Abstract
It is now clear that the root microbiome, which consists of bacteria, archaea, and fungi that colonize both the rhizosphere and the internal space of the root, is one of the most complex ecosystems in nature and is very important for root and plant health and function.
In this chapter we have focused on the role of the root microbiome functional traits in improvement of nutrient acquisition and abiotic stress tolerance, with a focus on drought stress, the biocontrol of root and shoot plant diseases, and the role of root-associated microbes in both producing plant growth-promoting hormones and impacting the plant hormone metabolism and signaling pathways to alter root growth. Additionally, we have also endeavored to give the readers an introduction into the rapid advances in this field, from the metagenomic analyses that now have become relatively routine for the study of “what is there” in the root microbiome, regarding microbial composition, diversity, and abundance, to nascent studies beginning to study the plant and microbial molecular and physiological mechanisms and processes that underlie how the microbiome is assembled, and how the microbiome confers improved functional crop traits. Furthermore, given the incredible complexity of this ecosystem, we discuss the recent research involving systems biology analysis of the root microbiome, which will be critical in deciphering the trait–function links and interactions between roots and soil microbes. Finally, we also discuss the agricultural and genetic interventions that are being employed to modify the root microbiome via inoculation of the seed and plant with potentially beneficial soil microbes, as well as the studies looking at the role of plant genetic and molecular variation in impacting the composition and function of the microbiome.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
3.1 Introduction
Diverse microbial communities are an integral part of plant and animal life. A root–microbial ecosystem that consists of soil microbes that colonize and inhabit the soil at the root surface, the rhizosphere, and live and function in the root, comprise what plant biologists commonly refer to as the root microbiome.
The composition, diversity, and microbial species structure of the root microbiome can be plant species-specific and contain both beneficial and harmful microorganisms (Bressan et al. 2009; Lugtenberg and Kamilova 2009; Takeuchi et al. 1996). Root microbiome composition is shaped by the host plant and, often to a greater degree, the soil microflora, and many of the microorganisms in this biome are beneficial for host plant and root health and function. Research findings have shown that beneficial microbes in the root microbiome can increase the solubility and uptake of soil macro- and micronutrients such as phosphorus, nitrogen, and iron (Yadav et al. 2021).
Several soil-based abiotic stresses such as drought and salinity can impact the composition of the root microbiome, resulting in microbiome shifts that may confer increased tolerance to these stresses (Chen et al. 2017; Marasco et al. 2012; Vurukonda et al. 2016; Giri et al. 2018). Beneficial microorganisms in the root microbiome can also enhance plant growth and development by both synthesizing growth phytohormones and altering plant hormone metabolism required for plant growth and thereby increase root system growth and crop productivity (Arkhipova et al. 2005; Duca et al. 2018; Kudoyarova et al. 2014; Ping and Boland 2004; Prasad and Zhang 2022). On the other hand, there are microbial species in the root microbiome that are harmful and can negatively affect plant and human health such as disease-causing bacteria and fungi (Takeuchi et al. 1996). A saying that “nothing is free” also stands true for root and soil microbial interactions as plant roots and root exudates serve as a source of carbon and sugars that are required for the survival, growth, and replication of the microorganisms in the root microbiome (Bais et al. 2006; Foster 1986). Furthermore, specific root exudates play roles as signaling molecules between the root and soil microbes, influencing the microbial composition of the root microbiome. The root microbiome is one of the largest and most complex biomes in nature and plays significant roles in the maintenance and growth of plants.
The exponential growth of the human population and diminishing agricultural lands and input resources (e.g., water, fertilizer) required for efficient agricultural output, warrants novel and sustainable ways to achieve food security. Employment of genetically narrow crop germplasm (seeds or tissue specific to a species, genotype, or population maintained for plant breeding purposes) and intensive selection techniques in current breeding programs have enhanced the vulnerability to agricultural pests and diseases (Hammons 1976). Significant recent advances in molecular-based plant breeding include tools such as genomic selection, which has been shown to improve genetic gain via prediction of crop performance without phenotypic analysis of novel germplasm. Further improvements to genomic selection is enabling plant breeders to improve genetic gain while not losing genetic diversity (see, for example, Daetwyler et al. 2015). However powerful these advances in plant breeding are, they have yet to be intensively applied to the “hidden half” of the plant—the root, and especially the root microbiome.
It is important now to focus on the role of the root microbiome in functional plant traits to continue to improve crop resiliency and sustainability. For example, application of inorganic fertilizers and pesticides has significantly improved crop yields since the dawn of the Green Revolution. However, increasing pesticide and pathogen resistance, the increasing cost of fertilizers, and especially the finite availability of phosphorous fertilizer, as well as environmental degradation due to leaching of a significant portion of applied fertilizer into surface and ground waters, make these approaches unsustainable in the future (Denholm et al. 1998; Savci 2012). To address these complex problems, a systems approach integrating advances in precision fertilizer and water management with fundamental research innovations resulting in more nutrient and water-efficient crop varieties is needed (Macintosh et al. 2019). A critical component of the research aimed at improving crop nutrient and water acquisition will be the investigations that enhance our understanding of the development and function of the root microbiome. This will provide new strategies for improving crop yields and agricultural sustainability through modification of the root microbiome in part via use of biofertilizers that enhance the availability and acquisition of essential mineral nutrients and also through microbial-based biocontrol approaches to enhance disease and pest resistance.
It has been known for well over 100 years that certain root–microbe interactions, specifically N2-fixing bacteria in legume root nodules as well as arbuscular mycorrhizae (AM), play key roles in root N and P acquisition from the soil (Beijerinck 1901; Frank 1885; Hellriegel and Wilfarth 1888). In recent years with the realization that microbiomes associated with eukaryotic organisms play key roles in that organism’s health, well-being, and function, research on the root microbiome is clearly demonstrating that in addition to N2-fixing bacteria and AM, many other free-living or more intimately root-associated bacteria and fungi play roles in improving mineral nutrient availability in the rhizosphere and enhance biotic and abiotic stress tolerance. One example of this is microbial-mediated solubilization in the rhizosphere of sparingly soluble essential minerals such as P, Fe, and Zn (Fabiańska et al. 2019; Gururani et al. 2013; Harbort et al. 2020; Hiruma et al. 2016; Weiß et al. 2016). However, the molecular basis and physiological mechanisms underlying these improvements in plant function are still quite poorly understood. To effect changes in the root microbiome as a strategy to improve crop resiliency, improve yields using less inputs, and enhance agricultural sustainability, it is of great importance to understand the genetic and functional (molecular and physiological) mechanisms and regulation underlying the structure and function of root microbiomes. Quite a few studies have been successful in the identification and the use of plant growth-promoting bacteria from the soil and plant tissues to improve crop production (Ji et al. 2014; Kloepper et al. 1980; Li et al. 2021).
Certainly, the most significant advance in microbiome research over the past decade has been the metagenomic analysis of microbiomes involving isolation of microbial genomic DNA from bulk soil, the rhizosphere, and the root endosphere, followed by amplification of specific highly variable regions of bacterial and fungal genomes (16S rRNA for bacteria and the internal transcribed spacer [ITS]) region for fungi. For a review, see Sczyrba et al. 2017. Next-generation sequencing of these amplified bacterial and fungal gDNA regions and subsequent computational analysis of the sequence is used to identify microbial operational taxonomic units, microbial structure, and diversity. Furthermore, these types of studies have made it possible to begin to probe possible functions of the root microbiome community and to postulate about gene-function links that may be useful in identifying or designing specific root-associated microbes or communities of microbes that can be used in crop improvement (Chen et al. 2017; Naylor and Coleman-Derr 2018; Shulse et al. 2019; Xu et al. 2018, 2021). Comparatively recent advances in genome-resolved metagenomic and holo-omics approaches are finally enabling the researchers to identify the changes in host plant metabolomes and possibly important causal interactions between the root and microbes that could enhance nutrient uptake, microbial phytohormone production resulting in root growth promotion, and greater tolerance to abiotic and biotic stresses (Xu et al. 2021). In this chapter, we focus on possible advantages the microbiome could confer to the root systems in terms of acquiring resources for better plant growth, improving resistance to pathogens, and microbiome gene-functional links that have been and can be exploited further for these purposes.
3.2 Overview of the Root Microbiome
Dynamic and diverse groups of root-associated microorganisms, which include plant-beneficial microbes, are recruited by plant roots via signaling between the root and soil microbes (Hartman and Tringe 2019), to comprise the extremely complex microhabitat, that is, the root microbiome. This ecosystem can have impacts on plant health, growth, and function via direct or indirect pathways. The root microbiome consists of bacteria and archaea, algae, fungi, and protozoa, with bacteria being the most abundant component. The root microbiome can be quite diverse, and can consist of microbiota with as many as 33,000 different bacterial and archaea species (Mendes et al. 2011). Despite this amazing diversity, it has been reported that there are primarily two bacterial phyla, Actinobacteria and Proteobacteria, that dominate the global soil microbiome and root microbiome, for example, in disease suppressive soils (Delgado-Baquerizo et al. 2018; Mendes et al. 2011).
The root microbiome is actively recruited by plant roots and specific microbial communities are certainly influenced by the unique chemical composition of root exudates (Berendsen et al. 2012; Doornbos et al. 2012). Plants acquire the majority of their nutrients and water from the roots and, in return, low-molecular-weight organic compounds such as organic acids, sugars, phenolics, and amino acids are secreted as root exudates (Antoun 2013). Published research findings have shown that there can be an enrichment, for example, of unique organic acids in the root exudates in different plant species that can play an important role in shaping the root microbiome (Cotton et al. 2019; Hu et al. 2018; Huang et al. 2019; Tan et al. 2013; Wang et al. 2021). Kamilova et al. (2006) demonstrated that tomato root-tip-specific colonizing microbes had better growth and enrichment than other rhizobial microbes, when tomatoes were grown on minimal media with citrate, a major tomato root-tip organic exudate, as the primary carbon source. Furthermore, by introducing the stable carbon isotope, 13CO2, into the rhizosphere of wheat, maize, rape, and barrel clover plants, it was shown that there were differences in the sources of carbon released from roots and used by different groups of microbes in the rhizosphere (el Zahar Haichar et al. 2008).
Root exudate metabolites often play a beneficial role by modifying the rhizobiome, which in turn can alter plant hormonal content and function. For example, Hu et al. (2018) identified the secondary metabolite, benzoxazinoids, which is released by maize roots and can shape the rhizobiome community. In turn, this can favor plant defenses by increasing the production of the plant defense hormone, jasmonic acid, conferring protection against herbivores. Differences in the genotype of a plant species, which can significantly impact the composition of root exudates, are another factor that not surprisingly influences the microbial composition of the root microbiome. It has been reported that transgenic Arabidopsis thaliana plants expressing the sorghum CYP79A1 gene, resulted in the accumulation of high levels of a derivative of the sulfur secondary metabolite, glucosinolate, p-hydroxybenzylglucosinolate. This transgenic line exhibited significant alterations in the profile of the root exudation of glucosinolate compounds, which altered the microbial composition of root microbiome. This study showed that even small modifications in root metabolism can have significant effects on root exudates and the microbial composition of the root microbiome (Bressan et al. 2009).
There is a specific nomenclature used to define the microbiome on versus inside the root. Root microbes residing inside the root tissue are known as endophytes whereas the rhizomicrobiome is defined as microbes inhabiting the rhizosphere, the thin layer of soil intimately associated with the root surface (Bulgarelli et al. 2012; Edwards et al. 2015; Lundberg et al. 2012; Schlaeppi et al. 2014). Bulgarelli et al. (2012) demonstrated that the endosphere microbiome is distinct and does not have the same variation in microbial composition as does the rhizosphere, which is certainly more strongly influenced by the microbial composition of the bulk soil surrounding the root. Later, it was shown that the microbes residing on the surface of roots, sometimes termed the rhizoplane microbiome, are also distinct from the other two root microbiomes (the endomicrobiome and the rhizomicrobiome; Edwards et al. 2015).
3.3 Functional Traits to Enhance Plant Productivity
3.3.1 Biofertilizers that Impact Mineral Nutrient Availability and Acquisition by Roots
3.3.1.1 Nitrogen Fixation
Nitrogen is one of the most important mineral nutrients required for plant growth and can be a significant limiting factor to crop yields. Of course, nitrogen is essential for synthesis of amino acids, proteins, and enzymes that are prerequisite for many plant physiological processes (Novoa and Loomis 1981). Furthermore, nitrogen is the core component of chlorophyll that is required for photosynthesis, which provides the fixed carbon that underpins plant growth. Due to absence of large amounts of bioavailable nitrogen in the soil, most agricultural crop production relies upon the application of nitrogen fertilizers, usually as ammonia or urea. However, the increasing costs of nitrogen fertilizers and the significant environmental costs associated with the leaching of as much as 50–60% of N fertilizer before the plant roots can absorb it, often lead to nitrate pollution of ground waters and pose a greater risk to the environment (McCasland et al. 1985). Biological nitrogen fixation (BNF) is the process by which N2 gas in the atmosphere is converted to NH3 by nitrogenase enzyme activity in microbial diazotrophs, which are prokaryotes that have the ability to fix N2 gas to ammonia (Kim and Rees 1994). BNF has the potential to be a sustainable alternative to fulfill the nitrogen requirement of plants. However, only a few species of plants have the ability to be colonized by nitrogen-fixing bacteria as symbiotic microbes. There are two types of diazotrophs defined based on their habitat: (1) symbiotic N2 fixers which live within root nodules of primarily leguminous crop species, which include bacteria in the genera Rhizobium and Frankia; and (2) non-nodular diazotrophic bacteria that can establish either associative or free-living relationships with roots where the bacteria reside on or near the root surface (epiphytes). Other species of non-nodular N2-fixing bacteria form endophytic relationships with a wide range of non-legumes, where the bacteria colonize inner plant tissues and reside within root and even shoot tissues. These free-living diazotrophic bacteria include species within the genera Azospirillum, Azoarcus, and Herbaspirillum (Santi et al. 2013).
Most current root-nodulated crops obtain fixed nitrogen by the activity of a molybdenum (Mo)-dependent nitrogenase complex (Boyd et al. 2011; Rubio and Ludden 2008). It has been shown that only three genes—nifH, nifD, and nifK—are required to encode the structural subunits of nitrogenase enzyme (Seefeldt et al. 2009; Yang et al. 2018). Mo-dependent nitrogenase (Nif) complexes are a two-component enzyme system. The dinitrogenase reductase component is a homodimeric iron (Fe) protein encoded by the NifH gene that donates electrons, and a dinitrogenase or heterotetrameric Mo–Fe protein component encoded by NifDK that contains the Fe–Mo cofactor that serves as the substrate reduction site, accepting electrons from the Fe–S electron transfer protein, ferredoxin (Bulen and LeComte 1966). Apart from these two components, maturation of the nitrogenase enzyme complex for its activity also involves some regulatory proteins encoded by nifE, nifN, and nifB genes. Due to high similarity in 16S rRNA phylogeny and conserved nature of nif sequences, this similarity has been quite useful in identifying nitrogen-fixing bacteria from a soil sample. Quite a few studies have employed nifH as a phylogenetic marker to identify bacterial strains that make functional contributions to N2 fixation (Boyd et al. 2011; Bürgmann et al. 2004; Coelho et al. 2009; Seefeldt et al. 2009).
Our understanding of the mechanisms and regulation of nitrogenase N2 fixation and the nature of how and why these diazotrophs form these symbiotic relationships with the host plants open potential opportunities to enable non-leguminous crops to benefit from BNF. As cereals are the most widely grown food crops and are the source of the largest proportion of calories consumed by the human population, there has been considerable engineering biology research investigating and developing the tools to transfer nodule-based symbiotic nitrogen fixation to cereal crops (Burén et al. 2017; Ryu et al. 2020). This approach has been taken in the Burén et al. (2017) publication from the Voight Lab at MIT. The focus is to express the genes in the bacterial nitrogenase-dependent nitrogen-fixing pathway genes in the mitochondria or chloroplasts of eukaryotes, as these organelles have an ancient bacterial origin and thus are better suited for expression of bacterial genes than the nucleus of plant cells. In the Burén et al. (2017) publication, they have reengineered the 16 gene nitrogenase pathway from an N2-fixing bacterium to remove its native regulation and replaced it with well-understood synthetic genetic parts. They have ultimately been able to express an important and functional section of the bacterial N2-fixing pathway in the mitochondria eukaryotic model system, Saccharomyces cerevisiae, which is an important first step in generating a functional nitrogenase enzyme in plant cells.
A second approach to enhancing crop N nutrition via N2-fixing soil microbes involves research focusing on free-living N2-fixing bacteria. There are different approaches that are being explored for this and one of them is to transfer nitrogen-fixation ability from non-native diazotrophs to plant-host-colonizing rhizobacteria by introducing genomic islands that can encode the nitrogenase activity into free-living bacteria that readily colonize plant roots. Fox et al. (2016) demonstrated that transfer of X940 genomic island from Pseudomonas A1501 to the aerobic root-associated beneficial bacterium, Pseudomonas protegens Pf-5, followed by the inoculation of maize and wheat plants with this genetically modified bacterium, enabled the host plant’s root surface (rhizoplane) and rhizosphere to be colonized by Pf-5, providing enough radiolabeled fixed nitrogen to the roots to confer higher grain and biomass yields.
3.3.1.2 Phosphorus Bioavailability and Uptake
Phosphorus is the second most important mineral nutrient for the plant (after N), and can be a major limiting factor in plant growth as P deficiency is important to a wide range of plant processes, including cell division, root and shoot development, biomass production, photosynthesis, and reproduction (Hu and Schmidhalter 2005; López-Arredondo et al. 2014; Razaq et al. 2017). Even if soils are not deficient in total P, inorganic phosphate, which is the primary inorganic form of P absorbed by plant roots, will readily bind to the Fe and Al oxides/hydroxides on the surface of soil clay minerals, rendering them unavailable for root P absorption (López-Arredondo et al. 2014). Root P acquisition is relatively inefficient, and only approximately 20–25% of the applied P fertilizer is taken up by the plant during the first season after fertilizer application (Roberts and Johnston 2015). Much of the remaining phosphate fertilizer is either fixed to soil minerals, or is lost as runoff into surface waters, resulting in environmental damage including algal blooms in lakes and streams, which are costly to remediate.
The availability of inorganic phosphate to plant roots also is highly dependent on soil pH (Marschner 1995). In acidic soils (pH < 5.5), as mentioned above, the phosphate anion binds to aluminum and iron in clay minerals, whereas at higher soil pH values, insoluble calcium phosphate is formed that cannot be absorbed by roots (Hinsinger 2001). Currently, more than half of the arable lands worldwide consist of either acidic or alkaline soil and are deficient in inorganic phosphate (López-Arredondo et al. 2014). Soil microbes can solubilize the phosphate bound to Fe and Al in the soil, as they can release organic acids such as citric and malic acids which are strong chelators of Fe and Al, which then will release the bound phosphate into the soil solution for absorption by plant roots. Phosphorus also accumulates in soil in relatively unavailable organic forms, thus being tied up in soil organisms and plant litter. For example, phytic acid or phytate, an organic storage form of P, accumulates in soil and plants and some rhizobacteria can release the enzyme, phytase, which hydrolyzes phytate, releasing inorganic phosphate into the soil solution where it also can be absorbed by roots (Ke et al. 2021; López-Arredondo et al. 2014; Shulse et al. 2019). These P-solubilizing soil microbes open additional avenues of research that may enable the development of sustainable microbial-based strategies to improve phosphate availability, enhancing root P acquisition efficiency in agricultural crops. Biofertilization is likely not sufficient to meet the complete phosphorus requirement of crops. However, it clearly could be a strategy to significantly improve P bioavailability from applied phosphate fertilizer, increasing farmer’s yields per unit of P fertilizer applied, and reduce the environmental costs associated with remediating P pollution of waterways and ground water.
Research aimed at engineering rhizobacteria to increase P bioavailability was recently conducted by Shulse et al. (2019). In this study, they demonstrated that multiple rhizobacteria species (Pseudomonas and Ralstonia sp.), when transformed with phytase genes, significantly increased the release of soluble phosphate from soil phytate. In this study, a total of 6674 metagenomes were screened and 82 phylogenetically diverse phytase genes were selected and their sequences optimized for high gene expression in three species of rhizobacteria. The researchers identified 12 strains across three bacterial species that generated a significant increase in growth using Arabidopsis thaliana grown on phytate as the sole phosphate source. However, to take advantage of research findings such as the findings from Shulse et al. (2019) using genetically engineered soil microbes, it will be necessary to develop a more comprehensive regulatory framework to foster the more facile movement of inventions such as genetically modified soil microbes from the laboratory to the marketplace. This will likely require the development of real partnerships between academia, government, and industry to produce a science-based regulatory system which is predictable and straightforward to navigate.
In another study, to investigate the microbial community structure of the root microbiome, metagenomic analysis of the root microbiome of Lotus japonicus, a wild legume that has a history of reasonable growth and yield without phosphate fertilizer, was done. It was found that specific bacterial phyla, including Bacteroidetes, Betaproteobacteria, Chlorobi, Dehalococcoidetes, and Methanobacteria, were specifically abundant in the root microbiome (Unno and Shinano 2013). These rhizobacterial genera include bacterial strains that could enhance phytic acid utilization, promoting plant growth using phytate as a P source. Furthermore, several gene clusters possibly involved in phytic acid utilization, including alkaline phosphatase and citrate synthase, were investigated by Chhabra et al. (2013) who characterized the mineral phytate as well as phosphate solubilization trait employing functional metagenomics of the barley rhizosphere. They discovered that mineral phosphate solubilization screening of fosmid clones in E. coli identified genes/operons related to phosphorus mineralization and uptake. Further, Yadav et al. (2021) discovered that genes involved in gluconic acid synthesis are involved in phosphate solubilization by creating transposon insertion mutant libraries of Mesorhizobium ciceri Ca181, which is a symbiotic rhizobacteria colonized with chickpea roots. Clearly, recent research in this area has identified several rhizobacterial species that could play a role in root microbiome-induced enhanced solubilization of P for root uptake from either fixed inorganic phosphate in the soil or from organic P released from phytate.
3.3.1.3 Increasing Soil Iron Bioavailability via Bacterial Siderophores
Iron is an essential micronutrient required for several significant cellular processes including chlorophyll synthesis, photosynthesis, respiration, nitrogen fixation, and hormone production in plants (Vert et al. 2002). Iron is available in the soil as ferric (Fe3+) and ferrous (Fe2+) ions depending on soil pH, and soil aeration and redox potential. In aerobic soils, ferric iron predominates, and it is quite insoluble across most soil pH values, except in highly acidic soils. Hence, most of the soluble ferrous iron exists as ligands chelated by compounds such as organic acids, phenolics, humic acids, and bacterial siderophores (Guerinot and Yi 1994). Root microbes play an important role in soil iron solubility and uptake by releasing siderophores, which are low-molecular-weight organic Fe3+ chelators that form strong ligands via high-affinity binding with Fe3+ ions (Das et al. 2007). These Fe3+ siderophore complexes are taken up into bacterial cells by specific bacterial membrane transporters under iron-limiting conditions (Neilands 1984). The major classes of compounds that function as bacterial siderophores include catecholates (also known as phenolates), hydroxamates, and carboxylates (derivatives of citric acid).
Plants have also evolved their own mechanisms to acquire Fe3+ from the soil. In dicots and non-gramineous monocots, they employ a root cell plasma membrane ferric reductase to reduce extracellular chelated Fe(III) iron and the resulting Fe2+ ions are released by the ferric chelate and rapidly transported into the root cell by iron-regulated transporter 1 (IRT1), a root cell plasma membrane Fe2+ transporter (Guerinot and Yi 1994; Marschner 1995). Grass species use their own phytosiderophore process, releasing non-protein amino acids in response to Fe deficiency, which chelate ferric (Fe3+) ions in the soil and the Fe(III)-phytosiderophore complex is transported in toto across the root cell plasma membrane. Both types of plant Fe acquisition systems can absorb Fe chelated by bacterial siderophores. Using the reductase-based system, dicots and non-grass monocots can reduce the iron in the Fe(III)-siderophore complex at the root cell plasma membrane and then the released Fe2+ ion is transported into root cells via the IRT1 transporter. In grasses, it has been suggested that the uptake of Fe(III) complexes by bacterial siderophores can occur by Fe exchange between siderophores and phytosiderophores, or that the Fe-siderophore complex may be transported by the plant root plasma membrane transporter specifically functioning to absorb Fe(III)-phytosiderophore complexes (Bar-Ness et al. 1992; Crowley et al. 1988; Wang et al. 1993).
In addition to using bacterial siderophore release to enhance crop Fe acquisition and nutrition, the agronomic use of siderophore-producing plant-beneficial microbes can also be part of a strategy for biocontrol of plant pathogenic microbes. For example, the fluorescent Pseudomonads contain a number of plant-beneficial bacterial species. These fluorescent Pseudomonads release very high affinity Fe-binding siderophores which cannot generally be used by pathogenic bacteria. Hence, by decreasing the iron availability to phytopathogenic bacteria by chelating most of iron, these Pseudomonads promote plant growth by acting as a biocontrol agent against plant pathogens (Smarrelli and Castignetti 1986). Furthermore, fluorescent Pseudomonas sp. is one of initial bacterial strains explored that produced siderophores that have resulted in enhanced iron uptake in oat and mungbean (Crowley et al. 1988; Sharma et al. 2003). Sharma et al. (2003) showed that mungbean plants inoculated with a siderophore-producing Pseudomonas strain GRP3, exhibited reduced chlorosis and increased chlorophyll content under low Fe growth conditions. The rhizobacterial species, Rhizobium and Bradyrhizobuim, have also been reported to produce siderophores under iron-deficient conditions (Nambiar and Sivaramakrishnan 1987). Molecular microbiologists are dissecting the gene pathways involved in regulating siderophore synthesis and release in response to Fe-limiting conditions. One key regulator of these processes is the ferric uptake regulator, or Fur protein, that regulates siderophore production in variable iron conditions (Hassan and Troxell 2013). As the understanding of the regulation of these processes advances, more molecular tools are becoming available that will better facilitate the modification of rhizobacterial genomes, to enable agricultural researchers to improve the root microbiome to enhance crop Fe nutrition in Fe-limiting soils, and to provide microbial-based biocontrol against certain species of plant pathogenic bacteria.
3.3.2 Drought Tolerance
Drought stress is the most important abiotic stress limiting the yield of agricultural crops worldwide (Daryanto et al. 2016; Zipper et al. 2016). Additionally, climate change associated with global warming has exacerbated both the severity and frequency of droughts with more dire impacts on food crops, and increases the need for the identification of more sustainable approaches to improve crop performance under drought, mitigating drought-induced yield losses. Applications of beneficial rhizobacteria to both soils and via plant inoculation have been studied in various crop species to investigate the role of soil microbes and the root microbiome in enhanced resistance to drought stress, and have demonstrated some promising prospects for coping with drought (Chen et al. 2017; Marasco et al. 2012; Vurukonda et al. 2016). Beneficial rhizobacterial inoculants have been suggested to enhance performance under drought in different ways. First is production of enzymes that can catabolize stress-responsive phytohormones such as ACC deaminase enzyme that can degrade the ethylene precursor, ACC (1-aminocyclopropane-1-carboxylic acid), and thereby reduce ethylene production which at high levels inhibits root growth (Mayak et al. 2004). Another possible drought-resistance strategy involving beneficial rhizobacteria involves alterations in the levels of exopolysaccharides and the drought-associated amino acid, proline (Vardharajula et al. 2011). Proline acts as an osmolyte and when its concentration increases in the cell symplasm under drought, this creates a water potential gradient directed into the cell and allows the water to flow into the cell even though the water potential outside the cell has dropped due to drought. This confers the ability to increase the host plant’s drought tolerance by increasing cellular osmotic and thus water potential, thus maintaining cell water content under drought stress. Also, proline can play a role in protecting cell membranes and other cell components against damage caused by free radical production during drought. Exopolysaccharides produced by rhizobacteria during drought stress can enhance soil aggregation and structure and thereby retain soil water near the roots during drought stress (Khan et al. 2017; Naseem and Bano 2014; Yoshiba et al. 1997).
Additionally, inoculation of plants with certain rhizobacteria can alter hormone-mediated drought responses including changes in root system architecture via modulation of phytohormones to increase lateral root growth and biomass, thereby enhancing the host plant’s ability to acquire water from soils under drought (Arzanesh et al. 2011; Shakir et al. 2012; Zahir et al. 2008). Another example of this type of rhizobacterial drought response was a report about rhizobacterial-induced drought-resistance mechanisms that involved the production of volatile metabolites that led to a systemic response inducing stomatal closure and reduced water loss during drought stress. This systemic response appears to involve the volatile compound inducing interplay between ABA, jasmonic acid, and ethylene (Cho et al. 2008).
Although a few published studies have shown that colonization of roots with certain types of rhizobacteria resulted in improved plant performance under drought, the functional physiological and molecular mechanisms that drive the recruitment of a drought-responsive microbiome and the host plant responses remain poorly understood. Research based on 16S rRNA sequencing and metagenomic analysis has shed light on dynamic changes in the composition and function of the root microbiome under drought, by enrichment of bacterial phyla such as Actinobacteria, primarily in the root endosphere, but also in the rhizosphere (Edwards et al. 2015; Naylor and Coleman-Derr 2018; Naylor et al. 2017; Xu et al. 2018). In the Xu et al. (2018) study, a research approach integrating genome-resolved metagenomics, transcriptomics, and plant root metabolomics was conducted in sorghum during response to drought. The authors found that drought increased the abundance and activity of monoderm bacteria (Actinobacteria, Firmicutes, and Chloroflexi), which only have a single outer cell membrane and have thick cell walls. They also found that drought increased the production of certain metabolites in the root, including certain carbohydrates (especially glycerol-3-P) and amino acids, and that the monoderm bacteria showed a concomitant increase in the expression of transporters for these metabolites. Furthermore, inoculation of sorghum roots with monoderm isolates suggested that increased abundance of monoderm bacteria in the root microbiome increased plant growth during drought. These findings suggest that production of specific root metabolites by drought and their transfer to monoderm bacteria may play a role in reshaping the microbiome bacterial composition, to enhance plant growth under drought. These findings indicate that it may be possible to develop agricultural and/or genetic interventions that reshape the root microbiome to enhance crop performance under drought in agricultural crops.
3.3.3 Biocontrol of Plant Diseases
Crop diseases caused by pathogens (bacteria and fungi) can result in up to 30% yield loss in economically important crops (Soko et al. 2018; Tirnaz and Batley 2019). Hence, the impact of plant pathogens on crop production is a major challenge to global food security. For decades, chemical control measures have been applied to control plant diseases; however, the lack of rapid rates of breakdown of these chemicals into relatively safer constituents makes this approach detrimental to the environment (Gilden et al. 2010). Additionally, rapid trends in the evolution of pesticide resistance in plant pathogens have warranted the search for alternative and environmentally sustainable approaches, such as biocontrol (Lucas et al. 2015), which could be part of a broader strategy integrating biocontrol with plant genetics and breeding, better agronomic practices, and new classes of pesticides. Therefore, as the emphasis on environmentally friendly biocontrol strategies increases, over the past 20 years there have been an increasing number of publications on the use of beneficial bacteria to help control plant pathogens (Akgül and Mirik 2008; Girish and Umesha 2005; Mishra and Arora 2018; Sang et al. 2008). Even more recently, root and soil microbiome metagenomic studies have been published and these types of studies should ultimately increase our understanding of how the root microbiome operates as a system to protect especially against root diseases (Wille et al. 2019). For example, Lee et al. (2008) demonstrated the biocontrol of root diseases employing Bacillus subtilis extracted from rhizosphere soil to control Phytophthora blight of pepper caused by Phytophthora capsici, when seeds of pepper plants were inoculated with Bacillus subtilis before planting. There have been reports of several types of antibiotics produced by rhizobacteria that have been used to control plant root pathogens such as: pyrrolnitrin, phloroglucinols, phenazines, cyclic lipopeptides, and hydrogen cyanide (Haas and Défago 2005). Some of these antibiotics have been characterized functionally, including bacterial cyanide production and soil antifungal activity against root fungal pathogens that are both mediated by the GacA/GacS quorum sensing system in Pseudomonas sp. (Heeb and Haas 2001). The GacA/GacS two-component system is involved in the synthesis and release of secondary metabolites that have antifungal activity. The same GacA/GacS system is used by the beneficial rhizobacterium Pseudomonas fluorescens F113, and is responsible for HCN synthesis that enables P. fluorescens F113 to control the pathogenic fungal species, Pythium ultimum, which causes damping off disease in important agricultural crops (Aarons et al. 2000).
Another example of quorum sensing involved in production and release of antifungal compounds was shown in a study on the PhzR/PhzI system in the plant-beneficial bacterium, Pseudomonas chlororaphis 30-84. The PhzR/PhzI quorum sensing system regulates the synthesis and release of phenazine derivatives that are antibiotics with antifungal activity. The synthesis of phenazine is mediated by PhzR, whereas PhzI encodes for acyl-homoserine-lactone (AHL) synthase. AHL is the signal that activates PhzR to synthesize phenazine, which is then released into the soil. Using this two-component system, Pseudomonas chlororaphis 30-84 was effective at controlling the serious fungal disease, Take-all, in wheat (Chin-A-Woeng et al. 2003; Pierson and Pierson 1996; Zhang and Pierson 2001).
In other publications, Bacillus amyloliquefaciens SQR9 has been reported to control Fusarium wilt disease in cucumber, and Fusarium head blight in wheat has been shown to be controlled by use of Bacillus amyloliquefaciens AS 43.3 (Cao et al. 2011; Dunlap et al. 2013). It has been shown that regulation of bacterial chemotaxis can impact biocontrol effectiveness. For example, Weng et al. (2013) reported on a mutation in the AbrB gene in Bacillus amyloliquefaciens SQR9 that regulates chemotaxis and biofilm formation around root, which resulted in the enhancement of Bacillus amyloliquefaciens SQR9 chemotaxis and biofilm formation, thereby improving root colonization and biocontrol efficacy. Dunlap et al. (2013) showed that whole-genome sequencing and analysis of Bacillus amyloliquefaciens AS 43.3 identified a number of biosynthetic gene clusters (ribosomal and non-ribosomal) for synthesis of secondary metabolites that act as antibiotics such as surfactin, difficidin, and plantazolicin. Finally, recent advances in genome-resolved metagenomics have enabled researchers to identify novel soil bacteria that can be resources for genes involved in the synthesis of important secondary metabolites (Crits-Christoph et al. 2018). In Crits-Christoph et al. (2018), the authors reconstructed hundreds of near-complete genomes of bacteria isolated from a natural grassland soil ecosystem and identified members of understudied bacterial phyla such as Acidobacteria, Rokubacteria, Gemmatimonadetes, and Verrucomicrobia that contain novel gene clusters encoding important plant-beneficial bacterial biosynthetic pathways. In this study, they found members of these phyla contained genes that encode diverse polyketides and non-ribosomal peptides, and these classes of compounds include many antibiotics, antifungals, and siderophores that thus could be used as novel beneficial rhizobacteria.
3.3.4 Plant Hormone-Producing Bacteria
Optimal situation-dependent levels of plant hormones such as indole-3-acetic acid (IAA), gibberellic acid (GA), the direct precursor of the hormone ethylene, 1-aminocyclopropane-1-carboxylic acid (ACC) deaminase, ethylene, and cytokinin are required for adequate plant growth due to their significant roles in all the stages of plant growth development from embryogenesis to seed development (Chen et al. 2014; Davies 2010). Certain taxa of plant growth and performance promoting bacteria enhance root and/or shoot growth by inducing alterations in plant hormonal homeostasis that can lead to improved overall plant growth, root growth, abiotic stress tolerance, and, ultimately, improved yield (Duca et al. 2018; Miransari 2014; Vessey 2003). Although quite a few species of rhizobacteria have been reported to produce phytohormones, Pseudomonas and Bacillus spp. have been studied more extensively than other bacterial species with regard to microbial biosynthesis of plant hormones (Duca et al. 2018; Patten and Glick 2002b; Shilev 2013; Vessey 2003).
3.3.4.1 Indole-3-Acetic Acid (IAA)
Plant root exudates often contain tryptophan, a precursor in the biosynthesis of the predominant class of auxin compounds, indole-3-acetic acid (IAA) via the indolepyruvic acid pathway (Patten and Glick 2002a). Root soil colonization in tea (Camellia sinensis) with the IAA-producing rhizobacteria, Bacillus megaterium DE BARY TRS-4, resulted in significantly improved tea plant growth and decreased level of the fungal disease, brown root rot (Chakraborty et al. 2006). Gene sequences and genetic pathways for IAA biosynthesis have been identified in Pseudomonas sp. UW4 in Duan et al. (2013), and later this information was used by Duca et al. (2018) to transform Pseudomonas sp. UW4, overexpressing four native IAA biosynthesis genes: ami, nit, nthAB, and phe. Overexpression of all four genes individually in Pseudomonas sp. UW4, resulted in significant increases in bacterial IAA concentrations. Canola seed inoculation with one of the four bacterial overexpression lines and subsequent canola growth demonstrated that all four overexpression lines had greater root growth in plants 10 days post inoculation. The Pseudomonas sp. UW4 nit overexpression line had the greatest stimulation in root growth (Duca et al. 2018). The authors also measured the activity of ACC (1-aminocyclopropane-1-carboxylic acid) deaminase in the transgenic and wildtype bacterial lines and found significant decreases (30–70%) in enzyme activity in the overexpression lines. As this enzyme decreases the amount of ACC, the direct precursor to ethylene, they speculated that the root growth increase could be due to a direct effect of IAA or one of the other auxin compounds released from the transgenic overexpression lines colonizing canola roots, or reduction in root ACC and ethylene by the increased levels of ACC deaminase could also increase root growth.
Low levels of auxin are often associated with lower plant growth; however, excessively high auxin levels can affect shoot and root growth adversely (Thimann 1939). Therefore, there have been a number of studies on bacterial genes and genetic pathways that enhance degradation of IAA and their impact on plant growth (Costacurta and Vanderleyden 1995; Patten and Glick 1996; Spaepen et al. 2007). Leveau and Gerards (2008) identified and characterized a putative IAA degrading iac gene cluster by using the Pseudomonas putida 1290 strain originally isolated from pear tree foliage (Leveau and Lindow 2005). The iac gene cluster was shown to introduce IAA degradability in the P. putida KT2440 sp., which does not have the ability to degrade IAA, demonstrating the likeliness that some of the genes in this operon are involved in IAA catabolism (Leveau and Gerards 2008). Subsequently, Scott et al. (2013) conducted insertional inactivation of each of the genes in the iac cluster, and expression of the altered gene cluster in E. coli were combined with MS-based auxin metabolite analysis to demonstrate that iac-based degradation of IAA involved the first gene in the cluster, iacA, and transcript profiling of a knockout of another gene in the cluster, iacR, which encodes for a repressor of iacA expression, and this repression is overcome by exposure to IAA. As high levels of IAA also inhibit Pseudomonas putida 1290 growth as they can with regard to plant growth, the presence of this IAA degradation pathway in Pseudomonas putida might allow it to better colonize root tips where higher levels of root-synthesized IAA might occur.
3.3.4.2 Cytokinin
Cytokinin is an essential phytohormone of paramount importance for plant growth regulatory processes and cell division (Skoog and Armstrong 1970). Cytokinin production has been reported by quite a few rhizobacteria such as members of the genera Azospirillum, Rhizobium, Pseudomonas, and Bacillus (Cacciari et al. 1989; García de Salamone et al. 2001; Grover et al. 2021; Timmusk et al. 1999). It has been shown that inoculating plants with cytokinin-producing rhizobacteria can enhance plant growth and yield (Arkhipova et al. 2005; Kudoyarova et al. 2014; Ping and Boland 2004; Wang et al. 2018). Wang et al. (2018) showed that inoculation of Arabidopsis cytokinin receptor knockout mutants with cytokinin-producing Bacillus sp. LZR216 alters shoot growth and root system architecture, significantly stimulates lateral root number and length, and inhibits tap root length, while increases leaf surface area and shoot biomass. Bacillus sp. LZR216 treatment upregulated the gene expression of the Arabidopsis cytokinin signaling genes AHK3/AHK4, AHP1/AHP3, and ARR4/5/7/10/12/15 in root tips, suggesting that root-cytokinin-regulated genes may play a role in interactions and signaling between the root and cytokinin-producing bacteria such as Bacillus sp. LZR216.
3.3.4.3 ACC Deaminase Activity and Ethylene
Ethylene and its precursor 1-aminocyclopropane-1-carboxylate (ACC) play important roles not only in plant developmental but also in plant defense and symbiotic programs. Thus, it is likely that ethylene and ACC play a central role in the regulation of bacterial colonization and formation of root and shoot microbiomes. A number of rhizobacteria can produce significant amounts of the enzyme, 1-aminocyclopropane-1-carboxylate (ACC) deaminase, enabling rhizobacteria to degrade the ACC produced and exuded by plant roots and thereby reduce ethylene levels in roots (Glick et al. 1998; Jacobson et al. 1994). ACC is an immediate precursor to ethylene biosynthesis from methionine in plants (Adams and Yang 1979), and triggering of a surge in ethylene production can lead to root growth inhibition (Jackson 1991). Although the catalyzing function of ACC deaminase was first studied in free-living Pseudomonas spp., subsequently its activity was identified in many other bacteria including rhizobacteria from the genera Rhizobium, Bacillus pumilus, and Rhodococcus, and Burkholderia phytofirmans sp. Nov. (Glick 2005). The ACC deaminase gene, acdS, is well characterized in the beneficial rhizobacteria, Pseudomonas putida UW4, and it has been shown to be transcriptionally regulated by an acdR gene that encodes the leucine-responsive regulatory protein (Lrp). Lrp proteins have been found immediately upstream of many bacteria ACC deaminase genes, suggesting transcriptional regulation of these deaminase genes by Lrp’s is a key feature in the regulation of bacterial ACC deaminases. On the other hand, the nifA gene, which is a master regulator of nitrogen fixation, has also been identified as a regulator of an acdS gene in Mesorhizobium loti MAFF303099 (Kaneko et al. 2000). These results suggest evolution of parallel pathways of regulation of acdS gene in different classes of proteobacteria that form symbiotic relationships with plant roots.
ACC deaminases in rhizobacteria, which apparently enable the bacteria to modulate the levels of root ethylene in the host organ and increase or decrease root growth where the bacteria reside, likely indicate this enzyme plays an important role in root–rhizobacterial interactions. The ability to enzymatically modify root ethylene levels is also a potential tool for agricultural researchers attempting to modulate the composition and function of the root microbiome. But in order to effectively use this information to facilitate root-microbiome-mediated enhancement of crop functional traits, it will be necessary to more deeply and completely understand the relationship between ACC deaminase containing rhizobacteria and the plant root.
3.4 Conclusions
3.4.1 Genome-Level Investigations of the Root Microbiome and Holo-Omics Are Required to Fully Exploit Microbiome Functional Traits
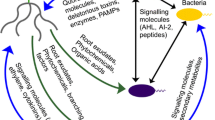
Plant growth and performance promoting microbes (PGPPM) in the root microbiome have the potential to be used as important alternative strategies to plant breeding or agronomic methods to enhance crop productivity, by enhancing nutrient acquisition, producing plant growth hormones, and enhancing disease and pest resistance. The genome of the root microbiome can be thought of as a second plant genome that needs to be decoded to provide the knowledge necessary to use natural and genetically modified root microbes to improve crop yields by enhancing root mineral nutrient and water acquisition, abiotic and biotic stress tolerance, and root system growth and vigor via altered plant hormone homeostasis (Fig. 3.1). Despite having great potential for the utilization of microbial functional traits, efficient root colonization with beneficial microbes is certainly one of the major challenges in enabling modification of root microbiomes to enhance crop functional traits. There have been major advances in this research area that ranges from the use of microbe-specific structural and functional markers to multi-omics approaches that combine two or more omics technologies such as metagenomics, transcriptomics, metabolomics, and proteomics to study the root microbiome at the genome level. However, functional studies of the root and its associated microbiome are still in their infancy, as scientists are developing and improving techniques to study the microbiome. As these types of studies advance and mature, they will enable the next steps, which will be to study the root microbiome as a functional ecosystem, using approaches such as deep-sequencing-based genome-resolved metagenomics, holo-omics, and microbiome/crop pan genome approaches. Using more systems-based approaches will be necessary to gain a deeper understanding of the detailed genetic, biochemical, and physiological networks underlying complex PGPPM-mediated processes and interactions of the root microbiome with the host plant roots and exudates, required for the microbial recruitment and root colonization resulting in enhancement of important crop functional traits. It is imperative that more of our efforts now focus on understanding the functional aspects of root–microbe interactions, which will be necessary to more effectively and efficiently use root microbiome modifications as a sustainable tool in our crop improvement toolbox to enhance plant resiliency and productivity, to help ensure global food security.
Sustainable improvement in crop productivity via root microbiome-related functional traits. Genomic DNA or RNA is extracted from the root microbiome for meta-genomic and meta-transcriptomic analyses. Plant metabolites are extracted from the root and/or collected as root exudates (very difficult to currently do for roots in soil) for metabolomic analysis. The holo-omics approaches involves combining and analyzing the different meta-omics data from root microbes, root tissue, and root exudates, which will be necessary for a more informative functional characterization of microbial plant performance promoting traits and to identify gene-functional links and networks. Functional information will be important to identify root beneficial microbes and to genetically manipulate naturally occurring bacteria that can be used as plant growth and performance promoting microbes (PGPPM). Inoculation with naturally occurring beneficial or genetically engineered rhizobacteria can possibly alter the root microbiome composition (if the introduced microbial species can compete and be established in this ecosystem), to increase plant productivity by different mechanisms such as biofertilization, abiotic and biotic stress tolerance, and plant hormone homeostasis. PGPPM can provide the major macronutrient, nitrogen, in the form of NH3 by fixing atmospheric N2 via nitrogenase in nodular and free-living root-associated N2 fixing microbes. Inorganic phosphates that tend to be fixed in the soil, reducing their availability to roots, can be solubilized by PGPPM to provide bioavailable phosphate to the roots. Micronutrient iron can be provided to plant roots by siderophores released by bacteria in the root microbiome. Exopolysaccharides produced by PGPPM and microbe-induced increases in proline in plants can act as soil aggregants (exopolysaccharides) and root and shoot osmolytes (proline) to contribute to drought stress resistance. Additionally, microbiome-induced changes in plant hormone metabolism can alter root system architecture via increases in the number and length of lateral roots to promote increases in water and mineral nutrient acquisition efficiency. Antibiotics secreted by the root microbiome can act as biocontrol agents for fungal and bacterial soil-borne diseases, and competition by PGPPM with pathogenic bacteria for carbon, Fe, and other nutrients essential for microbial growth can also reduce the abundance of harmful microbes via competition within the root microbiome. Various plant hormones such as cytokinin and IAA required for adequate plant growth can be synthesized by specific members of the root microbiome and enhance plant growth, whereas reduction in concentrations of root growth inhibiting hormones such as ethylene by rhizobacterial-produced ACC deaminase can also promote root growth and plant productivity
References
Aarons S, Abbas A, Adams C, Fenton A, O’Gara F (2000) A regulatory RNA (PrrB RNA) modulates expression of secondary metabolite genes in Pseudomonas fluorescens F113. J Bacteriol 182(14):3913–3919
Adams D, Yang SF (1979) Ethylene biosynthesis: identification of 1-aminocyclopropane-1-carboxylic acid as an intermediate in the conversion of methionine to ethylene. Proc Natl Acad Sci 76(1):170–174
Akgül D, Mirik M (2008) Biocontrol of Phytophthora capsici on pepper plants by Bacillus megaterium strains. J Plant Pathol 90:29–34
Antoun H (2013) Plant-growth-promoting rhizobacteria. Brenner’s encyclopedia of genetics, vol 5, 2nd edn. Elsevier, Amsterdam
Arkhipova T, Veselov S, Melentiev A, Martynenko E, Kudoyarova G (2005) Ability of bacterium Bacillus subtilis to produce cytokinins and to influence the growth and endogenous hormone content of lettuce plants. Plant Soil 272(1):201–209
Arzanesh MH, Alikhani H, Khavazi K, Rahimian H, Miransari M (2011) Wheat (Triticum aestivum L.) growth enhancement by Azospirillum sp. under drought stress. World J Microbiol Biotechnol 27(2):197–205
Bais HP, Weir TL, Perry LG, Gilroy S, Vivanco JM (2006) The role of root exudates in rhizosphere interactions with plants and other organisms. Annu Rev Plant Biol 57:233–266
Bar-Ness E, Hadar Y, Chen Y, Shanzer A, Libman J (1992) Iron uptake by plants from microbial siderophores: a study with 7-nitrobenz-2 oxa-1, 3-diazole-desferrioxamine as fluorescent ferrioxamine B analog. Plant Physiol 99(4):1329–1335
Beijerinck M (1901) Uber oligonitrophile mikroben. Zentralbl Bakterol Parasitenkd Infektionskr Hyg Abt II 7:561–582
Berendsen RL, Pieterse CM, Bakker PA (2012) The rhizosphere microbiome and plant health. Trends Plant Sci 17(8):478–486
Boyd E, Anbar A, Miller S, Hamilton T, Lavin M, Peters J (2011) A late methanogen origin for molybdenum-dependent nitrogenase. Geobiology 9(3):221–232
Bressan M, Roncato M-A, Bellvert F, Comte G, el Zahar Haichar F, Achouak W, Berge O (2009) Exogenous glucosinolate produced by Arabidopsis thaliana has an impact on microbes in the rhizosphere and plant roots. ISME J 3(11):1243–1257
Bulen W, LeComte J (1966) The nitrogenase system from Azotobacter: two-enzyme requirement for N2 reduction, ATP-dependent H2 evolution, and ATP hydrolysis. Proc Natl Acad Sci U S A 56(3):979
Bulgarelli D, Rott M, Schlaeppi K, van Themaat EVL, Ahmadinejad N, Assenza F, Rauf P, Huettel B, Reinhardt R, Schmelzer E (2012) Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488(7409):91–95
Burén S, Jiang X, López-Torrejón G, Echavarri-Erasun C, Rubio LM (2017) Purification and in vitro activity of mitochondria targeted nitrogenase cofactor maturase NifB. Front Plant Sci 8:1567
Bürgmann H, Widmer F, Von Sigler W, Zeyer J (2004) New molecular screening tools for analysis of free-living diazotrophs in soil. Appl Environ Microbiol 70(1):240–247
Cacciari I, Lippi D, Pietrosanti T, Pietrosanti W (1989) Phytohormone-like substances produced by single and mixed diazotrophic cultures of Azospirillum and Arthrobacter. Plant Soil 115(1):151–153
Cao Y, Zhang Z, Ling N, Yuan Y, Zheng X, Shen B, Shen Q (2011) Bacillus subtilis SQR 9 can control Fusarium wilt in cucumber by colonizing plant roots. Biol Fertil Soils 47(5):495–506
Chakraborty U, Chakraborty B, Basnet M (2006) Plant growth promotion and induction of resistance in Camellia sinensis by Bacillus megaterium. J Basic Microbiol 46(3):186–195
Chen J, Lausser A, Dresselhaus T (2014) Hormonal responses during early embryogenesis in maize. Portland Press, London
Chen C, Xin K, Liu H, Cheng J, Shen X, Wang Y, Zhang L (2017) Pantoea alhagi, a novel endophytic bacterium with ability to improve growth and drought tolerance in wheat. Sci Rep 7(1):1–14
Chhabra S, Brazil D, Morrissey J, Burke JI, O’Gara F, Dowling DN (2013) Characterization of mineral phosphate solubilization traits from a barley rhizosphere soil functional metagenome. Microbiology 2(5):717–724
Chin-A-Woeng TF, Bloemberg GV, Lugtenberg BJ (2003) Phenazines and their role in biocontrol by Pseudomonas bacteria. New Phytol 157(3):503–523
Cho SM, Kang BR, Han SH, Anderson AJ, Park J-Y, Lee Y-H, Cho BH, Yang K-Y, Ryu C-M, Kim YC (2008) 2R, 3R-butanediol, a bacterial volatile produced by Pseudomonas chlororaphis O6, is involved in induction of systemic tolerance to drought in Arabidopsis thaliana. Mol Plant-Microbe Interact 21(8):1067–1075
Coelho MR, Marriel IE, Jenkins SN, Lanyon CV, Seldin L, O’Donnell AG (2009) Molecular detection and quantification of nifH gene sequences in the rhizosphere of sorghum (Sorghum bicolor) sown with two levels of nitrogen fertilizer. Appl Soil Ecol 42(1):48–53
Costacurta A, Vanderleyden J (1995) Synthesis of phytohormones by plant-associated bacteria. Crit Rev Microbiol 21(1):1–18
Cotton TA, Pétriacq P, Cameron DD, Al Meselmani M, Schwarzenbacher R, Rolfe SA, Ton J (2019) Metabolic regulation of the maize rhizobiome by benzoxazinoids. ISME J 13(7):1647–1658
Crits-Christoph A, Diamond S, Butterfield CN, Thomas BC, Banfield JF (2018) Novel soil bacteria possess diverse genes for secondary metabolite biosynthesis. Nature 558(7710):440–444
Crowley DE, Reid CP, Szaniszlo PJ (1988) Utilization of microbial siderophores in iron acquisition by oat. Plant Physiol 87(3):680–685
Daetwyler HD, Hayden MJ, Spangenberg GC, Hayes BJ (2015) Selection on optimal haploid value increases genetic gain and preserves more genetic diversity relative to genomic selection. Genetics 200(4):1341–1348
Daryanto S, Wang L, Jacinthe P-A (2016) Global synthesis of drought effects on maize and wheat production. PLoS One 11(5):e0156362
Das A, Prasad R, Srivastava A, Giang PH, Bhatnagar K, Varma A (2007) Fungal siderophores: structure, functions and regulations. In: Varma A, Chincholkar SB (eds) Microbial siderophores. Springer-Verlag, Berlin, pp 1–42
Davies PJ (2010) The plant hormones: their nature, occurrence, and functions. In: Plant hormones. Springer, New York, pp 1–15
Delgado-Baquerizo M, Oliverio AM, Brewer TE, Benavent-González A, Eldridge DJ, Bardgett RD, Maestre FT, Singh BK, Fierer N (2018) A global atlas of the dominant bacteria found in soil. Science 359(6373):320–325
Denholm I, Cahill M, Dennehy T, Horowitz A (1998) Challenges with managing insecticide resistance in agricultural pests, exemplisfied by the whitefly Bemisia tabaci. Philos Trans R Soc Lond B Biol Sci 353(1376):1757–1767
Doornbos RF, van Loon LC, Bakker PA (2012) Impact of root exudates and plant defense signaling on bacterial communities in the rhizosphere. A review. Agron Sustain Dev 32(1):227–243
Duan J, Jiang W, Cheng Z, Heikkila JJ, Glick BR (2013) The complete genome sequence of the plant growth-promoting bacterium Pseudomonas sp. UW4. PLoS One 8(3):e58640
Duca DR, Rose DR, Glick BR (2018) Indole acetic acid overproduction transformants of the rhizobacterium Pseudomonas sp. UW4. Antonie Van Leeuwenhoek 111(9):1645–1660
Dunlap CA, Bowman MJ, Schisler DA (2013) Genomic analysis and secondary metabolite production in Bacillus amyloliquefaciens AS 43.3: a biocontrol antagonist of Fusarium head blight. Biol Control 64(2):166–175
Edwards J, Johnson C, Santos-Medellín C, Lurie E, Podishetty NK, Bhatnagar S, Eisen JA, Sundaresan V (2015) Structure, variation, and assembly of the root-associated microbiomes of rice. Proc Natl Acad Sci 112(8):E911–E920
el Zahar Haichar F, Marol C, Berge O, Rangel-Castro JI, Prosser JI, Balesdent J, Heulin T, Achouak W (2008) Plant host habitat and root exudates shape soil bacterial community structure. ISME J 2(12):1221–1230
Fabiańska I, Gerlach N, Almario J, Bucher M (2019) Plant-mediated effects of soil phosphorus on the root-associated fungal microbiota in Arabidopsis thaliana. New Phytol 221(4):2123–2137
Foster R (1986) The ultrastructure of the rhizoplane and rhizosphere. Annu Rev Phytopathol 24(1):211–234
Fox AR, Soto G, Valverde C, Russo D, Lagares A Jr, Zorreguieta Á, Alleva K, Pascuan C, Frare R, Mercado-Blanco J (2016) Major cereal crops benefit from biological nitrogen fixation when inoculated with the nitrogen-fixing bacterium Pseudomonas protegens Pf-5 X940. Environ Microbiol 18(10):3522–3534
Frank B (1885) Über die auf Wurzelsymbiose beruhende Ernährung gewisser Bäume durch unterirdische Pilze. Plant Biol 3(4):128–145
García de Salamone IE, Hynes RK, Nelson LM (2001) Cytokinin production by plant growth promoting rhizobacteria and selected mutants. Can J Microbiol 47(5):404–411
Gilden RC, Huffling K, Sattler B (2010) Pesticides and health risks. J Obstet Gynecol Neonatal Nurs 39(1):103–110
Giri B, Prasad R, Varma A (2018) Root biology. Springer International Publishing. ISBN 978-3-319-75910-4. https://www.springer.com/us/book/9783319759098
Girish N, Umesha S (2005) Effect of plant growth promoting rhizobacteria on bacterial canker of tomato. Arch Phytopathol Plant Protect 38(3):235–243
Glick BR (2005) Modulation of plant ethylene levels by the bacterial enzyme ACC deaminase. FEMS Microbiol Lett 251(1):1–7
Glick BR, Penrose DM, Li J (1998) A model for the lowering of plant ethylene concentrations by plant growth-promoting bacteria. J Theor Biol 190(1):63–68
Grover M, Bodhankar S, Sharma A, Sharma P, Singh J, Nain L (2021) PGPR mediated alterations in root traits: way toward sustainable crop production. Front Sustain Food Syst 4:287
Guerinot ML, Yi Y (1994) Iron: nutritious, noxious, and not readily available. Plant Physiol 104(3):815
Gururani MA, Upadhyaya CP, Baskar V, Venkatesh J, Nookaraju A, Park SW (2013) Plant growth-promoting rhizobacteria enhance abiotic stress tolerance in Solanum tuberosum through inducing changes in the expression of ROS-scavenging enzymes and improved photosynthetic performance. J Plant Growth Regul 32(2):245–258
Haas D, Défago G (2005) Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat Rev Microbiol 3(4):307–319
Hammons RO (1976) Peanuts: genetic vulnerability and breeding strategy 1. Crop Sci 16(4):527–530
Harbort CJ, Hashimoto M, Inoue H, Niu Y, Guan R, Rombolà AD, Kopriva S, Voges MJ, Sattely ES, Garrido-Oter R (2020) Root-secreted coumarins and the microbiota interact to improve iron nutrition in Arabidopsis. Cell Host Microbe 28(6):825–837.e826
Hartman K, Tringe SG (2019) Interactions between plants and soil shaping the root microbiome under abiotic stress. Biochem J 476(19):2705–2724
Hassan HM, Troxell B (2013) Transcriptional regulation by ferric uptake regulator (FUR) in pathogenic bacteria. Front Cell Infect Microbiol 3:59
Heeb S, Haas D (2001) Regulatory roles of the GacS/GacA two-component system in plant-associated and other gram-negative bacteria. Mol Plant-Microbe Interact 14(12):1351–1363
Hellriegel H, Wilfarth H (1888) Untersuchungen uber die Stickstoffnahrung der Gramineen und Leguminosen. Buchdruckerei der "Post" Kayssler, Berlin
Hinsinger P (2001) Bioavailability of soil inorganic P in the rhizosphere as affected by root-induced chemical changes: a review. Plant Soil 237(2):173–195
Hiruma K, Gerlach N, Sacristán S, Nakano RT, Hacquard S, Kracher B, Neumann U, Ramírez D, Bucher M, O’Connell RJ (2016) Root endophyte Colletotrichum tofieldiae confers plant fitness benefits that are phosphate status dependent. Cell 165(2):464–474
Hu Y, Schmidhalter U (2005) Drought and salinity: a comparison of their effects on mineral nutrition of plants. J Plant Nutr Soil Sci 168(4):541–549
Hu L, Robert CA, Cadot S, Zhang X, Ye M, Li B, Manzo D, Chervet N, Steinger T, Van Der Heijden MG (2018) Root exudate metabolites drive plant-soil feedbacks on growth and defense by shaping the rhizosphere microbiota. Nat Commun 9(1):1–13
Huang AC, Jiang T, Liu Y-X, Bai Y-C, Reed J, Qu B, Goossens A, Nützmann H-W, Bai Y, Osbourn A (2019) A specialized metabolic network selectively modulates Arabidopsis root microbiota. Science 364(6440):eaau6389
Jackson M (1991) Ethylene in root growth and development. In: Matoo AK, Suttle JC (eds) The plant hormone ethylene. CRC Press, Boca Raton, FL
Jacobson C, Pasternak J, Glick B (1994) Partial purification and characterization of ACC deaminase from the plant growth-promoting rhizobacterium Pseudomonas putida GR12-2. Can J Microbiol 40(1):19–1025
Ji SH, Gururani MA, Chun S-C (2014) Isolation and characterization of plant growth promoting endophytic diazotrophic bacteria from Korean rice cultivars. Microbiol Res 169(1):83–98
Kamilova F, Kravchenko LV, Shaposhnikov AI, Azarova T, Makarova N, Lugtenberg B (2006) Organic acids, sugars, and L-tryptophane in exudates of vegetables growing on stonewool and their effects on activities of rhizosphere bacteria. Mol Plant-Microbe Interact 19(3):250–256
Kaneko T, Nakamura Y, Sato S, Asamizu E, Kato T, Sasamoto S, Watanabe A, Idesawa K, Ishikawa A, Kawashima K (2000) Complete genome structure of the nitrogen-fixing symbiotic bacterium Mesorhizobium loti. DNA Res 7(6):331–338
Ke J, Wang B, Yoshikuni Y (2021) Microbiome engineering: synthetic biology of plant-associated microbiomes in sustainable agriculture. Trends Biotechnol 39(3):244–261
Khan N, Bano A, Babar M (2017) The root growth of wheat plants, the water conservation and fertility status of sandy soils influenced by plant growth promoting rhizobacteria. Symbiosis 72(3):195–205
Kim J, Rees DC (1994) Nitrogenase and biological nitrogen fixation. Biochemistry 33(2):389–397
Kloepper JW, Leong J, Teintze M, Schroth MN (1980) Enhanced plant growth by siderophores produced by plant growth-promoting rhizobacteria. Nature 286(5776):885–886
Kudoyarova GR, Melentiev AI, Martynenko EV, Timergalina LN, Arkhipova TN, Shendel GV, Kuz'mina LY, Dodd IC, Veselov SY (2014) Cytokinin producing bacteria stimulate amino acid deposition by wheat roots. Plant Physiol Biochem 83:285–291
Lee KJ, Kamala-Kannan S, Sub HS, Seong CK, Lee GW (2008) Biological control of Phytophthora blight in red pepper (Capsicum annuum L.) using Bacillus subtilis. World J Microbiol Biotechnol 24(7):1139–1145
Leveau JH, Gerards S (2008) Discovery of a bacterial gene cluster for catabolism of the plant hormone indole 3-acetic acid. FEMS Microbiol Ecol 65(2):238–250
Leveau JH, Lindow SE (2005) Utilization of the plant hormone indole-3-acetic acid for growth by Pseudomonas putida strain 1290. Appl Environ Microbiol 71(5):2365–2371
Li M, Wang J, Yao T, Wang Z, Zhang H, Li C (2021) Isolation and characterization of cold-adapted PGPB and their effect on plant growth promotion. J Microbiol Biotechnol 31(9):1218–1230
López-Arredondo DL, Leyva-González MA, González-Morales SI, López-Bucio J, Herrera-Estrella L (2014) Phosphate nutrition: improving low-phosphate tolerance in crops. Annu Rev Plant Biol 65:95–123
Lucas JA, Hawkins NJ, Fraaije BA (2015) The evolution of fungicide resistance. Adv Appl Microbiol 90:29–92
Lugtenberg B, Kamilova F (2009) Plant-growth-promoting rhizobacteria. Annu Rev Microbiol 63:541–556
Lundberg DS, Lebeis SL, Paredes SH, Yourstone S, Gehring J, Malfatti S, Tremblay J, Engelbrektson A, Kunin V, Del Rio TG (2012) Defining the core Arabidopsis thaliana root microbiome. Nature 488(7409):86–90
Macintosh KA, Doody DG, Withers PJ, McDowell RW, Smith DR, Johnson LT, Bruulsema TW, O’Flaherty V, McGrath JW (2019) Transforming soil phosphorus fertility management strategies to support the delivery of multiple ecosystem services from agricultural systems. Sci Total Environ 649:90–98
Marasco R, Rolli E, Ettoumi B, Vigani G, Mapelli F, Borin S, Abou-Hadid AF, El-Behairy UA, Sorlini C, Cherif A (2012) A drought resistance-promoting microbiome is selected by root system under desert farming. PLoS One 7(10):e48479
Marschner H (1995) Adaptation of plants to adverse chemical soil conditions. In: Mineral nutrition of higher plants. Academic, New York
Mayak S, Tirosh T, Glick BR (2004) Plant growth-promoting bacteria that confer resistance to water stress in tomatoes and peppers. Plant Sci 166(2):525–530
McCasland M, Trautmann NM, Wagenet RJ (1985) Nitrate: health effects in drinking water. Cornell Cooperative Extension, Ithaca, NY
Mendes R, Kruijt M, De Bruijn I, Dekkers E, van der Voort M, Schneider JH, Piceno YM, DeSantis TZ, Andersen GL, Bakker PA (2011) Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332(6033):1097–1100
Miransari M (2014) Plant growth promoting rhizobacteria. J Plant Nutr 37(14):2227–2235
Mishra J, Arora NK (2018) Secondary metabolites of fluorescent pseudomonads in biocontrol of phytopathogens for sustainable agriculture. Appl Soil Ecol 125:35–45
Nambiar P, Sivaramakrishnan S (1987) Detection and assay of siderophores in cowpea rhizobia (Bradyrhizobium) using radioactive Fe (59Fe). Lett Appl Microbiol 4(2):37–40
Naseem H, Bano A (2014) Role of plant growth-promoting rhizobacteria and their exopolysaccharide in drought tolerance of maize. J Plant Interact 9(1):689–701
Naylor D, Coleman-Derr D (2018) Drought stress and root-associated bacterial communities. Front Plant Sci 8:2223
Naylor D, DeGraaf S, Purdom E, Coleman-Derr D (2017) Drought and host selection influence bacterial community dynamics in the grass root microbiome. ISME J 11(12):2691–2704
Neilands JB (1984) Methodology of siderophores. In: Siderophores from microorganisms and plants. structure and bonding, vol 58. Springer, Berlin, Heidelberg. https://doi.org/10.1007/BFb0111309
Novoa R, Loomis R (1981) Nitrogen and plant production. Plant Soil 58(1):177–204
Patten CL, Glick BR (1996) Bacterial biosynthesis of indole-3-acetic acid. Can J Microbiol 42(3):207–220
Patten CL, Glick BR (2002a) Regulation of indoleacetic acid production in Pseudomonas putida GR12-2 by tryptophan and the stationary-phase sigma factor RpoS. Can J Microbiol 48(7):635–642
Patten CL, Glick BR (2002b) Role of Pseudomonas putida indoleacetic acid in development of the host plant root system. Appl Environ Microbiol 68(8):3795–3801
Pierson LS, Pierson EA (1996) Phenazine antibiotic production in Pseudomonas aureofaciens: role in rhizosphere ecology and pathogen suppression. FEMS Microbiol Lett 136(2):101–108
Ping L, Boland W (2004) Signals from the underground: bacterial volatiles promote growth in Arabidopsis. Trends Plant Sci 9(6):263–266
Prasad R, Zhang S-H (2022) Benefical microorganisms in agriculture. Springer, Singapore. https://springerlink.bibliotecabuap.elogim.com/book/9789811907326
Razaq M, Zhang P, Shen H-l (2017) Influence of nitrogen and phosphorous on the growth and root morphology of Acer mono. PLoS One 12(2):e0171321
Roberts TL, Johnston AE (2015) Phosphorus use efficiency and management in agriculture. Resour Conserv Recycl 105:275–281
Rubio LM, Ludden PW (2008) Biosynthesis of the iron-molybdenum cofactor of nitrogenase. Annu Rev Microbiol 62:93–111. https://doi.org/10.1146/annurev.micro.62.081307.162737
Ryu M-H, Zhang J, Toth T, Khokhani D, Geddes BA, Mus F, Garcia-Costas A, Peters JW, Poole PS, Ané J-M (2020) Control of nitrogen fixation in bacteria that associate with cereals. Nat Microbiol 5(2):314–330
Sang MK, Chun S-C, Kim KD (2008) Biological control of Phytophthora blight of pepper by antagonistic rhizobacteria selected from a sequential screening procedure. Biol Control 46(3):424–433
Santi C, Bogusz D, Franche C (2013) Biological nitrogen fixation in non-legume plants. Ann Bot 111(5):743–767
Savci S (2012) An agricultural pollutant: chemical fertilizer. Int J Environ Sci Dev 3(1):73
Schlaeppi K, Dombrowski N, Oter RG, van Themaat EVL, Schulze-Lefert P (2014) Quantitative divergence of the bacterial root microbiota in Arabidopsis thaliana relatives. Proc Natl Acad Sci 111(2):585–592
Scott JC, Greenhut IV, Leveau JH (2013) Functional characterization of the bacterial iac genes for degradation of the plant hormone indole-3-acetic acid. J Chem Ecol 39(7):942–951
Sczyrba A, Hofmann P, Belmann P, Koslicki D, Janssen S, Dröge J, Gregor I, Majda S, Fiedler J, Dahms E (2017) Critical assessment of metagenome interpretation—a benchmark of metagenomics software. Nat Methods 14(11):1063–1071
Seefeldt LC, Hoffman BM, Dean DR (2009) Mechanism of Mo-dependent nitrogenase. Annu Rev Biochem 78:701–722
Shakir MA, Asghari B, Muhammad A (2012) Rhizosphere bacteria containing ACC-deaminase conferred drought tolerance in wheat grown under semi-arid climate. Soil Environ 31(1):108–112
Sharma A, Johri B, Sharma A, Glick B (2003) Plant growth-promoting bacterium Pseudomonas sp. strain GRP3 influences iron acquisition in mung bean (Vigna radiata L. Wilzeck). Soil Biol Biochem 35(7):887–894
Shilev S (2013) Soil rhizobacteria regulating the uptake of nutrients and undesirable elements by plants. In: Plant microbe symbiosis: fundamentals and advances. Springer, New York, pp 147–167
Shulse CN, Chovatia M, Agosto C, Wang G, Hamilton M, Deutsch S, Yoshikuni Y, Blow MJ (2019) Engineered root bacteria release plant-available phosphate from phytate. Appl Environ Microbiol 85(18):e01210–e01219
Skoog F, Armstrong DJ (1970) Cytokinins. Ann Rev Plant Physiol 21(1):359–384
Smarrelli J Jr, Castignetti D (1986) Iron acquisition by plants: the reduction of ferrisiderophores by higher plant NADH: nitrate reductase. Biochim Biophys Acta 882(3):337–342
Soko T, Bender CM, Prins R, Pretorius ZA (2018) Yield loss associated with different levels of stem rust resistance in bread wheat. Plant Dis 102(12):2531–2538
Spaepen S, Vanderleyden J, Remans R (2007) Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol Rev 31(4):425–448
Takeuchi T, Sawada H, Tanaka F, Matsuda I (1996) Phylogenetic analysis of Streptomyces spp. causing potato scab based on 16S rRNA sequences. Int J Syst Evol Microbiol 46(2):476–479
Tan S, Yang C, Mei X, Shen S, Raza W, Shen Q, Xu Y (2013) The effect of organic acids from tomato root exudates on rhizosphere colonization of Bacillus amyloliquefaciens T-5. Appl Soil Ecol 64:15–22
Thimann KV (1939) Auxins and the inhibition of plant growth. Biol Rev 14(3):314–337
Timmusk S, Nicander B, Granhall U, Tillberg E (1999) Cytokinin production by Paenibacillus polymyxa. Soil Biol Biochem 31(13):1847–1852
Tirnaz S, Batley J (2019) DNA methylation: toward crop disease resistance improvement. Trends Plant Sci 24(12):1137–1150
Unno Y, Shinano T (2013) Metagenomic analysis of the rhizosphere soil microbial community. Mol Micro Ecol Rhizosp 1:1099–1103
Vardharajula S, Zulfikar Ali S, Grover M, Reddy G, Bandi V (2011) Drought-tolerant plant growth promoting Bacillus spp.: effect on growth, osmolytes, and antioxidant status of maize under drought stress. J Plant Interact 6(1):1–14
Vert G, Grotz N, Dédaldéchamp F, Gaymard F, Guerinot ML, Briat J-F, Curie C (2002) IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 14(6):1223–1233
Vessey JK (2003) Plant growth promoting rhizobacteria as biofertilizers. Plant Soil 255(2):571–586
Vurukonda SSKP, Vardharajula S, Shrivastava M, SkZ A (2016) Enhancement of drought stress tolerance in crops by plant growth promoting rhizobacteria. Microbiol Res 184:13–24
Wang Y, Brown H, Crowley D, Szaniszlo P (1993) Evidence for direct utilization of a siderophore, ferrioxamine B, in axenically grown cucumber. Plant Cell Environ 16(5):579–585
Wang J, Zhang Y, Jin J, Li Q, Zhao C, Nan W, Wang X, Ma R, Bi Y (2018) An intact cytokinin-signaling pathway is required for Bacillus sp. LZR216-promoted plant growth and root system architecture altereation in Arabidopsis thaliana seedlings. Plant Growth Regul 84(3):507–518
Wang P, Chai YN, Roston R, Dayan FE, Schachtman DP (2021) The Sorghum bicolor root exudate Sorgoleone shapes bacterial communities and delays network formation. mSystems 6(2):e00749-20
Weiß M, Waller F, Zuccaro A, Selosse MA (2016) Sebacinales–one thousand and one interactions with land plants. New Phytol 211(1):20–40
Weng J, Wang Y, Li J, Shen Q, Zhang R (2013) Enhanced root colonization and biocontrol activity of Bacillus amyloliquefaciens SQR9 by abrB gene disruption. Appl Microbiol Biotechnol 97(19):8823–8830
Wille L, Messmer MM, Studer B, Hohmann P (2019) Insights to plant–microbe interactions provide opportunities to improve resistance breeding against root diseases in grain legumes. Plant Cell Environ 42(1):20–40
Xu L, Naylor D, Dong Z, Simmons T, Pierroz G, Hixson KK, Kim Y-M, Zink EM, Engbrecht KM, Wang Y (2018) Drought delays development of the sorghum root microbiome and enriches for monoderm bacteria. Proc Natl Acad Sci 115(18):E4284–E4293
Xu L, Pierroz G, Wipf HM-L, Gao C, Taylor JW, Lemaux PG, Coleman-Derr D (2021) Holo-omics for deciphering plant-microbiome interactions. Microbiome 9(1):1–11
Yadav A, Singh RP, Singh AL, Singh M (2021) Identification of genes involved in phosphate solubilization and drought stress tolerance in chickpea symbiont Mesorhizobium ciceri Ca181. Arch Microbiol 203(3):1167–1174
Yang J, Xie X, Xiang N, Tian Z-X, Dixon R, Wang Y-P (2018) Polyprotein strategy for stoichiometric assembly of nitrogen fixation components for synthetic biology. Proc Natl Acad Sci 115(36):E8509–E8517
Yoshiba Y, Kiyosue T, Nakashima K, Yamaguchi-Shinozaki K, Shinozaki K (1997) Regulation of levels of proline as an osmolyte in plants under water stress. Plant Cell Physiol 38(10):1095–1102
Zahir Z, Munir A, Asghar H, Shaharoona B, Arshad M (2008) Effectiveness of rhizobacteria containing ACC deaminase for growth promotion of peas (Pisum sativum) under drought conditions. J Microbiol Biotechnol 18(5):958–963
Zhang Z, Pierson LS (2001) A second quorum-sensing system regulates cell surface properties but not phenazine antibiotic production in Pseudomonas aureofaciens. Appl Environ Microbiol 67(9):4305–4315
Zipper SC, Qiu J, Kucharik CJ (2016) Drought effects on US maize and soybean production: spatiotemporal patterns and historical changes. Environ Res Lett 11(9):094021
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Chandnani, R., Kochian, L.V. (2023). The Role of the Root Microbiome in the Utilization of Functional Traits for Increasing Plant Productivity. In: Chhabra, S., Prasad, R., Maddela, N.R., Tuteja, N. (eds) Plant Microbiome for Plant Productivity and Sustainable Agriculture . Microorganisms for Sustainability, vol 37. Springer, Singapore. https://doi.org/10.1007/978-981-19-5029-2_3
Download citation
DOI: https://doi.org/10.1007/978-981-19-5029-2_3
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-19-5028-5
Online ISBN: 978-981-19-5029-2
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)