Abstract
Key message
A grain weight locus from Agropyron cristatum chromosome 5P increases grain weight in different wheat backgrounds and is localized to 5PL (bin 7–12).
Abstract
Thousand-grain weight is an important trait in wheat breeding, with a narrow genetic basis being the main factor limiting improvement. Agropyron cristatum, a wild relative of wheat, harbors many desirable genes for wheat improvement. Here, we found that the introduction of the 5P chromosome from A. cristatum into wheat significantly increased the thousand-grain weight by 2.55–7.10 g, and grain length was the main contributor to grain weight. An increase in grain weight was demonstrated in two commercial wheat varieties, indicating that the grain weight locus was not affected by the wheat background. To identify the chromosome segment harboring the grain weight locus, three A. cristatum 5P deletion lines, two wheat–A. cristatum 5P translocation lines and genetic populations of these lines were used to evaluate agronomic traits. We found that the translocation lines harboring the long arm of A. cristatum chromosome 5P (5PL) exhibited high grain weight and grain length, and the genetic locus associated with increased grain weight was mapped to 5PL (bin 7–12). An increase in grain weight did not adversely affect other agronomic traits in translocation line 5PT2, which is a valuable germplasm resource. Overall, we identified a grain weight locus from chromosome 5PL and provided valuable germplasm for improving wheat grain weight.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Common wheat (Triticum aestivum L., AABBDD, 2n = 6x = 42) is one of the world's most important food crops, providing approximately 20% of the protein and calories in the human diet globally (Reynolds et al. 2009; Shiferaw et al. 2013). However, continued human population growth, climate change, and stagnant crop yields pose great challenges to global food security (Tilman et al. 2011). Wheat yield is affected by three major factors: the number of spikes per unit area, the number of grains per spike, and the thousand-grain weight (TGW) (Kuchel et al. 2007). Among the yield components, grain weight is most affected by genetic characteristics, and the general heritability can reach 59–80% (Zhuang 2003). Breeding practices have shown that genetic improvement in relation to TGW is the most significant, and it can be seen from the development of new wheat varieties over time in China that the average TGW increased from 31.4 g in the 1950s to 42.2 g in the 1980s (Zhuang 2003). It is estimated that an increase in yield of 140–160 kg ha−1 can be obtained by just a 1.0 g increase in TGW (Tian et al. 2006). Therefore, TGW is an important trait for achieving high yield in wheat, and the discovery of important genes controlling grain weight is a prerequisite for molecular wheat breeding.
Quantitative trait loci (QTLs) controlling TGW and grain size (GS) have been identified on almost all 21 chromosomes of wheat, as demonstrated by QTL mapping and association mapping conducted on wheat of different genetic backgrounds and from different environmental conditions (Cao et al. 2020). Notably, GS improvement has effectively promoted grain yield in rice (Zuo and Li 2014). The major QTLs and regulatory genes have been cloned mainly by forward genetics approaches. The genes controlling the size and shape of rice grains are mainly associated with three major genetic pathways, i.e., degradation of proteins through the ubiquitin‒proteasome pathway, signaling mediated by G protein/protein kinase, and plant phytohormone signal transduction mediated by auxin, cytokinin, and brassinosteroids (Li and Yang 2017).
Studies on comparative genomics have shown high synteny and collinearity between different graminoid crop genomes, such as those of rice and wheat (Valluru et al. 2014). Based on a conserved genetic network that regulates TGW/GS in rice and wheat, many homologs associated with grain yield, such as TaGW2 (Su et al. 2011), TaDA1 (Liu et al. 2020b), TaGS5 (Ma et al. 2016), TaBT1 (Wang et al. 2019), TaCKX6-D1 (Zhang et al. 2012), and TaSTT3b (Zhu et al. 2022), have been identified by comparative genomics approaches in common wheat (Li and Yang 2017). However, few genes controlling TGW/GS have been identified by map-based cloning. VEGETATIVE TO REPRODUCTIVE TRANSITION 2 (VRT2), keto-acyl thiolase 2B (KAT-2B), Tasg-D1, TabHLH489, and TaGL1-B1 associated with wheat grain size were identified via forward genetics approaches (Adamski et al. 2021; Chen et al. 2020; Cheng et al. 2020; Liu et al. 2021; Lyu et al. 2024; Niaz et al. 2023; Xiao et al. 2021, 2022). VRT2 encodes a member of the MADS-box gene family, and ectopic expression of VRT2 in Polish wheat elongates the glume and grain, leading to an increase in grain size (Adamski et al. 2021; Liu et al. 2021; Xiao et al. 2021). KAT-2B is involved in β-oxidation during jasmonic acid (JA) synthesis, which increases grain width and weight through the JA pathway (Chen et al. 2020). Tasg-D1 encodes the Ser/Thr protein kinase glycogen synthase kinase 3 (STKc_GSK3) and negatively regulates grain size by repressing brassinosteroid (BR) signaling (Cheng et al. 2020). Notably, TaGL1-B1 encodes prolycopene isomerase, which increases carotenoid and chlorophyll content through interaction with TaPAP6, a key gene involved in the JA pathway, thereby affecting wheat grain length and improving wheat yield (Niaz et al. 2023).
The wild relatives of wheat contain abundant genetic variation and desirable genes (Zhang et al. 2017a). The introduction of many excellent alien genes into wheat through distant hybridization has greatly promoted the genetic improvement of wheat (Mujeeb-Kazi et al. 2013). In the history of worldwide wheat breeding, the outstanding contribution of alien translocation lines has benefited from the utilization of only a few disease resistance genes (Liu et al. 2020a). However, studies on yield-related genes in wild relatives, especially those related to grain weight or grain size, are rare. Some wild relatives of wheat are important resources for improving wheat yield traits. For example, the T1BL·1RS translocation line harbors yield-related genes in addition to their resistance genes (Carver and Rayburn 1994; Villareal et al. 1998, 1991). Therefore, nearly 40% of the cultivated varieties in China came to carry T1BL·1RS after the 1980s (Zhou et al. 2004). The T6VS·6AL translocation line has been shown to increase wheat grain weight (Li et al. 2007). Additionally, the grain weight and yield of synthetic hexaploid wheat were found to be greatly improved due to the contribution of beneficial genes from Aegilops species (Hao et al. 2020; Yan et al. 2018).
Agropyron Gaertn., a perennial genus of the tribe Triticeae, contains one basic P genome in three ploidy levels: diploid, tetraploid, and hexaploid (Dewey 1984). It possesses many useful traits, including resistance to many wheat diseases, such as rusts and powdery mildew; tolerance to drought, cold, and salt; and characteristics associated with high yields, such as the production of multiple spikelets, florets, and fertile tillers. Therefore, this genus is an important contributor to wheat improvement (Dewey 1984; Dong et al. 1992). To transfer its desirable genes to common wheat, the distant hybridization of the common wheat cultivar Fukuhokomugi (Fukuho hereafter) with tetraploid A. cristatum (accession Z559) was achieved via embryo rescue (Li et al. 1997, 1998). Subsequently, a series of wheat–A. cristatum addition, substitution, deletion, and translocation lines involving seven homoeologous groups with critical phenotypic differences were produced (Han et al. 2023; Jiang et al. 2018; Kuang et al. 2023; Li et al. 2017; Song et al. 2016; Sun et al. 2021; Wang et al. 2022; Wu et al. 2006; Yang et al. 2010; Zhang et al. 2019, 2017b). The desirable genes on each P chromosome have been extensively investigated in the past decade. For example, the genes for resistance to powdery mildew and leaf rust were located on chromosome 2P by Jiang et al. (2018) and Li et al. (2017). Similarly, the genes for resistance to powdery mildew, leaf rust, and stripe rust, as well as those for enhanced grain number per spike (GNS) and TGW, have been identified on chromosome 6P (Lin et al. 2022; Song et al. 2016; Wu et al. 2006; Zhang et al. 2019, 2017b). The genes for enhanced TGW were mapped onto chromosome 7P (Sun et al. 2021). The discovery of these yield-related genes greatly broadened the genetic basis of wheat improvement, and the production of many translocation lines has benefited wheat breeding (Lin et al. 2023; Xu et al. 2022; Zhang et al. 2016, 2015, 2017b).
In this study, our objective was to investigate the genetic effect of chromosome 5P on agronomic traits in wheat. Our genetic analysis revealed that chromosome 5P increased the TGW in different wheat backgrounds, primarily due to grain length (GL). We then aimed to precisely locate the grain weight gene and to elucidate the possible genetic effects of different fragments of chromosome 5P through the utilization of deletion lines and translocation lines with different fragment sizes. We expected these findings to lay a foundation for the future application of desirable genes from the 5P chromosome to increase wheat yield.
Materials and methods
Plant materials
The wheat–A. cristatum 5P disomic addition line II-11-1b (2n = 44) was derived from the distant hybridization between Triticum aestivum cv. Fukuho (2n = 6x = 42, AABBDD) and A. cristatum accession Z559 (2n = 4x = 28, PPPP, from Xinjiang, China). To investigate the genetic effect of chromosome 5P, five segregated populations of different generations were developed from a cross between the wheat–A. cristatum 5P addition line II-11-1b and Fukuho, and the other two segregated populations were derived from a cross between II-11-1b and two commercial wheat varieties, i.e., Xinong 979 and Kenong 199. Three wheat–A. cristatum 5P deletion lines 5PD1, 5PD2, and 5PD3 and translocation line 5PT1 (T5PL-2AS·2AL) were previously developed by Han et al. (2023), and a new translocation line 5PT2 (T4AL·5PL) identified in this study (Table 1) was used to create genetic populations with Fukuho as the recurrent parent to analyze the effects of different segments of chromosome 5P.
Cytogenetic analysis
Seeds were germinated on moistened filter paper in Petri dishes. The actively growing roots were removed from the plants, subjected to nitrous oxide treatment for 2 h, fixed in 90% acetic acid for 8 min, and stored in 70% v/v ethanol. Chromosome preparation was performed following Kato et al. (2004) and Han et al. (2006). Cytological observations were performed under a phase-contrast microscope.
Genomic in situ hybridization (GISH) was performed with A. cristatum genomic DNA as a probe and Fukuho genomic DNA as a blocker. Genomic DNA was extracted from young leaf tissue of A. cristatum and Fukuho plants using a modified CTAB method (Feng et al. 2017). Nondenaturing fluorescence in situ hybridization (ND-FISH) with oligo-pTa535-2 and oligo-pSc119.2–2 as probes was performed according to Tang et al. (2014). The oligonucleotide sequences used were 5’- or 3’-end labeled with 6-carboxyfluorescein or 6-carboxytetramethylrhodamine. All images were acquired with an Olympus AX80 fluorescence microscope (Olympus Corp., Tokyo, Japan) and processed with Adobe Photoshop CS 3.0 (Adobe, San Jose, CA, USA).
Reads coverage analysis
The clean RNA-seq reads obtained from Han et al. (2023) were mapped to the integrated reference sequences of Chinese Spring RefSeqv2.1 (Zhu et al. 2021) and A. cristatum Z559 (unpublished) using default parameters in HISAT2 (Pertea et al. 2016). PCR duplicates were detected and removed using Picard’s MarkDuplicates (https://broadinstitute.github.io/picard/). The unique reads were then extracted with the SAM tag "NH:i:1" and not "ZS:i" using samtools and grep commands. Subsequently, bedtools (Quinlan and Hall 2010) was used to calculate the whole-genome exon coverage utilizing the command "bedtools coverage -a BAM -b exon.bed -sorted" with the exon.bed file sourced from the integrated annotation file from Refseq2.1 and Z559. Considering the transcriptome data, exon coverage was utilized as an indicator of identification. The visualization of coverage was performed by the ggplot2 package of R (Maag 2018).
Molecular marker genotyping for different segments of chromosome 5P
A. cristatum-specific sequence-tagged site (STS) markers were developed from expressed sequence tag (EST) sequences identified in A. cristatum transcriptome sequences (Feng et al. 2017), and 56 markers were mapped onto the physical map of A. cristatum chromosome 5P (Han et al. 2023). Five 5P-specific markers and one P genome-specific marker, AcPR3a (Han et al. 2017), were used to detect wheat–A. cristatum 5P addition line, deletion lines, and translocation lines and their corresponding segregated populations. PCR amplification was carried out as previously described by Zhang et al. (2017a, b). The amplified products were separated by using 8% polyacrylamide or 1.5% agarose gel electrophoresis.
Evaluation of agronomic traits and statistical analysis of the data
All the materials were planted in fields at the Xinxiang Experimental Station of the Chinese Academy of Agricultural Sciences (35°18′13.71″N, 113°55′15.05″E, Henan Province, China). The BC5F1, BC6F1, BC6F2, BC7F1, and BC8F1 populations of II-11-1b in the Fukuho background were planted during the 2017–2018, 2018–2019, 2019–2020, and 2021–2022 growing seasons, respectively. The F2 populations of II-11-1b with Xinong 979 and Kenong 199 and the five segregated populations of different deletion lines and translocation lines with Fukuho were planted during the 2022–2023 growing season. Twenty grains were evenly planted in 2.0 m rows spaced 0.3 m apart. The agronomic trait evaluations of the segregated populations included plant height (PH), effective tiller number (ETN), spike length (SL), spikelet number per spike (SNS), grain number per spikelet (GNPS), GNS, TGW, grain length (GL), and grain width (GW). The grain traits, including TGW, GL, and GW, were assessed using a thousand-kernel-weight analyzer (Model SC-G; Hangzhou Wanshen Detection Technology Co., Ltd., Hangzhou, China). The plants in each population were classified into two types based on molecular marker profiles: plants with or without chromosome 5P. The statistical analysis of the phenotypic data was performed with Student’s t test or one-way ANOVA using the SAS software package (V8.1, SAS Institute, Inc., Cary, NC, USA).
Results
Chromosome 5P contains a gene(s) that increases the TGW of wheat
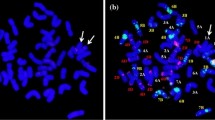
II-11-1b is a wheat–A. cristatum chromosome 5P disomic addition line, which was confirmed by GISH and transcriptomic analysis (Fig. 1). We observed that the trends and densities of the 5P chromosome reads were similar to those on wheat chromosomes based on the exon coverage analysis of II-11-1b, while other P-genome chromosomes exhibited distinct patterns (rare reads), possibly because differences between II-11-1b and the Chinese Spring reference genome led to mismatches between homoeologous genes (Fig. 1b). Combining these observations with the results of GISH (Fig. 1a), II-11-1b was verified to be wheat–A. cristatum chromosome 5P disomic addition line. Several important traits of II-11-1b and its wheat parent Fukuho were investigated, and the TGW of II-11-1b was approximately 5.0 g greater than that of Fukuho. The increase in grain weight was dependent mainly on the increase in grain length (Fig. S1).
GISH and RNA-seq identification of the wheat–A. cristatum 5P addition line. a GISH pattern of the wheat–A. cristatum 5P addition line obtained using the A. cristatum P genome (red) as the probe; b reads coverage analysis of the wheat–A. cristatum 5P addition line using RNA-seq data. The horizontal coordinate represents the chromosome position. The ordinate represents the read coverage at the corresponding location
To confirm the presence of TGW-related genes on chromosome 5P in the wheat background, genetic populations of 5P addition lines (BC5F1, BC6F1, BC6F2, BC7F1, and BC8F1) in the Fukuho background were developed and evaluated across four years (2017–2018, 2018–2019, 2019–2020, and 2021–2022) in Xinxiang, Henan Province. Molecular markers specific to 5PS and 5PL (Han et al. 2023) were used to detect the 5P chromosome of the five populations. The results of the statistical analysis suggested that there were significant differences in TGW and GL between the positive and negative plants of the 5P addition line and that there was an increase in GW in only two populations (BC5F1 and BC8F1), with no significant difference in the other populations. These findings indicated that the effect of 5P on enhanced GL was genetically stable across different years, while the effect of 5P on enhanced GW was less stable (Fig. 2). According to four years of field data, introducing 5P increased the wheat TGW by 2.55–7.10 g and the grain length by 0.30–0.50 mm, corresponding to increases of 7.62–19.29% and 4.49–8.93%, respectively (P < 0.01) (Table S1). In the BC8F1 population of II-11-1b in 2022–2023, the TGW of plants carrying A. cristatum 5P (43.91 ± 2.12 g) was 7.10 g higher than that of plants lacking this chromosome, representing the largest increase in TGW. The GL of plants with A. cristatum 5P was approximately 5.98 ± 0.13 mm, which was approximately 0.50 mm longer than that of plants without this chromosome. In the BC7F1 population of II-11-1b in 2019–2020, the TGW of the positive plants (36.03 ± 3.63 g) was 2.55 g higher than that of the negative plants, representing the least increase in TGW. The grain length of plants with A. cristatum 5P also increased by 0.40 mm. These findings confirmed that A. cristatum chromosome 5P contains an environmentally stable gene that enhances wheat TGW.
Comparison of grain traits (TGW, GL, and GW) in different genetic populations derived from the cross between II-11-1b and Fukuho. a Grain morphology comparison between the recurrent parents Fukuho, 5P-positive plants (5P +), and 5P-negative plants (5P-) from the BC8F1 segregated population. b Comparison of TGW between 5P-positive plants and 5P-negative plants in different genetic populations. c Comparison of GL between 5P-positive plants and 5P-negative plants in different genetic populations. d Comparison of GW between 5P-positive plants and 5P-negative plants in different genetic populations. Scale bar = 1 cm
Chromosome 5P increases the TGW in different wheat backgrounds
To evaluate the genetic effects of A. cristatum chromosome 5P in different wheat backgrounds, we crossed II-11-1b with Xinong 979 and Kenong 199 to develop F2 populations. Xinong 979 and Kenong 199 are two commercial varieties in China. The two F2 populations were planted in Xinxiang during the 2022–2023 growing season. A total of 78 positive plants and 70 negative plants in the II-11-1b/Xinong 979 F2 population and 99 positive plants and 81 negative plants in the II-11-1b/Kenong 199 F2 population were identified by chromosome 5P-specific molecular markers. Compared with the TGW, GL, and GW of plants without 5P, the TGW, GL, and GW of plants with 5P in the II-11-1b/Xinong 979 F2 population significantly increased by 6.22 g, 0.46 mm, and 0.11 mm, respectively, representing increases of 17.89%, 7.55%, and 4.40%, respectively (P < 0.01) (Fig. 3; Table S2). Similarly, compared with the TGW, GL, and GW of plants without 5P, the TGW, GL, and GW of plants with 5P in the II-11-1b/Kenong 199 F2 population significantly increased by 6.58 g, 0.44 mm, and 0.14 mm, corresponding to increases of 20.23%, 8.10%, and 5.49%, respectively (P < 0.01) (Fig. 3; Table S2). The above results showed that chromosome 5P increased TGW in different wheat backgrounds, indicating dominant inheritance independent of the wheat background, and that grain length in the Xinong 979 and Kenong 199 backgrounds significantly increased, consistent with that in the Fukuho background. However, grain width significantly increased in these two wheat backgrounds.
Comparison of grain traits (TGW, GL, and GW) in F2 populations derived from crosses between II-11-1b and Xinong 979 or Kenong 199. a Comparison of grain morphology between 5P-positive plants (XN_5P ( +)) and 5P-negative plants (XN_5P (-)) from the F2 population derived from II-11-1b and Xinong 979. b Comparison of grain morphology between 5P-positive plants (KN_5P ( +)) and 5P-negative plants (KN_5P (-)) from the F2 population derived from II-11-1b and Kenong 199. c Comparison of TGW between 5P-positive plants and 5P-negative plants in the two F2 populations. d Comparison of GL between 5P-positive plants and 5P-negative plants in the two F2 populations. e Comparison of GW between 5P-positive plants and 5P-negative plants in the two F2 populations. Scale bar = 1 cm
Chromosomal segmental localization of the TGW-enhancing locus
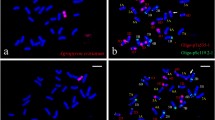
To identify which chromosomal arm of A. cristatum 5P carried the enhanced-TGW locus, three deletion lines (5PD1, 5PD2, and 5PD3) and two translocation lines (5PT1 and 5PT2), which carried the 5P short arm (5PS), 5PS and bin 1–2 of the 5P long arm (5PS + 5PL (bin 1–2)), 5PS and bin 1–6 of 5PL (5PS + 5PL (bin 1–6)), 5P long arm (5PL), and bin 1–6 of 5PL (5PL (bin 1–6)), respectively (Fig. 4; Fig. S2), were developed into segregated populations of different generations. The plants of the five segregated populations were identified by using chromosome 5P-specific molecular markers and evaluated for agronomic traits. The analytical grain trait results revealed that, compared with those of the plants without the 5P segment in the 5PD1 and 5PD3 BC3F2 populations, the GL of the plants with the 5P segment increased by 0.18 and 0.15 mm (P < 0.01), respectively. Additionally, there was a decrease in the grain width by 0.05 mm (P < 0.01 and P < 0.05, respectively), while there was no difference in TGW (Fig. 5a, c; Table 2).
Comparison of grain morphology in segregated populations developed from three deletion lines and two translocation lines. a, b, c Grain morphology comparison between 5P-positive plants and 5P-negative plants from the BC3F2 population derived from the 5PD1, 5PD2, and 5PD3 deletion lines. d, e Grain morphology comparison between 5P-positive plants and 5P-negative plants from the BC1F9 and BC4F1 populations derived from translocation lines 5PT2 and 5PT1, respectively. Scale bar = 1 cm
In the 5PD2 BC3F2 population, the plants with the 5P segment also showed a 0.17 mm increase in GL, and no significant difference was observed in TGW compared with the plants without the 5P segment (Fig. 5b; Table 2). 5PD2 differed from the 5PD1 and 5PD3 deletion lines in that the GW did not decrease. However, in the population of 5PT2 BC1F9, we observed significant increases in GL and TGW among the translocation plants compared to the non-translocation plants. Specifically, the translocation plants exhibited a 0.44 mm increase in GL (P < 0.05) and a 6.86 g increase in TGW (P < 0.01), representing increases of 8.49% and 24.40%, respectively. In addition, GW remained unchanged, consistent with the results for the 5P addition lines (Fig. 5d; Table 2). However, in the 5PT1 BC4F1 population, there were no differences in TGW, GL, or GW between the translocation plants and non-translocation plants (Fig. 5e; Table 2). A comparison of the plants with the 5P segment in these five populations revealed that the GL, GW, and TGW of 5PD1, 5PD2, 5PD3, and 5PT1 were basically the same, while the TGW and GW of 5PT2 were significantly higher and wider than those of the other four lines (Fig. S3). Overall, both the 5PL and 5PS arms had genes that increased GL, while the effect of 5PL was larger than that of 5PS. Moreover, 5PS negatively affected GW, whereas 5PL did not show any significant effect. Therefore, the gene for increased GL and TGW was located in the 5PL (bin 7–12) segment (Fig. 6), and it did not negatively affect GW.
Chromosomal localization of the enhanced grain weight locus from A. cristatum chromosome 5P and genetic effects of different segments of chromosome 5P on agronomic traits. a Schematic representation of the wheat–A. cristatum 5P addition line and different deletion and translocation lines, and the TGW-enhancing locus was localized to 5PL (bin 7–12). b Evaluation of agronomic traits of the addition, deletion, and translocation lines corresponding to panel a. Arrows represent significance, dotted arrows indicate no significance, and short lines indicate no effect
The influence of chromosome 5P on other agronomic traits and the analysis of the genetic effects of different chromosome segments
To investigate the effects of introducing 5P on other yield components, the differences in yield-related traits between the positive and negative plants of the 5P addition line were investigated after harvest. Compared with the plants without chromosome 5P, 5P-introgressed plants had a shorter plant height (PH) and lower effective tiller number (ETN), grain number per spike (GNS), and grain number per spikelet (GNPS) in at least four populations of the Fukuho background (Fig. 6; Table S1). In the F2 populations of II-11-1b with Xinong 979 and Kenong 199, the 5P chromosome had the same negative effect on ETN, GNS, and GNPS but had no effect on PH (Fig. 6; Table S2), indicating that the effect of the 5P chromosome on PH was influenced by the wheat background.
To analyze the effects of different segments of chromosome 5P on yield components, we compared differences in the yield-related traits of different 5P segments (Table 2; Fig. 6). The introduction of chromosomes 5PS, 5PS + 5PL (bin 1–2), and 5PS + 5PL (bin 1–6) reduced GNS, KNPS, and spikelet number per spike (SNS) but had no marked effect on PH or ETN. However, the introduction of chromosomes 5PL and 5PL (bin 1–6) had no observable effect on PH, ETN, GNS, KNPS, or SNS. Taken together, these findings indicated that the negative effects on GNS, KNPS, and SNS were from 5PS, not from 5PL, and the effects on reducing PH and ETN in plants harboring different 5P chromosome fragments were significantly weakened compared with those in 5P addition lines, suggesting that interactions between 5PL and 5PS might significantly reduce PH and ETN. The 5PT2 translocation line exhibited an increase in grain weight and grain length and did not adversely affect other agronomic traits. Therefore, this line could serve as a valuable germplasm for improving wheat grain weight (Fig. 6; Table 2).
Collectively, the genetic effects on yield-related traits showed that 5PS increased GL, decreased GW, maintained TGW, reduced GNS, and had no effect on PH or ETN, while 5PL (bin 1–6) had almost no effect on yield-related traits. 5PL (bin 7–12) was associated with increased GL, unchanged GW, and increased TGW and had minimal effects on PH, ETN, and GNS.
Discussion
The potential usefulness of the grain weight gene on chromosome 5P
The wild relatives of wheat harbor many novel and valuable genes that can serve as gene pools for wheat breeding (Zhang et al. 2017a). However, due to insufficient compensation or genetic drag, alien genes with important roles in wheat breeding include only a few disease resistance genes. These included Lr24/Sr24 and Sr26 from Thinopyrum elongatum, Sr36/Pm6 from T. timopheevii, Lr26/Sr31/Yr9/Pm8 and Gb2/Pm17 from Secale cereale, Yr17/Lr37/Sr38/Cre5 from Aegilops ventricose, and Pm21 from Dasypyrum villosum. These genes have greatly contributed to the cultivation and utilization of new cultivars with excellent alien genes in the history of wheat breeding worldwide (Friebe et al. 1996; Gill et al. 2006). However, studies on genes related to grain weight or grain size in wild relatives are rare. This may be related to the small genetic effect of grain weight as a quantitative trait in the wheat background. Due to the large wheat genome, the number of genes related to wheat grain weight or grain size cloned by forward genetics is also limited (Li and Yang 2017; Xiao et al. 2022). On the other hand, alien translocation often comes with genetic drag, introducing unfavorable genes along with beneficial ones (Jiang et al. 1994). The results in our study revealed that the alien gene on chromosome 5P significantly increased grain weight in different wheat backgrounds over different years. In addition, the negative effect of 5P was found to mainly result from 5PS, while the grain weight gene did not negatively affect other agronomic traits. This novel alien gene is therefore worthy of further research and utilization in breeding.
Comparison of genes related to grain weight on chromosome 5P and other grain weight-related genes in A. cristatum
A. cristatum possesses multiple genes that increase grain weight. Currently, two genetic loci that increase grain weight have been identified on chromosome 6P. One locus is located on the translocation segment of Pubing3035 (T1AS-6P-1AS·1AL) and can increase the TGW by 2.50 g in the Fukuho background (Zhang et al. 2015). The other locus is in the T6PS·6DL translocation line derived from the wheat–A. cristatum 6P substitution line 5106-DS, which can increase the TGW by approximately 4.00 g (Qi et al. 2021). The increase in the TGW of 5106-DS was mainly attributed to the increase in grain width, and the resulting grain phenotype was similar to that of the T6VS·6DL translocations (Feng et al. 2021). There is also a locus in the 7PS (bin 1–2) region of the pericentromeric region of chromosome 7P that can consistently increase TGW by approximately 2.00 g and increase grain weight in multiple wheat backgrounds, mainly due to the contribution of grain length (Sun et al. 2021). The grain weight locus on chromosome arm 5PL identified in this study was the fourth locus identified in A. cristatum that increased wheat grain weight. This locus increased grain weight by 2.55–7.10 g in different wheat backgrounds according to multiple years of phenotypic analysis. It also increased grain weight by increasing grain length, which was similar to the findings for the locus on chromosome 7P but different from those for the locus on chromosome 6P. The locus on chromosome 6P has not been evaluated in different wheat backgrounds. On the other hand, the grain weight locus of chromosome 7P is in the pericentromeric region, while the locus on chromosome 5P is located in the distal region of the long arm. The distal region is easier to recombine with wheat chromatin than the pericentromeric region (Luo et al. 2013; See et al. 2006). Therefore, this was an advantage of the grain weight gene on 5P, making it potentially easier to apply in future wheat breeding.
The production of translocation lines is an effective way to utilize alien genes
Since McFadden (1930) first transferred a stem rust resistance gene from T. dicoccum into common wheat, breeders and geneticists have created many translocation lines harboring favorable genes through chromosome engineering. A leaf rust resistance gene was transferred from Ae. umbellulata to wheat, resulting in the development of the "Transfer" line. Subsequently, the line was used to breed a series of disease-resistant varieties, such as Riley68 (Friebe et al. 1996). The wheat–S. cereale T1RS·1BL translocation line has greatly contributed to wheat breeding worldwide. This line has led to the production of many high-yield and disease-resistant wheat varieties, such as Neuzucht, Kavkaz, and Lovrin 13. Additionally, it has been used as a backbone parent in various breeding programs worldwide (Zhuang 2003). In China, the T1RS·1BL translocation lines from different sources were used to breed founder parents, such as Aimengniu and Zhou 8425B (Liu et al. 2020a). The T4AL·5PL translocation line, which was created in this study and characterized by large grain traits, exhibited good agronomic traits according to field evaluation data. Although the T4AL·5PL translocation was non-compensating, it increased grain length and grain weight without adversely affecting other agronomic traits, suggesting that it is potentially valuable for wheat yield breeding. An effective approach to enhance useability of alien genes in breeding is to produce small fragment translocations. For instance, Li et al. (2023) created translocations with smaller fragments to improve the utilization of the Fhb7 gene from Th. ponticum. Therefore, the next step is to create a 5PL small fragment translocation line carrying the enhanced grain weight gene. The genetic stocks and genomic tools developed from this study will facilitate the development of small fragment translocation lines.
Data availability
Data supporting the current study can be obtained by contacting the corresponding authors.
References
Adamski NM, Simmonds J, Brinton JF, Backhaus AE, Chen Y, Smedley M, Hayta S, Florio T, Crane P, Scott P, Pieri A, Hall O, Barclay JE, Clayton M, Doonan JH, Nibau C, Uauy C (2021) Ectopic expression of Triticum polonicum VRT-A2 underlies elongated glumes and grains in hexaploid wheat in a dosage-dependent manner. Plant Cell 33:2296–2319
Cao S, Xu D, Hanif M, Xia X, He Z (2020) Genetic architecture underpinning yield component traits in wheat. Theor Appl Genet 133:1811–1823
Carver BF, Rayburn AL (1994) Comparison of related wheat stocks possessing 1B or 1RS 1BL chromosomes: agronomic performance. Crop Sci 34:1505
Chen Y, Yan Y, Wu TT, Zhang GL, Yin H, Chen W, Wang S, Chang F, Gou JY (2020) Cloning of wheat keto-acyl thiolase 2B reveals a role of jasmonic acid in grain weight determination. Nat Commun 11:6266
Cheng X, Xin M, Xu R, Chen Z, Cai W, Chai L, Xu H, Jia L, Feng Z, Wang Z, Peng H, Yao Y, Hu Z, Guo W, Ni Z, Sun Q (2020) A single amino acid substitution in STKc_GSK3 kinase conferring semispherical grains and its implications for the origin of Triticum sphaerococcum. Plant Cell 32:923–934
Dewey DR (1984) The genomic system of classification as a guide to intergeneric hybridization with the perennial Triticeae. In: Gustafson JP (ed) Gene manipulation in plant improvement. In: Proceedings of 16th Stadler genetics symposium. Plenum Press, New York, 209–279
Dong YS, Zhou RH, Xu SJ, Li LH, Cauderon Y, Wang RR-C (1992) Desirable characteristics in perennial Triticeae collected in China for wheat improvement. Hereditas 116:175–178
Feng N, Song G, Guan J, Chen K, Jia M, Huang D, Wu J, Zhang L, Kong X, Geng S, Liu J, Li A, Mao L (2017) Transcriptome profiling of wheat inflorescence development from spikelet initiation to floral patterning identified stage-specific regulatory genes. Plant Physiol 174:1779–1794
Feng Z, Song L, Song W, Qi Z, Yuan J, Li R, Han H, Wang H, Chen Z, Guo W, Xin M, Liu J, Hu Z, Peng H, Yao Y, Sun Q, Ni Z, Xing J (2021) The decreased expression of GW2 homologous genes contributed to the increased grain width and thousand-grain weight in wheat-Dasypyrum villosum 6VS·6DL translocation lines. Theor Appl Genet 134:3873–3894
Friebe B, Jiang J, Raupp WJ, McIntosh RA, Gill BS (1996) Characterization of wheat-alien translocations conferring resistance to diseases and pests: Current status. Euphytica 91:59–87
Gill BS, Friebe B, Raupp WJ, Wilson DL, Cox TS, Sears RG, Brown-Guedira GL, Fritz AK (2006) Wheat genetics resource center: The first 25 years. Adv Agron 89:73–136
Han F, Lamb JC, Birchler JA (2006) High frequency of centromere inactivation resulting in stable dicentric chromosomes of maize. Proc Natl Acad Sci U S A 103:3238–3243
Han H, Liu W, Lu Y, Zhang J, Yang X, Li X, Hu Z, Li L (2017) Isolation and application of P genome-specific DNA sequences of Agropyron Gaertn. in Triticeae. Planta 245:425–437
Han H, Ma X, Wang Z, Qi K, Yang W, Liu W, Zhang J, Zhou S, Lu Y, Yang X, Li X, Li L (2023) Chromosome 5P of Agropyron cristatum induces chromosomal translocation by disturbing homologous chromosome pairing in a common wheat background. Crop J 11:228–237
Hao M, Zhang L, Ning S, Huang L, Yuan Z, Wu B, Yan Z, Dai S, Jiang B, Zheng Y, Liu D (2020) The resurgence of introgression breeding, as exemplified in wheat improvement. Front Plant Sci. https://doi.org/10.3389/fpls.2020.00252
Jiang J, Friebe B, Gill BS (1994) Recent advances in alien gene transfer in wheat. Euphytica 73:199–212
Jiang B, Liu T, Li H, Han H, Li L, Zhang J, Yang X, Zhou S, Li X, Liu W (2018) Physical mapping of a novel locus conferring leaf rust resistance on the long arm of Agropyron cristatum chromosome 2P. Front Plant Sci 9:817
Kato A, Lamb JC, Birchler JA (2004) Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize. Proc Natl Acad Sci U S A 101:13554–13559
Kuang Z, Ji X, Xu S, Han H, Zhang J, Zhou S, Yang X, Li X, Li L, Liu W (2023) Generation and identification of a wheat–Agropyron cristatum (L.) Gaertn 3P chromosome addition line and substitution line. Euphytica 219:23
Kuchel H, Williams KJ, Langridge P, Eagles HA, Jefferies SP (2007) Genetic dissection of grain yield in bread wheat. I QTL Analysis Theor Appl Genet 115:1029–1041
Li W, Yang B (2017) Translational genomics of grain size regulation in wheat. Theor Appl Genet 130:1765–1771
Li L, Li X, Li P, Dong Y, Zhao G (1997) Establishment of wheat-Agropyron cristatum alien addition lines. I. Cytology of F3, F2BC1, BC4, and BC3F1 progenies. Acta Genet Sin 24:154–159
Li L, Yang X, Zhou R, Li X, Dong Y (1998) Establishment of wheat-Agropyron cristatum alien addition lines II Identification of alien chromosomes and analysis of development approaches. Acta Genet Sin 25:538–544
Li G, Chen P, Zhang S, Wang X, He Z, Zhang Y, Zhao H, Huang H, Zhou X (2007) Effects of the 6VS.6AL translocation on agronomic traits and dough properties of wheat. Euphytica 155:305–313
Li H, Jiang B, Wang J, Lu Y, Zhang J, Pan C, Yang X, Li X, Liu W, Li L (2017) Mapping of novel powdery mildew resistance gene(s) from Agropyron cristatum chromosome 2P. Theor Appl Genet 130:109–121
Li X, Li D, Xuan Y, He Z, Zhao L, Hao Y, Ge W, Xu S, Hou B, Wang B, Guo J, Liu W, Li M, Han Y, Bo C, Bao Y, Qi Z, Xu SS, Bai G, Wang H, Kong L (2023) Elimination of the yellow pigment gene PSY-E2 tightly linked to the Fusarium head blight resistance gene Fhb7 from Thinopyrum ponticum. Crop J 11:957–962
Lin Y, Zhou S, Liang X, Guo B, Han B, Han H, Zhang J, Lu Y, Zhang Z, Yang X, Li X, Liu W, Li L (2022) Chromosomal mapping of a locus associated with adult-stage resistance to powdery mildew from Agropyron cristatum chromosome 6PL in wheat. Theor Appl Genet 135:2861–2873
Lin Y, Zhou S, Liang X, Han B, Yang J, Guo B, Zhang J, Han H, Liu W, Yang X, Li X, Li L (2023) Introgression of chromosome 6PL terminal segment from Agropyron cristatum to increase both grain number and grain weight in wheat. Crop J 11:878–886
Liu C, Han R, Wang X, Gong W, Cheng D, Cao X, Liu A, Li H, Liu J (2020a) Research progress of wheat wild hybridization, disease resistance genes transfer and utilization. Sci Agric Sin 53:1287–1308
Liu H, Li H, Hao C, Wang K, Wang Y, Qin L, An D, Li T, Zhang X (2020b) TaDA1, a conserved negative regulator of kernel size, has an additive effect with TaGW2 in common wheat (Triticum aestivum L.). Plant Biotechnol J 18:1330–1342
Liu J, Chen Z, Wang Z, Zhang Z, Xie X, Wang Z, Chai L, Song L, Cheng X, Feng M, Wang X, Liu Y, Hu Z, Xing J, Su Z, Peng H, Xin M, Yao Y, Guo W, Sun Q, Liu J, Ni Z (2021) Ectopic expression of VRT-A2 underlies the origin of Triticum polonicum and Triticum petropavlovskyi with long outer glumes and grains. Mol Plant 14:1472–1488
Luo MC, Gu YQ, You FM, Deal KR, Ma Y, Hu Y, Huo N, Wang Y, Wang J, Chen S, Jorgensen CM, Zhang Y, McGuire PE, Pasternak S, Stein JC, Ware D, Kramer M, McCombie WR, Kianian SF, Martis MM, Mayer KF, Sehgal SK, Li W, Gill BS, Bevan MW, Simkova H, Dolezel J, Weining S, Lazo GR, Anderson OD, Dvorak J (2013) A 4-gigabase physical map unlocks the structure and evolution of the complex genome of Aegilops tauschii, the wheat D-genome progenitor. Proc Natl Acad Sci U S A 110:7940–7945
Lyu J, Wang D, Sun N, Yang F, Li X, Mu J, Zhou R, Zheng G, Yang X, Zhang C, Han C, Xia G-M, Li G, Fan M, Xiao J, Bai M-Y (2024) The TaSnRK1-TabHLH489 module integrates brassinosteroid and sugar signalling to regulate the grain length in bread wheat. Plant Biotechnol J. https://doi.org/10.1111/pbi.14319
Ma L, Li T, Hao C, Wang Y, Chen X, Zhang X (2016) TaGS5-3A, a grain size gene selected during wheat improvement for larger kernel and yield. Plant Biotechnol J 14:1269–1280
Maag JLV (2018) gganatogram: An R package for modular visualisation of anatograms and tissues based on ggplot2. F1000Res 7:1576
McFadden ES (1930) A successful transfer of emmer characters to vulgare wheat. J Am Soc Agron 22:1020–1034
Mujeeb-Kazi A, Kazi AG, Dundas I, Rasheed A, Ogbonnaya F, Kishii M, Bonnett D, Wang RRC, Xu S, Chen PD, Mahmood T, Bux H, Farrakh S (2013) Genetic diversity for wheat improvement as a conduit to food security. Adv Agron 122:179–257
Niaz M, Zhang L, Lv G, Hu H, Yang X, Cheng Y, Zheng Y, Zhang B, Yan X, Htun AA, Zhao L, Sun C, Zhang N, Ren Y, Chen F (2023) Identification of TaGL1-B1 gene controlling grain length through regulation of jasmonic acid in common wheat. Plant Biotechnol J 21:979–989
Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL (2016) Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc 11:1650–1667
Qi K, Han H, Zhang J, Zhou S, Li X, Yang X, Liu W, Lu Y, Li L (2021) Development and characterization of novel Triticum aestivum-Agropyron cristatum 6P Robertsonian translocation lines. Mol Breeding 41:59
Quinlan AR, Hall IM (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842
Reynolds M, Foulkes MJ, Slafer GA, Berry P, Parry MAJ, Snape JW, Angus WJ (2009) Raising yield potential in wheat. J Exp Bot 60:1899–1918
See DR, Brooks S, Nelson JC, Brown-Guedira G, Friebe B, Gill BS (2006) Gene evolution at the ends of wheat chromosomes. Proc Natl Acad Sci USA 103:4162–4167
Shiferaw B, Smale M, Braun HJ, Duveiller E, Reynolds M, Muricho G (2013) Crops that feed the world 10. Past successes and future challenges to the role played by wheat in global food security. Food Secur 5:291–317
Song L, Lu Y, Zhang J, Pan C, Yang X, Li X, Liu W, Li L (2016) Physical mapping of Agropyron cristatum chromosome 6P using deletion lines in common wheat background. Theor Appl Genet 129:1023–1034
Su Z, Hao C, Wang L, Dong Y, Zhang X (2011) Identification and development of a functional marker of TaGW2 associated with grain weight in bread wheat (Triticum aestivum L.). Theor Appl Genet 122:211–223
Sun Y, Lyu M, Han H, Zhou S, Lu Y, Liu W, Yang X, Li X, Zhang J, Liu X, Li L (2021) Identification and fine mapping of alien fragments associated with enhanced grain weight from Agropyron cristatum chromosome 7P in common wheat backgrounds. Theor Appl Genet 134:3759–3772
Tang Z, Yang Z, Fu S (2014) Oligonucleotides replacing the roles of repetitive sequences pAs1, pSc119.2, pTa-535, pTa71, CCS1, and pAWRC.1 for FISH analysis. J Appl Genet 55:313–318
Tian J, Deng Z, Hu R, Wang Y (2006) Yield components of super wheat cultivars with different types and the path coefficient analysis on grain yield. Acta Agron Sin 32:1699–1705
Tilman D, Balzer C, Hill J, Befort BL (2011) Global food demand and the sustainable intensification of agriculture. Proc Natl Acad Sci U S A 108:20260–20264
Valluru R, Reynolds MP, Salse J (2014) Genetic and molecular bases of yield-associated traits: a translational biology approach between rice and wheat. Theor Appl Genet 127:1463–1489
Villareal RL, Rajaram S, Mujeeb-Kazi A, Del Toro E (1991) The effect of chromosome 1B/1R translocation on the yield potential of certain spring wheats (Triticum aestivum L.). Plant Breed 106:77–81
Villareal RL, Banuelos O, Mujeeb-Kazi A, Rajaram S (1998) Agronomic performance of chromosomes 1B and T1BL.1RS near-isolines in the spring bread wheat Seri M82. Euphytica 103:195–202
Wang Y, Hou J, Liu H, Li T, Wang K, Hao C, Liu H, Zhang X (2019) TaBT1, affecting starch synthesis and thousand kernel weight, underwent strong selection during wheat improvement. J Exp Bot 70:1497–1511
Wang X, Han B, Sun Y, Kang X, Zhang M, Han H, Zhou S, Liu W, Lu Y, Yang X, Li X, Zhang J, Liu X, Li L (2022) Introgression of chromosome 1P from Agropyron cristatum reduces leaf size and plant height to improve the plant architecture of common wheat. Theor Appl Genet 135:1951–1963
Wu J, Yang XM, Wang H, Li HJ, Li LH, Li XQ, Liu WH (2006) The introgression of chromosome 6P specifying for increased numbers of florets and kernels from Agropyron cristatum into wheat. Theor Appl Genet 114:13–20
Xiao J, Chen Y, Lu Y, Liu Z, Si D, Xu T, Sun L, Wang Z, Yuan C, Sun H, Zhang X, Wen M, Wei L, Zhang W, Wang H, Wang X (2021) A natural variation of an SVP MADS-box transcription factor in Triticum petropavlovskyi leads to its ectopic expression and contributes to elongated glume. Mol Plant 14:1408–1411
Xiao J, Liu B, Yao Y, Guo Z, Jia H, Kong L, Zhang A, Ma W, Ni Z, Xu S, Lu F, Jiao Y, Yang W, Lin X, Sun S, Lu Z, Gao L, Zhao G, Cao S, Chen Q, Zhang K, Wang M, Wang M, Hu Z, Guo W, Li G, Ma X, Li J, Han F, Fu X, Ma Z, Wang D, Zhang X, Ling HQ, Xia G, Tong Y, Liu Z, He Z, Jia J, Chong K (2022) Wheat genomic study for genetic improvement of traits in China. Sci China Life Sci 65:1718–1775
Xu S, Ji X, Sun S, Han H, Zhang J, Zhou S, Yang X, Li X, Li L, Liu W (2022) Production of new wheat–A. cristatum translocation lines with modified chromosome 2P coding for powdery mildew and leaf rust resistance. Mol Breeding 42:14
Yan L, Liu ZS, Xu HW, Zhang XP, Zhao AJ, Liang F, Xin MM, Peng HR, Yao YY, Sun QX, Ni ZF (2018) Transcriptome analysis reveals potential mechanisms for different grain size between natural and resynthesized allohexaploid wheats with near-identical AABB genomes. BMC Plant Biol 18:1–15
Yang GH, Yang XM, Wang RH, Gao AN, Li LH, Liu WH (2010) The inhibiting effect of 1.4 recombinant P chromosome of wheat-Agropyron cristatum addition line on the Ph gene. Chinese Sci Bull 55:153–157
Zhang L, Zhao Y-L, Gao L-F, Zhao G-Y, Zhou R-H, Zhang B-S, Jia J-Z (2012) TaCKX6-D1, the ortholog of rice OsCKX2, is associated with grain weight in hexaploid wheat. New Phytol 195:574–584
Zhang J, Zhang JP, Liu WH, Han HM, Lu YQ, Yang XM, Li XQ, Li LH (2015) Introgression of Agropyron cristatum 6P chromosome segment into common wheat for enhanced thousand-grain weight and spike length. Theor Appl Genet 128:1827–1837
Zhang J, Zhang J, Liu W, Wu X, Yang X, Li X, Lu Y, Li L (2016) An intercalary translocation from Agropyron cristatum 6P chromosome into common wheat confers enhanced kernel number per spike. Planta 244:853–864
Zhang H, Mittal N, Leamy LJ, Barazani O, Song B-H (2017a) Back into the wild—Apply untapped genetic diversity of wild relatives for crop improvement. Evol Appl 10:5–24
Zhang Z, Song L, Han H, Zhou S, Zhang J, Yang X, Li X, Liu W, Li L (2017b) Physical localization of a locus from Agropyron cristatum conferring resistance to stripe rust in common wheat. Int J Mol Sci 18:2043
Zhang Z, Han H, Liu W, Song L, Zhang J, Zhou S, Yang X, Li X, Li L (2019) Deletion mapping and verification of an enhanced-grain number per spike locus from the 6PL chromosome arm of Agropyron cristatum in common wheat. Theor Appl Genet 132:2815–2827
Zhou Y, He Z, Zhang G, Xia L, Chen X, Gao Y, Jing Z, Yu G (2004) Utilization of 1BL/1RS translocation in wheat breeding in China. Acta Agron Sin 30:531–535
Zhu T, Wang L, Rimbert H, Rodriguez JC, Deal KR, De Oliveira R, Choulet F, Keeble-Gagnere G, Tibbits J, Rogers J, Eversole K, Appels R, Gu YQ, Mascher M, Dvorak J, Luo MC (2021) Optical maps refine the bread wheat Triticum aestivum cv. Chin Spring Genome Assembly Plant J 107:303–314
Zhu X, Rong W, Wang K, Guo W, Zhou M, Wu J, Ye X, Wei X, Zhang Z (2022) Overexpression of TaSTT3b-2B improves resistance to sharp eyespot and increases grain weight in wheat. Plant Biotechnol J 20:777–793
Zhuang Q (2003) Chinese wheat improvement and pedigree analysis. China Agriculture Press, Beijing, China
Zuo J, Li J (2014) Molecular genetic dissection of quantitative trait loci regulating rice grain size. Annu Rev Genet 48:99–118
Funding
This work was financially supported by the National Key Research and Development Program of China (2021YFD1200605) and the Chinese Agriculture Research System (CARS-03).
Author information
Authors and Affiliations
Contributions
LHL and HMH conceived the research. HMH performed the research and wrote the paper. JLY, KQ, HYZ, and PQW collected the data. LHL and WHL produced the plant materials, and SHZ, JPZ, BJG, YQL, XMY, and XQL participated in the preparation of the reagents and materials for this study. All authors contributed to and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Communicated by Steven S. Xu.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
122_2024_4670_MOESM1_ESM.pdf
Supplementary file1 : Fig. S1 Comparison of grain traits (TGW, GL, and GW) between II-11-1b and Fukuho. a Comparison of TGW between II-11-1b and Fukuho. b Comparison of GL between II-11-1b and Fukuho. c Comparison of GW between II-11-1b and Fukuho. Fig. S2 GISH and ND-FISH identification of the wheat–A. cristatum chromosome 5P deletion and translocation lines. a, b, c, d, e GISH patterns of the deletion lines 5PD1, 5PD2, and 5PD3 and the translocation lines 5PT1 and 5PT2 obtained using the A. cristatum P genome (red) as the probe. f ND-FISH pattern of 5PT2 identified in this study using oligo-pTa535-2 (red) and oligo-pSc119.2-2 (green) as probes. Fig. S3 Comparison of grain traits (TGW, GL, and GW) between the three deletion lines and two translocation lines. a Comparison of TGW among three deletion lines and two translocation lines. b Comparison of GL between three deletion lines and two translocation lines. c Comparison of GW between three deletion lines and two translocation lines. 5PS+5PL (1-6): 5PD3; 5PS+5PL (1-2): 5PD2; 5PS: 5PD1; 5PL: 5PT2; 5PL (1-6): 5PT1
122_2024_4670_MOESM2_ESM.pdf
Supplementary file2 : Table S1 Evaluation of agronomic traits of genetic populations of the chromosome 5P addition line in the Fukuho background. Table S2 Evaluation of the agronomic traits of genetic populations derived from crosses of the chromosome 5P addition line and two commercial wheat varieties
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Han, H., Yang, J., Qi, K. et al. Introgression of chromosome 5P from Agropyron cristatum enhances grain weight in a wheat background. Theor Appl Genet 137, 165 (2024). https://doi.org/10.1007/s00122-024-04670-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00122-024-04670-5