, being the population of final MLF significantly different to the initial and middle MLF stages. Eleven LAB species were identified by PCR of isolated colonies from culture medium as it was described in a previous work [7]. In three of the 80 samples, colonies were not isolated (1-D (III), 2-C (I and II) (Online resource 1).
A total of 167 bands were present in PCR-16S rDNA/DGGE gels from must and wine samples although in ten of the samples this method did not provide results (Fig. 1). These bands were excised, sequenced, and identified as 19 LAB species belonging to the order Lactobacillales of Firmicutes class. The 65 % of the bands were identified as O. oeni. Precisely because of the relevance of this species, in the study a phylogenetic tree was constructed for the genus Oenococcus (Online resource 2). The clustering allowed differentiating two well-defined allelic groups: “A,” identical to the reference sequence PSU-1, and “B” different in one mutation (one thymine was substituted by a cytosine) respect to the PSU-1. Both allelic groups were present in all the wineries (except in H where only group “B” appeared) and simultaneously detected in 12 elaborations distributed in 3 vintages and in 7 out of the 10 wineries of this study. The O. oeni allele “A” appeared as the only profile in five wineries during two vintages. O. oeni allele “B” was the only profile in four wineries during the same years. In addition, 11 sequences were located between both O. oeni allelic groups (“A and B”) in the phylogenetic tree; showing differences in several nucleotide positions in comparison with both alleles. The presence of four bands identified as O. kitaharae can also be observed in the phylogram (Online resource 2). Besides the genus Oenococcus, all genera composing the family Leuconostocaceae were detected. For instance, the genus Weissella represented by Weissella sp., Weissella cibaria and Weissella soli; the genus Leuconostoc (Ln.) with the species Leuconostoc fallax, Leuconostoc citreum, Leuconostoc pseudomesenteroides and Leuconostoc mesenteroides; and Fructobacillus (F.) with the species Fructobacillus tropaeoli and Fructobacillus ficulneus, were detected. The family Lactobacillaceae was integrated by species belonging to genera Lactobacillus, as Lactobacillus mali, Lactobacillus plantarum, Lactobacillus pentosus, Lactobacillus rhamnosus, Lactobacillus brevis and Lactobacillus buchneri, and Pediococcus with two species, Pediococcus parvulus and Pediococcus pentosaceus. Finally, the family Streptococcaceae that was represented by the genus Lactococcus (Lc.) with Lactococcus lactis species. The Online resource 1 also shows 74 bands proceeding from PCR-DGGE rpoB gene gels identified as 5 LAB species, 4 out of them were detected also by the gene 16S rDNA and Weissella paramesenteroides was described only with the gene rpoB. In this approach, 21 samples did not allow the LAB species detection. The 72 % of the bands were identified as O. oeni. The allelic groups for rpoB gene “H” and “L” reported by Renouf et al. [13] were observed in this study. Five wineries (C, B, E, F and J) exhibited between their profiles both “H and L” alleles, whereas in two wineries (A and H), only allele “H” appeared and in 3 wineries (D, G and I) only allele “L” appeared. O. oeni alleles “H and L” coexisted only in a fermentation tank in winery F (data no shown).
Taking into consideration the results, two well-defined clusters were separated with a similarity rate of the 10 %. The first one included 14 samples belonging to 5 wineries and being isolated during the 3 years but only in AF. The 66 remaining samples were included in other cluster. Among the samples clustered in this second branch, 29 maintained a similarity percentage from 25 to 95 % approximately and were isolated in every sampled wineries, years, and stages, mainly in final AF and initial MLF. Another 37 samples, included in this cluster shared a similarity rate of the 100 % because all of them provided the unique identification of O. oeni species. These DNA samples were mainly extracted during the later stages of the MLF in the ten wineries and during the three sampled years.
LAB community distribution by Jack-knife statistical analysis
The mean and the random ERCC (%) calculated for each of the five groups defined for the grouping samples—winery, stage, year, subzone and fermentation type (Table 1) and the statistically comparison between both are shown in Table 2. The ERCC described for the “winery” was the 17.85 % what meant that more or least the 83.15 % of the samples could not correspond with the winery they were pre-assigned to. The ERCC for the “stage” was the 37.80 %, consequently, the 43.20 % of the samples assigned to a stage would probable not be perfectly pre-assigned. Very similar to the previous one and meaning exactly the same was the “year.” In the case of “subzones,” the described ERCC was the 41.25 % what indicated that the 59 % of the samples would not match with the pre-assigned year. For the fermentation type, the ERCC was the 89.44 % being only a 10 % of the samples mismatched with the pre-established fermentation type. Considering the assessed values of the random ERCC, significant differences could be determined for the percentages of the groups “stage” and “fermentation type.”
Data about detailed ERCC obtained for the group “fermentation type” are shown in Table 3. In AF, the 80 % of the banding patterns hosted the group and a 20 % of the samples could be more concordant with the MLF. Nevertheless, in the MLF case, only a 1.11 % of the samples could correspond with the samples pre-established as AF.
Discussion
The LAB community of Tempranillo Rioja wines was analyzed using 16S rDNA and rpoB/PCR-DGGE and culture-dependent approach. The diversity of LAB species by culture-dependent and independent methods showed 25 species detected from one to three years in the must and fermenting wines of ten wineries. Among these, ten species belonged to the genus Lactobacillus, four to the genus Weissella, and other four to genus Leuconostoc. Moreover, the genera Pediococcus, Fructobacillus, and Oenococcus were represented each one by two species, and Lc. lactis was the representative species of the genus Lactococcus.
The presence of certain species not usually detected in Tempranillo wines (O. oeni, O. kitaharae, Ln. fallax, L. mesenteroides, L. plantarum, L. buchneri, P. parvulus, P. pentosaceus and Lc. lactis) was corroborated [26]. In contrast, the description of other species not traditionally related to winemaking was for first time determined in the current research. Specifically, the species W. soli, W. cibaria, and Weissella sp. were only found in malting process [27] but not in winemaking, although W. paramesenteroides was described in the wine environment [28]. Furthermore, the species Ln. citreum and Ln. pseudomesenteroides related with silage fermentation [29] and other ones as F. tropaeoli and F. ficulneus associated with flowers environment [30] were noticed for the first time in the samples of this research. Curiously, L. rhamnosus characterized as probiotic and usually isolated from intestinal sources [31] was also found for the first time in winemaking environment in this study. Other species traditionally detected in wine by plating as L. mali, L. pentosus, and L. brevis [5] were also detected by PCR-DGGE. Similarly, sequences highly related to O. kitaharae belonging to genus Oenococcus were amplified from the DNA samples.
Successfully, it has been the first time in which two new allelic groups of O. oeni species were noticed by PCR-16S rDNA/DGGE. The allele “A” was approximately a 15 % more frequent than allele “B”, being present in the whole winemaking. In fact, both allelic groups appeared together in half of the wineries during the vinification. In relation with the allelic groups “H and L” detected by rpoB gene, they were mainly noticed during final AF and MLF stages with similar percentages but no together during the same winemaking. Other authors have suggested a more favorable adaptation of “L” strains to MLF [32, 33] but in the present study this distribution could not be determined.
The PCR-DGGE approach allowed the detection of 21 out of the 25 species noticed in the wine and must samples. Even when the counts were minor than 101 CFU/mL this culture-independent method provided results. Curiously, four out the 11 species described after culture-dependent methods were not noticed by PCR-DGGE, so the combination of both approaches was a completely successful tool to evaluate the LAB richness of wine and must samples [34–37]. Between the two genes employed the 16S rDNA gene allowed the detection of most of the LAB species described. Nevertheless, the rpoB gene was really interesting in the detection of non- O. oeni species during and furthermore it was able to notice the presence of W. paramesenteroides species while the gene 16S rDNA was not.
On the other note, it has been for first time demonstrated that the LAB communities of Tempranillo red wines could be consistently differentiated by Jack-knife method based on the six different winemaking stages, but especially on the two types of fermentation, AF and MLF. However, communities could not be distinguished by the subzone, year and winery of isolation. The ERCC for assignments based on winery, year and subzone was lower or equal than the random ERCC what meant that those three factors were not influencing the LAB population in wines. Similar results were reported when the clonal diversity of the species O. oeni was analyzed. In that case, O. oeni strains were likely to get adapted to the different fermenting conditions varying from year to year, from winery to winery and of course from one stage to other stage of the vinification [7].
The ERCC for assignments based on the winemaking stage of isolation was significantly higher than random but the value of the percentage was not consistent enough to consider the influence of the six separated sampled moments as the main factor influencing the LAB community distribution. Nevertheless, considering AF and MLF provided relevant ERCC values, thus the fermentation stage was the main factor affecting the distribution of the different LAB species in Tempranillo red wines. Moreover, this result was significantly different than random ERCC establishing statistical differences between LAB species isolated during AF and during MLF, independently of the winery, year and subzone of isolation.
Taking into consideration the correct grouping of samples in AF and MLF, a description of this two fermentation stages regarding LAB composition could be done. In the must samples plating counts were always minor than 102 CFU/mL and the rpoB gene did not report results, despite being considered operative in similar population level [38]. The PCR-DGGE detection limit was notoriously reduced with respect to the described so far [11] what was due to both the extraction with zirconium hydroxide and the employment of a high affinity DNA polymerase (PfuUltra II Fusion HS, Stratagene, Canada). Isolates identification provided complementary information to PCR-DGGE, being only three out of the 15 species described in musts samples (O. oeni, Ln. mesenteroides and L. plantarum) detected by both culture-dependent and culture-independent methods. This concordance between both strategies might be owing to the relevant presence of these species at initial fermentation stages [10]. During middle AF, 11 LAB species belonging to four genera were detected. In spite of the strong competition performed by yeasts in the wine at this fermentation stage, the species L. mali, L. plantarum, Ln. mesenteroides, and O. oeni were cultivable [39]. PCR-rpoB/DGGE allowed the description of three species in must samples, eight less than 16S rDNA gene what was in agreement with the difficulty of rpoB gene to detect minority LAB species [38]. The number of LAB species detected in the final stage of AF was the same as described for must samples. The Pediococcus genus recovered the grow ability and twelve LAB species were detected by 16S rDNA gene, reaching the greatest detection of non-cultivable species in the research. This great LAB species diversity might be due to the decreasing pressure exerted by yeast’s metabolism since middle AF. Probably, some yeast would have already died at this stage what supplied a growing nutrient source to the wine. For this reason, final AF stage could be favorable for the alive microorganisms that became able to develop their metabolism in a cultivable and non-cultivable way [40]. Other authors have described that bacterial diversity during the red wine AF was low because the phenolic compounds are involved in the selection of the most adapted species the red wine environment [41, 42]. Therefore, the results obtained in the present study were absolutely successful because of the high diversity of LAB species noticed at AF being the Lactobacillus genus was the predominant one before MLF beginning.
During initial MLF the LAB count was higher than 105 CFU/mL and, as it was expected, O. oeni was the predominant species. In contrast, Ln. mesenteroides, P. pentosaceus were also detected by PCR-DGGE and L. hilgardii by culture-dependent methods. During the middle MLF, LAB counts were important, and all the isolates were identified as O. oeni so that LAB diversity decreased even more. In effect, only O. oeni would have been detected if only plating had been carried out. Nevertheless, in the middle MLF, PCR-16S rDNA and rpoB/DGGE genes were able not only to find the majority O. oeni population but also to detect other secondary species such as O. kitaharae-like and P. parvulus. Curiously, Lactobacillus-species disappeared in this stage although species as L. plantarum have thought to be interesting in the development of MLF as starter culture because of its ethanol tolerance [43, 44]. Finally, the last stage of MLF was very similar to the middle MLF being the highest count result assessed at this last fermentation stage. Every isolate was again identified as O. oeni so that it was the majority species. Curiously, rpoB gene determined P. parvulus presence while 16S rDNA only noticed O. oeni. This was likely due to a high affinity of this gene to the detection of Pediococcus species or to an important population of these species at final MLF. Precisely, the detection of non-O. oeni species as P. parvulus had been previously described during MLF and at the end of MLF by Renouf et al. [45] as a consequence of O. oeni declination in wine what facilitated the presence of other species very well adapted to wine conditions and occasionally related with detrimental wine quality.
Conclusions
To sum up, it was the first time that such an exhaustive study was carried out in a region taking into account both culture-dependent and 16S rDNA/rpoB culture-independent results. Twenty-five species from seven genera and three families were described in the study. This research has for first time demonstrated that the distribution of the LAB community of Tempranillo red wine from the D. O. Rioja was significantly influenced only by the two fermentative stages during winemaking, AF and MLF, but no so much by sampling stage, vintage, winery and even subzone where the winery was located. Some species never before detected in wine samples were identified during the AF of some wineries. Furthermore, two novel O. oeni allelic groups (“A and B”) were established by using 16S rDNA-DGGE. Additionally, “L and H” rpoB O. oeni alleles were identified in this study but curiously with a totally different adaptation to the wine environment of Rioja region. Finally, it was possible to conclude that the direct DNA extraction method employed in combination with PCR-DGGE strategy could be considered the most adequate to describe LAB populations in must and red wine samples, though a supplementation with culture-dependent methods could be complementary in some cases. Further studies are required to analyze the correlation between A/B 16S rDNA and L/H rpoB O. oeni allelic groups and geographical distribution analyzing grapes or wines from a known origin.
References
Bartowsky EJ (2014) Encyclopedia of food microbiology. Encycl Food Microbiol. doi:10.1016/B978-0-12-384730-0.00357-8
Lonvaud-Funel A (2010) Managing wine quality. Manag Wine Qual. doi:10.1533/9781845699987.1.60
Renouf V, Gindreau E, Claisse O, Lonvaud-Funel A (2005) Microbial changes during malolatic fermentation in red wine elaboration. J Int Sci Vigne Vin 39:179–190
Petri A, Pfannebecker J, Fröhlich J et al (2013) Fast identification of wine related lactic acid bacteria by multiplex PCR. Food Microbiol 33:48–54. doi:10.1016/j.fm.2012.08.011
Mesas JM, Rodríguez MC, Alegre MT (2011) Characterization of lactic acid bacteria from musts and wines of three consecutive vintages of Ribeira Sacra. Lett Appl Microbiol 52:258–268. doi:10.1111/j.1472-765X.2010.02991.x
López R, Tenorio C, Gutiérrez ARAR et al (2012) Elaboration of Tempranillo wines at two different pHs. Influence on biogenic amine contents. Food Control 25:583–590. doi:10.1016/j.foodcont.2011.11.029
González-Arenzana L, Santamaría P, López R, López-Alfaro I (2013) Indigenous lactic acid bacteria communities in alcoholic and malolactic fermentations of Tempranillo wines elaborated in ten wineries of La Rioja (Spain). Food Res Int 50:438–445. doi:10.1016/j.foodres.2012.11.008
Lucena-Padrós H, Jiménes E, Maldonado-Barragán A et al (2015) PCR-DGGE assessment of the bacterial diversity in Spanish-style green table olives fermentations. Int J Food Microbiol 205:47–53. doi:10.1016/j.ijfoodmicro.2015.03.033
Bonetta S, Bonetta S, Carraro E et al (2008) Microbiological characterisation of Robiola di Roccaverano cheese using PCR-DGGE. Food Microbiol 25:786–792. doi:10.1016/j.fm.2008.04.013
Cocolin L, Alessandria V, Dolci P et al (2013) Culture independent methods to assess the diversity and dynamics of microbiota during food fermentation. Int J Food Microbiol 167:29–43. doi:10.1016/j.ijfoodmicro.2013.05.008
Cocolin L, Campolongo S, Alessandria V et al (2011) Culture independent analyses and wine fermentation: an overview of achievements 10 years after first application. Ann Microbiol 61:17–23. doi:10.1007/s13213-010-0076-6
Case RJ, Boucher Y, Dahllöf I et al (2007) Use of 16S rRNA and rpoB genes as molecular markers for microbial ecology studies. Appl Environ Microbiol 73:278–288. doi:10.1128/AEM.01177-06
Renouf V, Claisse O, Lonvaud-Funel A (2006) rpoB gene: a target for identification of LAB cocci by PCR-DGGE and melting curves analyses in real time PCR. J Microbiol Methods 67:162–170. doi:10.1016/j.mimet.2006.03.008
González-Arenzana L, López R, Santamaría P, López-Alfaro I (2012) Application of the Different Electrophoresis Techniques to the Detection and Identification of Lactic Acid Bacteria in Wines. In: Ghowsi K (ed) Electrophoresis. Intech, Croatia, pp 137–156
Lopez I, Ruiz-Larrea F, Cocolin L et al (2003) Design and evaluation of PCR primers for analysis of bacterial populations in wine by denaturing gradient gel electrophoresis. Appl Environ Microbiol 69:6801–6807. doi:10.1128/AEM.69.11.6801-6807.2003
González-Arenzana L, López R, Portu J et al (2014) Molecular analysis of Oenococcus oeni and the relationships among and between commercial and autochthonous strains. J Biosci Bioeng 118:272–276. doi:10.1016/j.jbiosc.2014.02.013
Lucore LA, Cullison MA, Jaykus L-AA (2000) Immobilization with metal hydroxides as a means to concentrate food-borne bacteria for detection by cultural and molecular methods. Appl Environ Microbiol 66:1769–1776. doi:10.1128/AEM.66.5.1769-1776.2000
Altschul SF, Gish W, Miller W et al (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi:10.1093/molbev/mst010
Posada D, Crandall KA (1998) MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817–818. doi:10.1093/bioinformatics/14.9.817
Guindon S, Dufayard J-FF, Lefort V et al (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi:10.1093/sysbio/syq010
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0 RID E-9283-2010. Mol Biol Evol 24:1596–1599. doi:10.1093/molbev/msm092
González-Arenzana L, Pérez-Martín F, Palop ML et al (2015) Genomic diversity of Oenococcus oeni populations from Castilla La Mancha and La Rioja Tempranillo red wines. Food Microbiol 49:82–94. doi:10.1016/j.fm.2015.02.001
Lucena-Padrós H, Jiménez E, Maldonado-Barragán A et al (2015) PCR-DGGE assessment of the bacterial diversity in Spanish-style green table-olive fermentations. Int J Food Microbiol 205:47–53. doi:10.1016/j.ijfoodmicro.2015.03.033
James JB, Sherman TD, Devereux R (2006) Analysis of bacterial communities in seagrass bed sediments by double-gradient denaturing gradient gel electrophoresis of PCR-Amplified 16S rRna genes. Microb Ecol 52:655–661. doi:10.1007/s00248-006-9075-3
González-Arenzana L, López R, Santamaría P et al (2012) Dynamics of indigenous lactic acid bacteria populations in wine fermentations from La Rioja (Spain) during three vintages. Microb Ecol 62:12–19. doi:10.1007/s00248-011-9911-y
Justé A, Malfliet S, Waud M et al (2014) Bacterial community dynamics during industrial malting, with an emphasis on lactic acid bacteria. Food Microbiol 39:39–46. doi:10.1016/j.fm.2013.10.010
Dicks LMT, Endo A (2009) Taxonomic status of lactic acid bacteria in wine and key characteristics to differentiate species. S Afr J Enol Vitic 30:72–90
Pang H, Qin G, Tan Z et al (2011) Natural populations of lactic acid bacteria associated with silage fermentation as determined by phenotype, 16S ribosomal RNA and recA gene analysis. Syst Appl Microbiol 34:235–241. doi:10.1016/j.syapm.2010.10.003
Endo A, Irisawa T, Futagawa-Endo Y et al (2011) Fructobacillus tropaeoli sp. nov., a fructophilic lactic acid bacterium isolated from a flower. Int J Syst Evol Microbiol 61:898–902. doi:10.1099/ijs.0.023838-0
Jia R, Chen H, Chen H, Ding W (2016) Effects of fermentation with Lactobacillus rhamnosus GG on product quality and fatty acids of goat milk yogurt. J Dairy Sci 99:221–227. doi:10.3168/jds.2015-10114
García-Ruiz A, Requena T, Peláez C et al (2013) Antimicrobial activity of lacticin 3147 against oenological lactic acid bacteria. Combined effect with other antimicrobial agents. Food Control 32:477–483. doi:10.1016/j.foodcont.2013.01.027
Renouf V, Vayssieres LC, Claisse O, Lonvaud-Funel A (2009) Genetic and phenotypic evidence for two groups of Oenococcus oeni strains and their prevalence during winemaking. Appl Microbiol Biotechnol 83:85–97. doi:10.1007/s00253-008-1843-1
Randazzo CL, Ribbera Á, Pitino I et al (2012) Diversity of bacterial population of table olives assessed by PCR-DGGE analysis. Food Microbiol 32:87–96. doi:10.1016/j.fm.2012.04.013
Bae S, Fleet GH, Heard GM (2006) Lactic acid bacteria associated with wine grapes from several Australian vineyards. J Appl Microbiol 100:712–727. doi:10.1111/j.1365-2672.2006.02890.x
Andorrà I, Landi S, Mas A et al (2010) Effect of fermentation temperature on microbial population evolution using culture-independent and dependent techniques. Food Res Int 43:773–779. doi:10.1016/j.foodres.2009.11.014
Rantsiou K, Urso R, Iacumin L et al (2005) Culture-dependent and -independent methods to investigate the microbial ecology of Italian fermented sausages. Appl Environ Microbiol 71:1977–1986. doi:10.1128/AEM.71.4.1977-1986.2005
Renouf V, Claisse O, Miot-Sertier C, Lonvaud-Funel A (2006) Lactic acid bacteria evolution during winemaking: use of rpoB gene as a target for PCR-DGGE analysis. Food Microbiol 23:136–145. doi:10.1016/j.fm.2005.01.019
Ruiz P, Seseña S, Izquierdo PM, Palop ML (2010) Bacterial biodiversity and dynamics during malolactic fermentation of Tempranillo wines as determined by a culture-independent method (PCR-DGGE). Appl Microbiol Biotechnol 86:1555–1562. doi:10.1007/s00253-010-2492-8
Ribéreau-Gayon P, Dubourdieu D, Donèche B, Lonvaud-Funel A (2007) Handbook of enology, the microbiology of wine and vinifications. Wiley, Sussex
Renouf V, Claisse O, Lonvaud-Funel A (2007) Inventory and monitoring of wine microbial consortia. Appl Microbiol Biotechnol 75:149–164. doi:10.1007/s00253-006-0798-3
Figueiredo AR, Campos F, de Freitas V et al (2008) Effect of phenolic aldehydes and flavonoids on growth and inactivation of Oenococcus oeni and Lactobacillus hilgardii. Food Microbiol 25:105–112. doi:10.1016/j.fm.2007.07.004
Cho G-S, Krauss S, Huch M et al (2011) Development of a quantitative PCR for detection of Lactobacillus plantarum starters during wine malolactic fermentation. J Microbiol Biotechnol 21:1280–1286. doi:10.4014/jmb.1107.07003
Pozo-Bayón MA, G-Alegría E, Polo MC et al (2005) Wine volatile and amino acid composition after malolactic fermentation: effect of Oenococcus oeni and Lactobacillus plantarum starter cultures. J Agric Food Chem 53:8729–8735
Renouf V, Strehaiano P, Lonvaud-Funel A (2007) Yeast and bacteria analysis of grape, wine and cellar equipments by PCR-DGGE. J Int Des Sci La Vigne Du Vin 41:51–61
Acknowledgments
This work was supported by funding and predoctoral grant (B.O.R. 6th March, 2009) of the Government of La Rioja, the I.N.I.A. Project RTA2007-00104-00-00 and it was possible thanks to the collaborating wineries.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Compliance with Ethics Requirements
This article does not contain any studies with human or animal subjects.
Electronic supplementary material
Below is the link to the electronic supplementary material.
217_2016_2720_MOESM1_ESM.doc
Online resource 1. Consensus dendrogram obtained by composite data set combining the results of the LAB species detected with culture-dependent methods and independent methods (PCR-16S rDNA/DGGE and PCR-rpoB/DGGE) showing cophenetic correlation values. Samples were labelled with a number corresponding with the isolation stage (1: must, 2: middle AF, 3: final AF, 4: initial MLF, 5: middle MLF and 6: final MLF), with a capital letter indicative of the winery (from A to J) and Roman numbers placed in parenthesis representing the consecutive isolation years (I, II and III). The identified species were labelled with lower Latin letters placed at the top of each representative band (a: O. oeni, b: O. kitaharae, c: L. citreum, d: L. mesenteroides, e: L. pseudomesenteroides, f: L. fallax, g: F, ficulneus, h: F. tropaeoli, i: W. cibaria, j: W. paramesenteroides, k: Weisella sp., l: W. solis, m: L. brevis, n: L. buchneri, o: L. coryniformis, p: L. mali, q: L. hilgardii, r: L. plantarum, s: L. pentosus, t: L. rhamnosus, u: Lactobacillus sp., v: L. uvarum, w: P. pentosaceus, x: P. parvulus, y: L. lactis). Supplementary material 1 (DOC 85 kb)
217_2016_2720_MOESM2_ESM.doc
Online resource 2. Phylogram for the 110 sequences identified as species belonging to genus Oenococcus, obtained from 16S rDNA PCR/DGGE. Each sequence is referred with the most accurate identification and the identity percentage (%), with the given accession number and with a code that means the isolation stage (from 1 to 6), winery (from A to J) and year (I, II or III). The evolutionary distances are in the units of the number of base substitutions per site. Supplementary material 2 (DOC 28 kb)
Rights and permissions
About this article
Cite this article
González-Arenzana, L., Santamaría, P., Gutiérrez, A.R. et al. Lactic acid bacteria communities in must, alcoholic and malolactic Tempranillo wine fermentations, by culture-dependent and culture-independent methods. Eur Food Res Technol 243, 41–48 (2017). https://doi.org/10.1007/s00217-016-2720-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00217-016-2720-2