Abstract
The powdery mildew resistance gene Pm6, transferred to common wheat from the tetraploid Triticum timopheevii, is effective in most epidemic areas for powdery mildew in China. RFLP probe BCD135 was previously associated with Pm6. In the present research, four STS primers (NAU/STSBCD135-1, NAU/STSBCD135-2, STS003 and STS004) were designed from the sequence data of BCD135. These primers were used for PCR amplification using the genomic DNA of resistant near-isogenic lines with Pm6 and their recurrent parent, cv. Prins. No polymorphic product was observed using primers STS003 and STS004; however, primers NAU/STSBCD135-1 and NAU/STSBCD135-2 amplified two and one bands, respectively, polymorphic between the resistant near-isogenic-lines and Prins. The two primers were then used to amplify the F2 population from the cross IGV1-465 (FAO163b/7*Prins) × Prins. The amplification and the powdery mildew resistance identification data were analyzed using the software Mapmaker 3.0. The results indicated that both NAU/STSBCD135-1 and NAU/STSBCD135-2 were closely linked to Pm6 with a genetic distance of 0.8 cM. A total of 175 commercial varieties without Pm6 from different ecological areas of China were tested using marker NAU/STSBCD135-2 and none of them amplified the 230 bp-specific band. This marker thus has high practicability and can be used in MAS of Pm6 in wheat breeding programs for powdery mildew resistance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Powdery mildew, caused by Blumeria graminis f.sp. tritici, is one of the most important diseases of wheat (T. aestivum L.) worldwide. Development and utilization of wheat varieties with powdery mildew resistance is the most economical and effective strategy for disease control. To date, 34 powdery mildew resistance gene loci have been identified in common wheat (McIntosh et al. 2003; Hsam et al. 2003; Zhu et al. 2005; Miranda et al. 2006). Accumulating multiple resistance genes within a single genotype, commonly referred to as “gene pyramiding,” is one strategy for increasing durability of resistance (Kameswara Rao et al. 2002). However, gene pyramiding can be difficult without utilysing markers. Restriction Fragment Length Polymorphism (RFLP) and Random Amplified Polymorphic DNA (RAPD) have been used to generate wheat genetic maps and to find molecular markers for resistance genes, but screening of RFLP probes for closely linked markers is laborious and time-consuming, even though the position of the target gene is known. Also, RAPD markers identified in one population are not always useful for another, and may lack reliability (Weeden et al. 1992).
To address the above problems, it is useful to convert a RFLP or RAPD polymorphic fragments into sequence tagged site (STS) or sequence-characterized amplified region (SCAR) markers (Olson et al. 1989). STSs are relatively easy to develop from DNA fragments identified by arbitrarily primed PCR, although they do not always result in detectable polymorphisms. STSs, as specific PCR products, are reproducible across a relatively wide range of reaction conditions in different laboratories thus overcoming the shortcomings of RAPDs.
Powdery mildew resistance gene Pm6 was introgressed from tetraploid wheat T. timopheevii into common wheat (T. aestivum) and mapped in the long arm of chromosome 2B (Nyquist 1963; Jorgensen and Jensen 1972, 1973). Pm6 has been widely used in wheat breeding for powdery mildew resistance, and is still one of the most effective resistance genes in many areas of the world, including China, Europe and North America, especially when it is used combined with other Pm genes, such as Pm2 (Jorgensen and Jensen 1972; Cai et al. 2005). Tao et al. (2000) defined the lengths of the introgressed T. timopheevii segments in six different introgression lines and identified several RFLP markers linked with Pm6. Among them, Xbcd135 was the closest, with a genetic distance of 1.6 ± 1.5 cM from Pm6. Ji et al. (2007) further discriminated the Pm6-carrying T. aestivum-T. timopheevii introgression lines using PCR-based molecular markers. The objectives of this study were to convert the RFLP marker Xbcd135 into STS markers, and to evaluate those markers for application in marker-assisted selection of Pm6 in wheat breeding programs.
Materials and methods
Plant materials
The Swedish Spring common wheat cultivar (cv.) Prins and eight introgression lines with Pm6 (listed in Table 1) were provided by Dr. J. MacKey, Swedish Agricultural University, Uppsala. The lines were developed through backcrossing for seven to eight generations with the recurrent parent Prins. The donor lines of Pm6 are listed in Table 1. Four wheat cultivars and lines carrying Pm6 (Coker 747, Coker 983, Timgalen, and T. timopheevii) (Nyquist 1963; Leath and Heun 1990; Bennett and Kints 1983) were also used to assess the applicability of the markers for identifying Pm6 in different wheat backgrounds. Prins was crossed with line IGV1-465. The resulting F1 population was selfed and an F2 population of 374 individuals was generated from which leaf tissue was harvested for genomic DNA extraction. The F2 plants were grown to maturity and selfed; the responses of the F3 families, viz. homozygous resistant, homozygous susceptible, or segregating for resistance were used to deduce F2 genotypes.
A total of 175 commercial wheat varieties from different ecological areas of China, kindly provided by Mr Ruiqi Zhang, Nanjing Agricultural University, were used to confirm that the STS markers would be useful for marker assisted selection.
Evaluation of powdery mildew response
Materials evaluated for powdery mildew response included the eight introgression lines, four wheat cultivars and lines carrying Pm6, the recurrent parent Prins, and F2 and F3 progenies of IGV1-465 × Prins, the F3 test comprising a progeny test of 20 seedlings from each F2 individual. All of the plants were inoculated with a local Blumeria graminis mixture at the four-leaf stage in the greenhouse, and inoculations were repeated after 48 h. Host responses were recorded 7–14 days after inoculation when the susceptible control showed obvious disease symptoms and the resistant control was powdery mildew-free. The reactions of individual seedlings were rated as either resistant or susceptible. Chi-squared tests for goodness of fit were used to test for deviation of observed data from theoretically expected segregation ratios.
STS primer design
Barley probe BCD135 (773K14.2) derived from barley BAC clone AF474072 (Rostoks et al. 2002; Park et al. 2004) was downloaded from the NCBI website (http://www.ncbi.nlm.nih.gov/), and analyzed using the computer program SSRHUNTER developed by Li and Wan (2005). Three SSRs (simple sequence repeats) were identified within the BCD135 sequence, and four STS primers were designed using Primer 3 Software (http://www.genome.wi.mit.Edu/cgi-bin/primer/primer3.cgi). The primers were synthesized at Genebase Biotechnology Co. Ltd., Shanghai. The sequences and annealing temperatures of the four primers are shown in Table 2.
Marker analysis
STS primers linked to Pm6 were identified by bulked segregant analysis (BSA). Resistant and susceptible bulks were made by pooling equal amounts of DNA from ten resistant and susceptible F2 plants, respectively.
DNA extraction was according to Qi et al. (1997) and PCR amplifications were performed in reaction mixture volumes of 25 μl, each containing 1 × PCR buffer, 1.8 mM MgCl2, 0.2 mM dNTPs, 0.24 μM of each STS primer, 80 ng template DNA and 1U Taq DNA polymerase (Takara Bio Inc., Japan). The PCR profile was: initial denaturation at 94°C for 4 min, 31 cycles at 94°C for 30 s, 56°C for 1 min, 72°C for 1 min, and a final extension at 72°C for 1 min. The PCR analyses were carried out in a PE 9600 thermal cycler (Perkin Elmer, Norwalk, CT, USA), and the products were separated in 8% non-denaturing polyacrylamide gels run in a 1 × TBE buffer.
Linkage analysis was performed using MAPMAKER/Exp Version 3.0b (Lincoln et al. 1993). Map distances were determined using the Kosambi mapping function (Kosambi, 1944).
Results
Evaluation of powdery mildew reactions
A total of 374 F2 individuals were obtained from the cross of (IGV1-465 × Prins), 279 were resistant, and 95 were susceptible, which was in agreement with the expected 3:1 segregation ratio for resistant and susceptible plants (χ2 3:1 = 0.03, P > 0.95). Based on the F3 data, the F2 genotypes were 101 Pm6Pm6, 178 Pm6pm6, and 95 pm6pm6, an acceptable fit to a 1:2:1 segregation (χ2 1:2:1 = 1.06, P > 0.50).
The recurrent parent Prins was susceptible, and the various introgression lines showed resistant reactions.
Linkage of the STS markers with Pm6
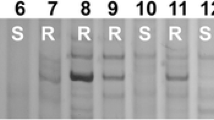
BSA was used to determine the relationship of the four markers with Pm6. No polymorphisms occurred between the two parents and the two pools using STS003 and STS004 as primers. However, primers NAU/STSBCD135-1 and NAU/STSBCD135-2 amplified bands polymorphic between IGV1-465 and Prins and between the two bulks. NAU/STSBCD135-1 amplified two bands (300 and 350 bp), present in the resistant lines and the resistant bulk, but absent in the susceptible lines and the susceptible bulk (Fig. 1). Primer NAU/STSBCD135-2 amplified one polymorphic band (230 bp), again present in the resistant lines and the resistant bulk, but absent in the susceptible lines and the susceptible bulk (Fig. 2).
PCR Results using primer NAU/STSBCD135-1 (arrows show the polymorphic bands) M: the size standard marker, to the left is the marker band size in bp, Lanes 1–15 represent Prins, S pools from (IGV1-465 × Prins) F2, R pools from (IGV1-465 × Prins) F2, IGV1-448, IGV1-458, IGV1-463, IGV1-464, IGV1-465, IGV1-466, IGV1-468, IGV1-474, Coker747, T. timopheevii, Coker983, Timgalen, respectively
PCR Results using primer NAU/STSBCD135-2 (arrows show the polymorphic bands) M: the size standard marker, to the left is the marker band size in bp, Lanes 1–15 represent Prins, S pools from (IGV1-465 × Prins) F2, R pools from (IGV1-465 × Prins) F2, IGV1-448, IGV1-458, IGV1-463, IGV1-464, IGV1-465, IGV1-466, IGV1-468, IGV1-474, Coker747, T. timopheevii, Coker983, Timgalen, respectively
To determine the genetic linkage of the STS markers with the Pm6 gene, NAU/STSBCD135-1 and NAU/STSBCD135-2 were further used to screen the entire F2 population (Fig 3). Both markers behaved identically. Bands were observed in all 278 homozygous resistant and heterozygous plants, and in two of the 95 homozygous susceptible plants. Linkage analysis indicated a genetic distance of 0.8 cM between the markers and Pm6.
Practicability of NAU/STSBCD135-2 in MAS for powdery mildew resistance
In order to evaluate the applicability of the STS markers for marker assisted selection of Pm6 in breeding programs, a total of 175 commercial varieties or elite lines were amplified using NAU/STSBCD135-2. The results indicated that none showed the specific 230 bp band, which was present in T. timopheevii and IGV1-465 (Fig. 4), thus indicating that this marker could be used in MAS of Pm6 over a wide range of genetic backgrounds.
PCR results using primer NAU/STSBCD135-2 on 28 commercial varieties or elite lines and controls.M: the size standard marker (lane 32), to the right is the marker band size in bp, Lanes 1–31: 1. Ailiduo, 2. Zhengzhou 24, 3. Nongda 183, 4. Youbao, 5. Jinan 12, 6. Funo, 7. Taishan 5, 8. Yannong 15, 9. Xiaoyan 6, 10. Shannongfu 63, 11. Lumai 10, 12. Xuzhou 21, 13.Yumai 21, 14. Shan 229, 15. Jinan 17, 16. Zheng 9023, 17. Huanyin 9628, 18. Xiaoyan 54, 19. Yuanfeng 898, 20. Jimai 20, 21. Xuzhou 438, 22. Shannong 6521, 23. Bainong 3217, 24. Nanyang 756, 25. Xiannong 151, 26. Chinese Spring, 27. Yangmai 5, 28. 92R137, 29. Prins, 30. IGV1-465, 31. T. timopheevii (arrow shows the polymorphic band. Arrow heads show the fragment size)
Discussion
Bennett (1984) reported that Pm6 was moderately effective and best expressed from the three-leaf stage onwards, but was also recognizable at the first-leaf stage. In preliminary experiments we inoculated the Prins materials at various growth stages from the second to fifth leaves and found that the fourth leaf stage was optimal for clear differences between resistant and susceptible genotypes. This growth stage was then used in our genetic experiments. Pm6 was inherited as a single Mendelian factor.
The precision of the measured genetic distance is related to the number of individuals in the mapping population. Primary genetic maps are typically based on 50–100 individuals, permitting the detection of recombination between markers 1 and 3 cM apart. For experiments requiring a higher degree of precision, additional individuals must be analyzed. Compared with Tao et al. (2000) and Wang et al. (2000), in which only 73 F2 individuals were analyzed, we used 374 F2 genotypes in the present research. Therefore our estimate of genetic distance should be more precise.
Pm6 was introduced into common wheat from T. timopheevii and its incorporation into the wheat genome involved recombination between related B and G chromosomes. Due to the differentiation of the 2G segment and the equivalent region of chromosome 2B, recombination near Pm6 may be reduced. Thus the physical distance between the marker and Pm6 might be larger than suggested by the recombination value. For map-based cloning of Pm6, a larger F2 mapping population (such as more than 1,000 F2 plants) could be generated by crossing the smallest introgression (IGV1-465) with Prins, followed by fine mapping for further recombinants using the STS marker isolated in the present study as one of the markers for Pm6. Even then, the remaining T. timopheevii segment may be physically too large and alternative procedures such as resistance gene analogues or subtractive hybridization might be more rewarding.
To facilitate the use of molecular markers in large-scale screening, it is necessary to convert RFLP markers into PCR-based markers. Talbert et al. (1994) successfully converted RFLP probes of wheat into STS markers. Later, many laboratories attempted to generate STS primers from either wheat genomic clones (Feuillet et al. 1995; Talbert et al. 1996; Seyfarth et al. 1999; Ma et al. 2004 ), RAPD PCR products (Hu et al. 1997; Naik et al. 1998; Dweikat et al. 2002) or AFLP bands (Parker and Langridge 2000; Prins et al. 2001; Smith et al. 2002). The conversion of heterologous probes, such as barley clones or oat clones, is more difficult because sequence polymorphisms may exist between the homoeologous loci of wheat and barley or oat. In addition, the amplified products may not originate from the same loci where the RFLP probes originally mapped. Byran et al. (1997) pointed out that PCR primers based on wheat cDNA have a tendency to produce several PCR products in similar size. In this paper, we firstly designed STS primers according to the three SSR loci found in the sequence of BCD135. This method for the STS primer design combined the advantages of the RFLP, SSR and STS.
Pm6 is an effective resistance gene in many powdery mildew epidemic areas of China, especially for adult resistance in the field. However, lines with Pm6 may be susceptible at the seedling stage, making it difficult to identify the presence of Pm6 in seedling tests. The strategy of marker-assisted selection is useful for combining resistance genes in a single genotype. Such a strategy is thought likely to increase the durability of resistance, but requires that the individual genes not to be used simultaneously in other cultivars. We found that the specific PCR products were only amplified in Pm6-carrying genotypes, indicating that NAU/STSBCD135-2 should be helpful for marker assisted selection of plants with Pm6.
References
Bennett FGA (1984) Resistance to powdery mildew in wheat: a review of its use in agricuture and breeding programmes. Plant Pathol 33:279–300
Bennett FGA, van Kints T (1983) Mildew of wheat U.K. Cereal Pathogen Virulence Survey Annual Report. National Institute of Agricultural Botany, Cambridge, UK, pp 7–21
Bryan GK, Collins AJ, Stephenson P, Orry A, Smith JB, Gale MD (1997) Isolation and characterization of microsatellites from hexaploid bread wheat. Theor Appl Genet 94:557–563
Cai SB, Cheng SH, Wu JZ, Yan W (2005) Evaluation, improvement and utilization of introducted wheat reserve resource resistant to powdery mildew. Acta Tritical Crops 25:116–120
Dweikat I, Zhang W, Ohm H (2002) Development of STS markers linked to Hessian fly resistance gene H6 in wheat. Theor Appl Genet 105:766–770
Feuillet C, Messmer M, Schacehermayr G, Keller B (1995) Genetic and physical characterization of the Lr1 leaf rust resistance locus in wheat (Triticum aestivum L.). Mol Gen Genet 248:553–562
Hsam SLK, Lapochkina IF, Zeller FJ (2003) Chromosomal location of genes for resistance to powdery mildew in common wheat (Triticum aestivum L. em Thell.). 8. Gene Pm32 in a wheat-Aegilops speltoides translocation line. Euphytica 133:367–370
Hu XY, Ohm HW, Dweikat I (1997) Identification of RAPD markers linked to the gene Pm1 for resistance to powdery mildew in wheat. Theor Appl Genet 94:832–840
Ji JH, CAO AZ, Wang HY, Qin B, Wang SL, Kong F, Chen PD, Liu DJ, Wang XE (2007) Discrimination of Triticum aestivum-T. timopheevii introgression lines using PCR-based molecular markers. Hereditas (Beijing) (In Press) (In Chinese with English abstract)
Jorgensen JH, Jensen CJ (1972) Genes for resistance to wheat powdery mildew in derivatives of Triticum timopheevi and T carthlicum. Euphytica 21:121–128
Jorgensen JH, Jensen CJ (1973) Gene Pm6 for resistance to powdery mildew in wheat. Euphytica 22:423
Kameswara Rao K, Lakshminarasu M, Jena KK (2002) DNA markers and marker-assisted breeding for durable resistance to bacterial blight disease in rice. Biotechnol Adv 20:33–47
Kosambi DD (1944) The estimation of map distances from recombination values. Ann Eugen 12:172–175
Leath S, Heun M (1990) Identification of powdery mildew resistance genes in cultivars of soft red winter wheats. Plant Dis 74:747–752
Li Q, Wan JM (2005) SSRHunter. Development of a local searching software for SSR sites. Hereditas (Beijing) 27:808–810
Lincoln SE, Daly MJ, Lander ES (1993) Constructing linkage maps with MAPMAKER/Exp Version 3.0. A tutorial reference manual, 3rd edn. Whitehead Institute for Medical Research, Cambridge, MA
Ma ZQ, Wei JB, Cheng SH (2004) PCR-based markers for the powdery mildew resistance gene Pm4a in wheat. Theor Appl Genet 109:140–145
McIntosh RA, Yamazaki Y, Devos KM, Dubcovsky J, Rogers WJ, Appels R (2003) Catalogue of gene symbols for wheat. In: Pogna NE, Romano M, Pogna EA, Galterio G (eds) Proc 10th Int Wheat Genet Symp vol 4. Instituto Sperimentale per la Cerealcoltura, Rome
Miranda LM, Murphy JP, Marshall D, Leath S (2006) Pm34: a new powdery mildew resistance gene transferred from Aegilops tauschii Coss. to common wheat (Triticum aestivum L.). Theor Appl Genet 113:1497–1504
Naik S, Gill KS, Prakasa Rao VS, Gupta VS, Tamhankar SA, Pujar S, Gill BS, Ranjekar PK (1998) Identification of a STS marker linked to the Aegilops speltoides-derived leaf rust resistance gene Lr28 in wheat. Theor Appl Genet 97:535–540
Nyquist WE (1963) Inheritance of powdery mildew resistance in hybrids involving a common wheat strain derived from Triticum timopheevi. Crop Sci 3:40–43
Olson M, Hood L, Cantor C, Botstein D (1989) A common language for physical mapping of the human genome. Science 245:1434–1435
Park YJ, Dixit A, Yoo JW, Bennetzen J (2004) Further evidence of microcollinearity between barley and rice genomes at two orthologous regions. Mol Cells 17:492–502
Parker GD, Langridge P (2000) Development of a STS marker linked to a major locus controlling flour colour in wheat (Triticum aestivum L.). Mol Breed 6:169–174
Prins R, Groenewald JZ, Marais GF, Snape JW, Koebner RMD (2001) AFLP and STS tagging of Lr19, a gene conferring resistance to leaf rust in wheat. Theor Appl Genet 103:618–624
Qi LL, Wang SL, Chen PD, Liu DJ, Friebe B, Gill BS (1997) Molecular cytogenetic analysis of Leymus racemosus chromosomes added to wheat. Theor Appl Genet 95:1084–1091
Rostoks N, Park YJ, Ramakrishna W, Ma J, Druka A, Shiloff BA, SanMiguel PJ, Jiang Z, Brueggeman R, Sandhu D, Gill K, Bennetzen JL, Kleinhofs A (2002) Genomic sequencing reveal gene content, genomic organization, and recombination relationships in barley. Funct Integr Genomics 2:51–59
Seyfarth R, Feuillet C, Schachermayr G, Winzeler M, Keller B (1999) Development of a molecular marker for the adult plant leaf rust resistance gene Lr35 in wheat. Theor Appl Genet 99:554–560
Smith PH, Koebner RMD, Boyd LA (2002) The development of a STS marker linked to a yellow rust resistance derived from the wheat cultivar Moro. Theor Appl Genet 104:1278–1282
Talbert LE, Blake NK, Chee PW, Blake TK, Magyar GM (1994) Evaluation of “sequence-tagged-site” PCR products as molecular markers in wheat. Theor Appl Genet 87:789–794
Talbert LE, Bruckner PL, Smith LY, Sears R, Martin TJ (1996) Development of PCR markers linked to resistance to wheat streak mosaic virus in wheat. Theor Appl Genet 93:463–467
Tao WJ, Liu JY, Liu DJ, Chen PD (2000) Genetic mapping of the powdery mildew resistance gene Pm6 in wheat by RFLP analysis. Theor Appl Genet 100:564–568
Wang XY, Qi ZJ, Chen PD, Liu DJ (2000) Identification of RAPD markers tightly linked to wheat powdery mildew resistance gene Pm6. Acta Genetica Sin 27:1072–1079
Weeden NF, Tierman GM, Hemmat M, Kneen BE, Lodhi MA (1992) Inheritance and reliability of RAPD markers. In: Hoisington D, McNab A (eds) Proc symp on application of RAPD technology to plant breeding (Joint Plant Breeding Symp. Ser.), CSSA, ASHS, AGA, Minneapolis, pp 31–35
Zhu ZD, Zhou RH, Kong XY, Dong YC, Jia JZ (2005) Microsatellite markers linked to two genes conferring resistance to powdery mildew in common wheat introgressed from Triticum carthlicum acc. PS5 Genome 48:585–590
Acknowledgments
The project was supported by the Specialized Research Fund for the Doctoral Program of Higher Education (SRFDP) (20020307011), the Chinese High Tech Program of China (2006AA10Z1F6, 2006AA100101) and the Program for Changjiang Scholars and Innovative Research Teams in Universities (No. 10418). We would like to thank Dr. R. McIntosh and Dr. Yiqun Weng for their critical reading of the manuscript. We are also grateful to Prof. Yilin Zhou and Prof. Xiayu Duan, Plant Protection Institute, Chinese Academy of Agricultural Sciences, for kindly supplying the Blumeria graminis isolates.
Author information
Authors and Affiliations
Corresponding author
Additional information
Jianhui Ji and Bi Qin contributed equally to this work.
Rights and permissions
About this article
Cite this article
Ji, J., Qin, B., Wang, H. et al. STS markers for powdery mildew resistance gene Pm6 in wheat. Euphytica 163, 159–165 (2008). https://doi.org/10.1007/s10681-007-9578-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-007-9578-0