Abstract
Bacteriophages are unique organisms with diverse genomic orientation and enriched with beneficial characteristics. Phage therapy was initially employed nearly a century ago, though it was undermined and neglected after the efficacious insertion of antibiotics. The therapeutic use of bacteriophages is undeniably the utmost explored tool and best suited to be part of the multidimensional strategies to fight against antibiotic resistance. Now, the pipeline for new antibiotics is almost dry with the upsurge of antibiotic resistance. So the forgotten cure is resurfacing with a well-deserved honor. Phage therapy primarily designates obligately lytic phages to kill their respective bacterial hosts, while parting the human cells unharmed and diminishing the shock on commensal bacteria that frequently result from antibiotic use.
This chapter depicts the prime points of bacteriophage genomic diversities, the evolutionary pattern, the calamity of antibiotic resistance that validates the phage use, their interface with the human immune system, and an evaluation within different antibiotic therapies. By going through peer reports of human clinical trials, this chapter will project the versatility of phage therapy. This informative writing will also cover orthodox methods and novel tactics, such as the use of phage-antibiotic permutations, phage-encoded enzymes, utilization of phage endurance mechanisms, and phage ergonomics. Potential and promising recent studies from all over the world define that the time has come to look more minutely at the prospective of phage therapy for future practice, both by strongly supporting new research and by scrutinizing the research already available, raising the optimism that this century-old magical cure will be an answer to multiple problems by varied invasive approaches.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Anti-phage antibodies
- Experimental therapy
- Immunomodulation
- Lysin
- Phage therapy
- Phage cocktails
- Prophage
- Synergy
- Macrophage
- Alternate medicine
- Dendritic cell
- Innate immunity
- Pathogen
1 Introduction
Bacteriophages are unique organisms with diverse elements in the biosphere. Since centuries, bacteriophages are used to treat humans. Antibiotic defiance is the prime threat to majority worldwide health issues in this current era. In each decade, a rising number of contagions carrying high fatality rate are a matter of concern and becoming harsher to treat in a cost-effective way. It is clearly evident from the past that phage treatment was scientifically employed approximately an epoch ago, but it was fetched to a cessation after the impressive application of antibiotics. Now, in the shadow of drug resistance, bacteriophage therapy is gaining a well-deserved resurgence. The enormous article lately published worldwide supports the concept and pragmatism of bacteriophage healing approach. This chapter concentrates to deliver a progressive view on bacteriophage cure, encompassing its exceptional potential. Bacteriophages are present not only in almost all the known biomes to human, from the gastrointestinal tract to the global ocean, but also in notable dwellings like oceanic basement (Nigro et al. 2017) and petrified stool specimen (Appelt et al. 2014). Phages exist as lysogens in the human gut and act as magnificent modulators in the gastrointestinal environment. This behavior of phages largely influences the physiology and metabolism of respective hosts (Kim and Bae 2018). In this chapter, strategic facts of the biotic and evolutionary doctrines of phages, genomic diversification in the light of metagenomics, their chemistry with the immune architecture, orthodox methodologies, novel stratagems (including the insertion of phage-antibiotic amalgamations), bacteriophage bioengineering, exploitation of bacteriophage resistance, phage-derived enzymes, and other emerging approaches are detailed.
1.1 Ancestry of Bacteriophages
Regardless of the extensive presence on earth, phage ancestry remains a myth. Swap of genetic elements is among the crucial mechanisms utilized by phages in the process of evolution. Irrespective of the genetic exchanges, there is huge scarcity of homology at the genome and amino acid level. It should be noted for most of the bacteriophages, a finite number of virion varieties occur in nature. So a definite question arises; whether the structural resemblances are the outcome of divergent or convergent evolution. A divergent evolution would signify a common ancestor with divergence beyond the noticeable homology in sequence, although they have maintained basic confirmation of their capsid proteins. On the other hand, a convergent evolution ensures about no common ancestor but rather has converged in the separate direction to form an optimum virion structure. Although both the evolution pathways can point to a single and very common pathway but with the accretion of equivalent knowledge about the capsid conformational characteristics, they clearly indicate toward divergent evolution hypothesis and, of course, from a common ancestor. The study findings that backed the divergent evolution are as follows: First, evidence fetched from a relation between double-stranded tailed phages, archaeal virus HSTV-1 (Pietilä et al. 2013), and herpes viruses (Baker et al. 2005). All these virion capsids contain a common fold called HK97. These virions also display similarity in their capsid assembly pathways (Rixon and Schmid 2014).
Tectiviridae family gives us the second evidence in the form of PRD1 phage, where the capsid protein is structurally equivalent to the archaeal virus Sulfolobus turreted icosahedral virus (STIV) (Khayat et al. 2005) and mammalian adenoviruses (Benson et al. 1999). Capsid protein architecture was made up of β-barrels. There are four different ways to fold β-barrels, but these viruses display only one common way of folding (Benson et al. 2004). Besides, adenoviruses and PRD1 embrace a linear double-stranded DNA (dsDNA) genome with inverted terminal repeats. Other viruses like phage PRD1 (infects gram-negative host) also display structure such as Tectiviridae, Corticoviridae, archeal, and other eukaryotic viruses (Krupovic et al. 2011).
The flag bearer for the third evidence is Cystoviridae phage phi6, which has a resemblance with the eukaryotic Reoviridae like blue tongue virus (BTV) and Totiviridae (El Omari et al. 2013). It was also evidenced that the inner coat protein of double-stranded viruses is similar in confirmation (Huiskonen et al. 2006). Besides, they also contain segmented genome enclosed within a double-layered capsid structure.
1.2 Resurrection of Phage Therapy
Continuous acquirement of insolence in pathogen toward antibiotics and the unstoppable rise in mortality are the prime reason for the resurrection of phage cure approach. Resistance in pathogens become evident in the form of multidrug-resistant (MDR), expansively drug-resistant, and certainly pan-drug-resistant (Magiorakos et al. 2012) bacteria. The flag bearer pathogens for drug resistance are Klebsiella pneumoniae, Pseudomonas aeruginosa, Acinetobacter baumannii, Enterococcus faecium, and Staphylococcus aureus. Though the pathogens drew enough attention of health researchers all over the world, the ultimate result is disappointing as there is no answer toward pathogen. The pipeline of a newer drug production is almost dry, resulting in the reemergence of the alternative treatment in the form of bacteriophage treatment. Immunocompromised and patients hospitalized with surgical, burn, or intensive care necessity are at an extremely high risk of catching multidrug-resistant infections. So if you consider post-antibiotic epoch, even simple infections can result in a reason of fatality (WHO 2014).
A recent study on the effect of multidrug-resistant bacteria on world economy and human civilization reveals ten million lives will cease annually, with a flashing US$100 trillion equivalent cost on world economy (O’Neill 2014). It is crystal clear that the global health is in grave crisis, regardless of age or socioeconomic status in the country (WHO 2015).
2 Genomic Diversity in Bacteriophages
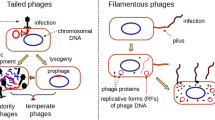
Phages display an array of structural morphologies in their genomic versions like double-stranded DNA (dsDNA), single-stranded DNA (ssDNA), single-stranded RNA (ssRNA), and double-stranded RNA (dsRNA) genome phages (Table 5.1).
Despite their tiny genomic organization, bacteriophages exhibit plethora of genomic abilities as found by viral metagenomics. The latest progresses in this field discovered several new phages belonging to non-tailed dsDNA (Brum et al. 2013; Kauffman et al. 2018) and ssDNA (Lim et al. 2015; Roux et al. 2019) family, and some of these bacteriophages are abundantly present in biomes.
In detail, metagenomic study additionally disclosed that genetic archeology of these phages. Remarkably, the sequencing data showed no such homology with reference to phage genomes, which proved the genome-level variety of phages (Paez-Espino et al. 2016; Gregory et al. 2019). Among the discovered phages, most of them contain dsDNA and a prominent tail (Ackermann 2007). The most common structural versions of bacteriophage are tailed phages rather than non-tailed phages. Bacteriophage classification was signified by viral archeology, target choice, and genome type (ssDNA, ssRNA, dsDNA, or dsRNA). In 2018, International Committee on Taxonomy of Viruses (ICTV) diversified bacteriophages into 5 families, 26 subfamilies, 363 genera, and 1320 species (https://talk.ictvonline.org/taxonomy/p/taxonomy_releases). Considering the evolution of genome-based techniques, it is expected that the list of bacteriophages will significantly increase in the future (Adriaenssens and Brister 2017).
2.1 Genome Size
Bacteriophage exhibits a varied range of genome lengths. Recently, megaphages were reported with the largest genome size of >540 kb. These phages were demonstrated to be resident of the human and animal gut and projected to infect Prevotella species as explored in a viral metagenomic study (Devoto et al. 2019). These megaphages are widespread as evident in baboons, pigs, and humans. Large genome size definitely helps these organisms in carrying genes accountable for replication and nucleotide metabolism, which are absent in small genome phages. Till date, the lowest phage genome to be known is Leuconostoc phage L5 (2435 bp). Certainly, L5 phages do have evolutionary advantage and disadvantages over the large-sized megaphages.
2.2 Role of Metagenomics in Bacteriophage
Genomic diversity of bacteriophages is hard to grasp, as phages do not contain a similar genetic indicator. Here comes the rescuer, viral metagenomics, which talks beyond the necessity of culture-based techniques and the dependence on marker genes. Metagenomics deals with the whole viral nucleic acid isolated from various environments. Metagenomic study helped to get the first human gut virome from a hale and hearty individual in 2003 (Breitbart et al. 2003). Phage infectivity toward a wide array of host cell makes the situation more complex as they bear minute or no sequence resemblance. Even a single host infecting distinct phages can exhibit significant sequence differences (Hatfull 2008; Krupovic et al. 2011). The use of statistical method like pairwise evaluation for genetic distance and gene content against 2333 phages displayed no homology for almost 97% cases. Besides, until now, metagenomics gave us 90 viromes from aquatic environments, 8 from soil, and 38 from the human gut. Furthermore, several studies that dealt with ulcerative colitis patient (Zuo et al. 2019), twins (Reyes et al. 2010), and healthy adults (Minot et al. 2011; Manrique et al. 2016) displayed interpersonal viral scenario in between health and ailments. The utmost modern discovery by metagenomics is the supreme abundance and widely distributed phage in the human gastrointestinal loci as crAssphage (Dutilh et al. 2014).
2.3 Single-Stranded DNA Phages
The most prominent family of these phages are Microviridae and the best discovered member is phiX174 (5386 bp) (McKenna et al. 1992). Microviridae family was widely ignored, until 258 new ssDNA phages were revealed in the gut environment of Ciona robusta (marine invertebrate). These phage groups contain small icosahedral capsid (26 nm) with β-barrel structure. Two subfamilies are enlisted under this group, named as Bullavirinae and Gokushovirinae. These two families are structurally quite different as Bullavirinae have pentameric major spike protein complexes at the end of each capsid apogee. In this occasion, Gokushovirinae “mushroom-like” protrusions can be observed that extend along the icosahedral axes of the capsid (Chipman et al. 1998).
Now, Inoviridae family also represents ssDNA genome. The virions in this family contain circular supercoiled ssDNA packed within a long filament, comprised of major capsid protein (MCP) (Ackermann 2012). The MCPs in the filamentous structure are structured uniquely along the genome of the bacteriophages. The MCPs constitute of α-helix with N-terminal signal peptide. This signal peptide helps this virus to translocate the membrane barrier (Xu et al. 2019). Recently, mining of Inoviridae phage family genome explored 10,295 Inovirus-like sequences. Among the vast diversified species network, only 5964 distinct species found were identified. This study strongly defines a hundredfold allocation of variety than previously described (57 genomes) within the Inoviridae family (Roux et al. 2019).
2.4 Double-Stranded DNA Phages
The most abundant and widely characterized phages contain double-stranded DNA (dsDNA) genome. These phages belong to Caudovirales order. This order can be further divided into five families: Myoviridae, Siphoviridae, Podoviridae, Ackermannviridae, and Herelleviridae. Together, an extensive variation in the capsid dimension can be witnessed among the members in this order. Capsid diameter arrays from 45 to 185 nm and perfectly corroborates with the genome size (Hua et al. 2017). The capsid proteins in these phages contain HK97 fold (Duda and Teschke 2019). The HK97 fold was first identified from phage HK97 through X-ray crystallography. The tail of the virions linked to the capsid by a connector complex comprised of portal protein coupled to connector proteins. Several structural studies have discovered that the portal complex is a dodecameric ring with an equivalent overall structure, though they have a low sequence resemblance (Lebedev et al. 2007). The glycoprotein 4 (gp4) (head to tail connector) protein of Podoviridae P22 has an equivalent structure to those present in siphophages SPP1 and HK97.
Myoviridae and Podoviridae exemplify 17% and 12% of the entire bacteriophages, respectively. Metagenomic studies from the Antarctic soil explored that the dsDNA tailed phages were dominant, and the occurrence in Myoviridae and Siphoviridae was inversely correlated (Adriaenssens and Brister 2017). Herelleviridae and Ackermannviridae are the furthermost fresh addition to Caudovirales order. It is evident that >85% of the bacteriophage genomes belong to Caudovirales order in the genome database. It has been perceived these two families are the utmost homogeneous family in view of genome analysis, which characterizes as Ackermannviridae (Myoviridae morphology with tail spikes at the base of the tail; e.g., AG3) and Siphoviridae (long noncontractile tail; e.g., phage lambda). Though Herelleviridae is an officially defined family, it reflects similar morphological pattern as Myoviridae.
In the case of non-tailed phages, we describe three families: First, Corticoviridae (circular dsDNA genome) family members enclosed within the capsid constituted of lipidic membrane (internal) enclosed by MCPs (e.g., PM2). Second, Tectiviridae (linear dsDNA genome) has an internal lipidic membrane inside an icosahedral capsid (e.g., PRD1). A signature feature of these two-phage family members is their specialized trimeric form of major capsid protein (MCP), comprised of a double β-barrel sheet structure. Structural analysis of PRD1 phage and their main capsid protein displays N-terminal α-helix formation. It can interact directly with the inner phage membrane and shares exclusive confirmational homology with adenovirus MCP (Benson et al. 1999). Besides, icosahedral apices of PRD1 phage and PM2 exceedingly corroborate structurally to N-terminal domain adenoviruses (that infect all human) (Abrescia et al. 2008). In this context, it should be noted that phage PRD1 do not contain a tail to transport its genome into its gram-negative host, but its membrane can perform as a proteo-lipidic tube, which can also pierce host envelopes (Peralta et al. 2013). Most pleomorphic phages belong to Plasmaviridae family, and it contains double-stranded circular DNA genome, which is surrounded by an envelope but without any capsid structure (e.g., MVL2).
2.5 Single-Stranded RNA Phages
Single-stranded RNA phages represent the group of Leviviridae. They are known to be the simplest known positive (+) sense single-stranded RNA. These genomes signify a size of 3.5–4.4 kilobases. Leviviridae, a ssRNA genome (e.g., MS2, R17) bearing phage have capsids with icosahedral and spherical geometry, with a span of 30 nm. The virions of these family encode mostly four proteins such as MCP, replicase, maturation, and lysis proteins. Curiously, MS2 was the first ssRNA virion whose whole genome sequence was decoded. These virions are made of MCP and an RNA-interacting protein of the replicase-encoding gene (Valegard et al. 1990). The most auspicious use of ssRNA phages is in vaccine development, which is a boon for human civilization.
2.6 Double-Stranded RNA Phages
Cystoviridae is the family that represents the dsRNA bacteriophages. These phages display an exclusive confirmational upgradation in the form of tri-segmented genome enclosed within a spherical capsid (e.g., phi6). These phages armored with three-layered structural integrity, two-layered inner capsid, and an exterior lipid membrane (Vidaver et al. 1973). The genome of Cystoviridae members transcribed into three segments: large (L), medium (M), and small (S). These three segments behave as an independent polycistronic mRNA and believed to be translated into 12 proteins with the help of host machinery.
3 Bacteriophages as a Therapeutic Agent
Some stringent features of bacteriophages like lytic in nature, target pathogen specific, cidal activity, and removal of bacterial endotoxins enhance the refinement of bacteriophages as a therapeutic agent. Besides, knowledge of phage-specific receptors on the host cells is technically essential for resisting phage resisting microbial cells (PRMCs). This knowledge will also guide the future strategy on using combination therapies.
As we know, bacteriophages signify two modes: lytic and lysogenic. From the point of treatment, lytic phages are the prime choice over the lysogenic phages as they can hamper the bactericidal activity, by inducing a prompt rise in homoimmunity and also by lysogenic conversion effect. Phage receptors present on the cell walls, capsules, pilli, and flagella (Bertozzi Silva et al. 2016) invite gram-positive (species or strain specific) or gram-negative (species or strain specific) phages. Attachment of bacteriophages to its highly specific receptors acts as lock and key mechanism. The receptor conformation can guide the phages against a widespread range of potential host. A growing knowledge of host receptors stimulates the research in host–phage interactions.
3.1 Why Bacteriophage Remedy Is Superior than Antibiotics
Phage therapy is undoubtedly a multidimensional, dynamic, and multidisciplinary option against rising resistance (Table 5.2). Whereas antibiotics are chemical compounds targeted against particular physiological process in bacterial cell, collateral damage is the common consequence of antibiotic treatment. Antibiotic treatment generally interrupts the microbiome of the host, in the form of antibiotic-associated diarrhea, metabolic or immunologic disorders, etc. (Langdon et al. 2016; Perepanova et al. 2020).
Bacteriophages are extremely precise to target, and this feature can sometimes act as a disadvantage (Table 5.3), as phages need precise etiological agent, occasionally to the strain level, and this methodology is resource and time consuming (Caliendo et al. 2013).
Target specificity also gives phages upper hand over the antibiotics as they don’t disturb the microbiome. It has been explored that early commencement of phage therapy can significantly reduce pathogen count (Jaiswal et al. 2013; Chhibber et al. 2008). Altogether, it is evident that collection of new phages should be in pipeline in a constant manner. Not only isolation but well-defined strain-level portrayal of bacteriophages should be refined to use it as potential phage candidate. These potential phage candidates should have short latency period, large burst size, and expansive host range (Nilsson 2014; Whittard et al. 2021). A current study in Australia validated safety and efficacy of a good manufacturing procedure where a three-phage cocktail inoculated intravenously to 13 patients with hostile S. aureus infections (Petrovic Fabijan et al. 2020).
The role of the immune system is crucial against these dual therapeutic techniques. The most lethal side effects of antibiotics are stimulation of hypersensitivity and mostly IgE-mediated anaphylaxis (Legendre et al. 2014). Relatively, there is no such documented occurrence of anaphylaxis with phage cure in humans (Sarker et al. 2012; Speck and Smithyman 2016). It should be noted that the human evidence-based study with bacteriophage remedy is small. It is to be noted that the immune system counterparts both the healing approaches.
Lytic phages are killers of their target, and still, they are not able to eradicate all the pathogen on their own. A well-known argument for the above statement is that eradication of host will result in cessation of the virus as well. That is why bacteriophages activate dynamic strategy as “kill the survivor,” (Campbell 1961; Maslov and Sneppen 2017) where host abundance is rapidly reduced to an assured extent that the immune system can take over the clearance protocol. The immune system assists the phages in clearing the pathogen, which can be an immunophage synergy (Roach et al. 2017). So these findings establish the theory that bacteriophages elicit both innate and acquired immune system. In view of all the valid points between phage therapy and antibiotics, there are some similarities as well (Table 5.4).
3.2 Antiphage Mechanism of Bacterial Host
Bacteriophages are not immune against resistance like antibiotics. Lytic phages employ an intense antimicrobial selective pressure, which generates rapid phage-resistant bacterial mutant. One of the first mechanisms employed by pathogens are (a) mutation in the phage receptors in the host so that phages are inhibited against and adsorption and superinfection step; (b) restriction modification system that is another strategy where host genome is kept protected, but the phage genome remains vulnerable by the target restriction enzymes; and (c) besides, clustered regularly interspaced short palindromic repeat (CRISPR)–Cas (Labrie et al. 2010) tactic where host cell keeps memory of the previously confronted foreign DNA and degrade again if it enters. Altogether, these mechanisms can be termed as prokaryotic immune system. In 2018, Doron et al. explored in research that beyond 45,000 bacterial and archaeal genomes have new antiphage mechanisms that are yet to be established.
3.3 Phages and the Human System
It is proved that phages in the human gut encode a population of hypervariable (large sequence variation) proteins (Minot et al. 2012). Most of these hypervariable proteins possess C-type lectin fold, and some of them displayed immunoglobulin (Ig)-like domains (Medhekar and Miller 2007). Interestingly, this fold was previously reported from Bordetella phage BPP-1 tail fibers. Ig-like domains are displayed in many structural proteins of phages (Fraser et al. 2006, 2007). In vitro growth conditions recommended that these Ig-like proteins aid in the attachment of their target prey under environmental conditions (McMahon et al. 2005; Fraser et al. 2007).
Phages are capable of crossing the mucosal barrier at a concentration where it can bypass and intermingle with the cellular epithelium. In vitro research findings using cell lines support the theory that phages can enter and cross epithelial cell layers by non-specific transcytosis mechanism, from apical to basal direction (Nguyen et al. 2017). Bacteriophage transcytosis phenomenon occurs across different types of epithelial cell (e.g., gut, lung, brain, liver, and kidney cells). This phenomenon works for diverse phage types and morphologies (e.g., Myoviridae, Siphoviridae, and Podoviridae). Critical experiments on transcytosis explored that merely 10% of the entire phages endocytosed through the epithelial cells, which found to be localized within the membrane-bound vesicles. These phage particles travel through the Golgi apparatus, before being exocytosed as functional units in the basal layer. This transcytosis process across epithelial layers certifies the manifestation of phages in the human system even in the nonappearance of disease or pathogen (Nguyen et al. 2017).
Phages are qualified of directly interacting with mammalian cells, as discovered by Bloch in 1940. This study validates the attachment of phages to cancer tissue, which primes the tumor inhibition. Later, it was reestablished that phage can not only bind to cancer cells in vitro and in vivo but also are quite capable of attaching lymphocyte plasma membrane (Wenger et al. 1978; Dąbrowska et al. 2014). Several studies started to understand the molecular basis phage–mammalian interaction. In 2003, Gorski et al. hypothesized that cross talk happens due to the existence of a tripeptide motif Lys-Gly-Asp (KGD). This motif found to be present on T4 capsid protein gp24 and verified to be acting as ligand for the β3 integrins on cells. In this framework, a significant study has to mentioned where phage capsid protein motif (WDC-2, containing a TRTKLPRLHLQS peptide motif) was modified. This amendment resulted in potential tumor-specific phages with 93% success rate (Eriksson et al. 2009).
3.4 Bacteriophage Interaction with the Immune System
The human body interacts with several viruses each second. A majority of the microorganisms settle first at body surfaces (intestine, skin, and respiratory tract) that are open to welcome microbes. Studies from the last decade explored that microflora of human beings also inducts viruses (White et al. 2012), especially bacteriophages. Metagenomic studies have discovered the hidden world of phages inside the human system (Minot et al. 2011).
It will be fascinating to discuss whether phage–host dynamic relation of the human intestine could significantly modify the composition of the normal flora and influence on the spread of pathogenic viruses (Duerkop and Hooper 2013). Niche occupation by phage and limiting pathogen colonization and resource use is the signature effect of host immunity. Few viruses target mammalian cells, but phages donate to the mainstream of the viral community (Cadwell 2015).
As we know, phages are daily commuters of the human body, but questions persist over the safety and immunogenicity regarding phage therapy (Cooper et al. 2016). It is evident now that bacteriophages have not only antibacterial properties but also a part of mucus layers and even migrate through the cell layers (Nguyen et al. 2017). When these phages interact with individual immune system, they induce both innate and adaptive immune responses (Van Belleghem et al. 2017; Majewska et al. 2015; Miernikiewicz et al. 2013; Hodyra-Stefaniak et al. 2015). No doubt bacteriophages adhere or attack nontarget tissues or cells to some point (Table 5.3). During portal entry, some phages are taken up by the gastrointestinal tract into the blood. This phenomenon ascertains phage to epithelium transition and reticuloendothelial system (Merril 2008; Górski et al. 2006).
In 2012, Gorski et al. discovered that purified phages show anti-inflammatory activities via the suppression of reactive oxygen species (ROS) and also NF-kB inhibition. Phages also affect the macrophage-mediated cytokine production (Van Belleghem et al. 2017). The immunomodulating effect of phages in the blood has been suggested by different studies of Górski et al. (2012, 2017). Besides, inhibition of inflammation caused by bacterial infections was also proved. A recent study explored that the immunomodulating effect of bacteriophages was regulated at the transcription level from monocyte-related immunity genes (Van Belleghem et al. 2017).
3.5 How Does the Immune System Work Against Bacteriophages?
3.5.1 Phagocytic Response
Mononuclear phagocytic cells were the prime members of innate immune system. Simultaneous inoculation of phage, together with host bacteria, undoubtedly stimulates bacterial phagocytosis, and this effect is ensured by certain opsonization of bacterial cells by phages. Besides, it was observed that phages stay functional and virulent to their host bacteria even after ingestion by granulocytes (Kaur et al. 2014). Therefore, some authors also endorsed that phages can be operative in lysing the phagocytosed bacteria, catalyzing the activity of phagocytic cells (Górski et al. 2012). It was speculated that phages might inhibit the platelet and T cell adhesion to fibrinogen (Kurzepa et al. 2009) and thus strengthens the critical role of bacteriophages in transplant rejection, metastasis, and angiogenesis. These phagocytic cells can effectively remove these viruses from the circulatory systems (Navarro and Muniesa 2017; Górski et al. 2012). Several antigenic combinations can perform as pathogen-associated molecular patterns (PAMPs), and in this case, phage nucleic acid can act as potential PAMP. These PAMPs are recognizable by cognate, toll-like receptors (TLRs) or TLR9 (responsible for recognition of DNA). Inside the cell, phage nucleic acid is unprotected and vulnerable to cellular defenses. Further, experiments on λ phage revealed phagocytic cells rapidly remove λ phage from the circulatory system in humans (Merril et al. 1973), but certain λ phage mutants can prevail for a longer period in the blood system than the wild-type version (Merril et al. 1996).
3.5.2 Cytokine Response Against Phage
Bacteriophages certainly stimulate cytokine response as evident by different studies. But most of the studies were performed with phages having bacterial endotoxins or proteins. A study conducted with 51 patients suffering from suppurative bacterial illnesses (antibiotic-resistant bacterial species) of numerous organs and tissues, who were treated with phages. Afterwards, blood sample was taken and analyzed for IL-6 and TNF-α. The authors witnessed a sharp decline in the illustration of these cytokines after a long-term treatment (i.e., 21 days). In another experiment, mice were remedied intraperitoneally for 5.5 h with phage T4 or capsid proteins (i.e., gp23, gp24, Hoc, Soc) and analyzed for cytokine expression. This experiment displayed no such inflammatory-mediating cytokines (Miernikiewicz et al. 2013).
Another instance, where phages and antibiotics were mutually used as a treatment measure in mice, showed that phage-cocktail-treated group had a gradual decrease in both IL-6 and TNF-α level for 3 days (Jaiswal et al. 2014). Astonishingly, phages also exhibit conserved anti-inflammatory properties. In a designated study, phage specific to Pseudomonas aeruginosa and Staphylococcus aureus has been shown to stimulate equivalent immune responses (Van Belleghem et al. 2017). Their expression levels regarding inflammatory markers like IL6, suppressor of cytokine signaling 3 (SOCS3), and IL-1 receptor antagonist (IL1RN) are quite similar. The above findings were also endorsed in murine models of xenografts (by Górski et al. 2016) where he described an immunosuppressive effect of phages.
3.5.3 Antibody Response Against Phages
It is obvious that phages do stimulate the manufacture of cognate antibodies as observed in humans and animals (Górski et al. 2012; Puig et al. 2001). It is the most potent barrier in using phages as therapeutics. Shortly, after the discovery of phages, antibodies against phages were also encountered (Jerne 1952, 1956).
One important observation regarding phages is that they don’t follow a simple rule of stimulation. Rather, the route of inoculation from individual to individual matters. Besides, the dose and application schedule individually also influence the antibody response (Dąbrowska et al. 2014; Łusiak-Szelachowska et al. 2014). The antibody response stimulated against phages can be detrimental (Huff et al. 2010), but as evident by scientific experiments, antiphage antibodies do not rule out a favorable therapeutic practice of phages in humans (Łusiak-Szelachowska et al. 2014). It is clearly known that humoral immunity can be stimulated by inherently occurring phages. So antiphage antibodies can be spotted in the serum of different species (e.g., human), even before any kind of phage treatment.
Since the 1970s, bacteriophages were used comprehensively to detect and track immune deficiencies in humans. One such example is the immunization of human with φX174 to gather detailed information about primary and secondary immunodeficiencies. This study elaborates no such adverse or severe antiphage antibodies in patients where phages were found to be present in the bloodstream for a prolonged period (Ochs et al. 1971; Rubinstein et al. 2000; Shearer et al. 2001). Earlier studies based on T4 phage confirmed that no antibodies were stimulated in volunteers at all (Bruttin and Brüssow 2005). These findings were further supported by Dąbrowska et al. (2014), where 50 healthy (never been subjected toward phage therapy or phage-related work) individuals were taken to observe phage T4 activity. Surprisingly, they showed 82% decreased activity against phage T4, whereas positive antiserum cases showed increased intensities of IgG antibodies against Hoc, Soc, gp23, and gp24 proteins. The antibody response was generated mainly due to the gp23 protein.
A study where 20 patients infected with S. aureus treated with MS1 phage cocktail by either oral route or local inoculation showed an elevated level of IgG, IgM, and IgA (Żaczek et al. 2016). Within them, few patients showed lower response, whereas some showed higher antibody response levels. This can be enlightened by several opinions. But ultimately the good news is that the presence of antiphage antibodies did not undermine the clinical effectivity of bacteriophage therapy.
3.6 Bacteriophage Pharmacokinetics
The route of administration of phages crosstalk with phage pharmacokinetics. There are variety of proven administration routes for phages. Several studies regarding topical (wound infection, eye infection, otitis), transectal (prostatitis), intravenous (septicemia), transnasal (lung infection), transurinary (bladder, kidney), or oral use of pages have been documented. It is evident when bacteriophages were directed by topical, transnasal, transurinary, and intravenous routes, phages can candidly interact with target pathogens and eradicate them. In contrast, when oral or transrectal routes are utilized, the phages need to travel from the gut environment to the blood and lymph by invading epithelial cell layers. Here, we will discuss briefly about the recent human experiments done on humans and their consequences from the pharmacokinetics angle.
3.6.1 Topical or Intranasal Applications
In Georgia, cystic fibrosis patient was treated for multidrug-resistant Achromobacter xylosoxidans. A mixture of two phages was administered orally and via inhalation through a nebulizer at a concentration of ~108 (plaque-forming units, pfu). This dose was continued for twice daily for 20 days, and the dose was repeated at 1,3, 6, and 12 months. This study showed a promising result against the infection and improved the patient clinically (Hoyle et al. 2018). In USA, Staphylococcus aureus–infected diabetic toe ulcer patients were treated with phage Sb-1. The bacteriophages were given to wound cavity with 0.1–0.5 ml phage suspension-soaked gauze. This treatment dose was given once in a week up to 7 weeks, and the patients recovered (Fish et al. 2016). In 2018, Fish et al. again successfully treated patient with digital staphylococcal osteomyelitis in diabetic foot ulcers infected with S. aureus. The soft tissues around the ulcer were injected with 0.7 ml once a week for 7 weeks and found significant recovery among patients. In Georgia, a young patient with Netherton syndrome (a rare disease) accompanied with severe S. aureus–infected skin disorder, treated with Sb-1 phage and other S. aureus cocktail phages. Patient limbs were sprayed and cream treated with around 107 phages. Patients also medicated with oral application of phages.
3.6.2 Intravenous Application of Therapeutic Phages
Here, phage particles were directly introduced into the bloodstream, so it doesn’t require any translocation to the host site. The safety of this administration route was experimentally proven by Speck and Smithyman (2016) to some extent.
In 2017, Schooley et al. did an experiment on multidrug-resistant Acinetobacter baumannii in diabetic patients with necrotizing tissues by percutaneous drainage catheters with phage φPC (a cocktail of phages AC4, C1P12, C2P21, and C2P24). In addition, the above phage φIV (a cocktail of phages AB-Navy1, AB-Navy4, ABNavy71, and AB-Navy97) was inoculated intravenously. After this treatment, the patient recovered from coma. This treatment was continued for 59 days, and the patients recovered with time. In 2017, patients infected by P. aeruginosa with acute kidney damage in Belgium were treated with 50 μl BFC1 cocktail (14/1 and PNM) and one S. aureus phage ISP. The dosage was 6 h intravenous infusion for 10 days, and also wound was irrigated with 50 ml of the cocktail every 8 h for 10 days. This treatment protocol resulted in a speedy recovery of the patients.
3.6.3 Oral and Intrarectal Administrations of Therapeutic Phages
The most challenging route of inoculation is oral and intrarectal where phages are not capable of interacting directly with the target pathogen, as the bacteriophages must transit from the gastrointestinal tract to the bloodstream. Though there are different successful studies with oral or rectal administration, the exact mechanism of bacteriophage translocation is still under the cloud. Oral administration has been explored to be effective against systemic infections (Alisky et al. 1998; Weber-Dąbrowska et al. 2000; Górski et al. 2006). It is highly essential to neutralize gastric acid before administering bacteriophage through oral route as low pH in the stomach may inactivate the bacteriophages (Alisky et al. 1998; Weber-Dąbrowska et al. 2000). Jaiswal et al. in 2014 showed neutralization of gastric acid prompted the phages to move from the stomach to the intestine and diminish the intestinal colonization of pathogenic bacteria in an animal model. Bacteriophage translocation from the stomach to the blood was experimentally proved in both neutropenic and healthy models (Matsuzaki et al. 2014; Międzybrodzki et al. 2017). Międzybrodzki showed that the transit of bacteriophages might highly influence the phage and animal species or characteristics. This experiment clarifies two animals (rat and mouse) and two bacteriophages (Escherichia coli phage T4 and S. aureus phage A5/80) were used. The very first observation was for phage T4 and A5/80 where these orally inoculated phages could travel from the gut to the small intestine faster if gastric acidity is neutralized. Second, it was detected that phage A5/80 effectively translocates from the gut to the blood in mouse, but considering T4 phage, the efficacy of transit was considerably lower. Interestingly, there was no translocation for both the phages in mouse. In this setting, Bari et al. in 2017 enlightened the possible reason behind the low-frequency translocation of T4 phage in mice. As explained by Bari et al. 2017, the reason may be the incidence of T4 head of immunoglobulin-like non-essential Hoc protein. In the intestine, T4 head attaches to the mucus layer via Hoc and leaves the tail tip free. So this adherence connection may be due to the probable missing link between lower or non-translocation of the bacteriophage from the stomach to the bloodstream.
Administration of phages through the intrarectal pathway against human enterococcal prostatitis has demonstrated to be efficient (Letkiewicz et al. 2009, 2010; Górski et al. 2018). The transit of phages in this situation was believed to be similar to that of the previous cases. Therapeutic phages have been used significantly in Georgia, Russia, and Poland for many years. Phages were applied through versatile administration routes like intravenous, oral, and transrectal routes. We have discussed successful human clinical trials around the world that emphasizes the effective and useful nature of phages irrespective of their method of administration, though it should be documented that oral path of administration is specially preferred due to its versatility. The precise mechanisms dealing with phage translocation into the blood are still under the lens of molecular biology and cytology experiments. Regrettably, however, there is very little evidence on the bacteriophage dynamics in the human body. So a detailed knowledge of phage pharmacodynamics is highly desirable to improve the suitability of bacteriophages as a perfect therapeutic agent.
3.7 Bacteriophage-Derived Lysins
Bacteriophage lysins are highly effective molecules, which facilitate release of its progeny phage during its replication cycle. These enzymes are specific to the five major bonds in peptidoglycan. It must be notable that lysins don’t have signal sequences, other than few exceptions. This factor bars the lysins to translocate through the cytoplasmic membrane and interact with their substrate in the peptidoglycan layer. The intermingle of lysins with their cognate substrate is tightly regulated by holing the lytic system (Wang et al. 2000). At a definite time during phage replication inside the target cell, holin molecules are injected into the bacterial cell membrane and create a patch and ultimately induce a localized membrane disruption (Wang et al. 2003). Now, the cytoplasmic lysins that were accumulated in the cytosol are free to interact on the peptidoglycan and cleave specific bonds. This causes the cell to lyse immediately. Unlike large DNA phages, small RNA and DNA phages engage a different strategy like interfering with the enzymes responsible for peptidoglycan synthesis (Bernhardt et al. 2001). It is clear now that phages evolved from several years to bear features like lysin production. Just like naturally occurring antibiotics in bacteria that help in combatting other organisms around them, bacteriophages do maintain their surrounding with the help of lysins.
It was proven that some lysins (isolated from gram-positive bacteria) can kill their targets within second of contact (Loeffler et al. 2001; Nelson et al. 2001). Such an example is nanogram amounts of lysins that could reduce 107 Streptococcus pyogenes by >6 log minutes after enzyme addition. This is a remarkable feature of lysin, as no biological compounds are found to kill bacteria at such speed other than chemicals. In 2001, Nelson et al. coined the phrase “enzybiotics” to designate these novels highly destructive antibacterial compound. It should be noted that until now, only two lysin-based ointments (polysporin and mupirocin) are extensively used to control colonization of pathogenic bacteria in mucus.
3.8 Lysin Structure and Mechanism of Action
3.8.1 Gram-Positive Specific Phage Lysin
Lysins secreted by DNA phage (specific to gram-positive bacteria) ranges between 25 and 40 kDa. Only one exception was discovered in the form of PlyC lysin (114 kDa), isolated from C1 phage of group C streptococci. A common trait of gram-positive host-specific phages is their two-domain structure. The two domains can be divided as catalytic domain or N-terminal domain and cell-binding domain (CBD) or C-terminal domain.
The activity of N-terminal domain can vary widely such as attack on (1) an N-acetylmuramoyl-l-alanine amidase (or amidase) that hydrolyzes the amide bond linking the glycan strand and peptide moieties (Loessner 2005), (2) an endo-β-N-acetylglucosaminidase or N-acetylmuramidase (lysozymes) that equally affects on the glycan moiety of the wall peptidoglycan, (3) an endopeptidase that cleaves the stem peptide moiety, or (4) a phage lysin with γ-d-glutaminyl-l-lysine endopeptidase efficiency that has also been discovered (Pritchard et al. 2007), but these enzymes are rare to find compared to others. It is also evident that in some bacterial phage (some staphylococcal and streptococcal phage), two to three types of catalytic features can be observed in a single domain (Cheng et al. 2005).
On the other hand, C-terminal or cell-binding domain (CBD) mostly targets cellular carbohydrates in the cell wall for attachment. Cell membrane–specific choline-binding lysins were discovered in pneumococcal phage lysin Cpl-1 (Hermoso et al. 2003). Later PlyL lysin and Ply21 lysins were discovered in Bacillus anthracis and B. cereus phage TP21, respectively. Interestingly, these phage lysins contain hairpin confirmation. This hairpin confirmation ensures that the catalytic domain and CBD interact with each other. This confirmation also defines that catalytic domain remains inactive until cell-binding domain interacts with its target molecule in the cell wall or membrane.
3.8.2 Gram-Negative Specific Phage Lysin
It is fascinating that lysin in gram-positive and gram-negative lysin behaves differently toward their target. Their characteristics are also different. Most gram-negative lysins behave like lysozymes. Gram-negative lysins are more complex as explored by Lood et al. (2015). He explored several lytic clones and found four distinct domains, unlike gram-positive bacteria. The four domains are (1) a TIGR02594 domain, (2) a catalytic domain, (3) a binding domain, and a (4) lysozyme domain. In this study, it was exclusively reported that TIGR02594 and lysozyme domain were flanked on a single side or both sides of the lysin with short positively charged amino acids. Thus, it helps the lysin both break the peptidoglycan with functional enzymatic domain and disrupt the outer membrane with charged domain, resulting in highly effective lysis of host cell (Thandar et al. 2016). It is established that gram-negative bacteria have lower internal turgor pressure (~3–5 atm) than their gram-positive (~15–25 atm) siblings. So in the case of gram-negative cells, there may be scarcity of sufficient pressure that supports the above theory.
3.9 Synergistic Approaches with Lysin
Several pneumococcal phage lysins were isolated, demonstrated, and categorized into two groups: amidases and lysozymes. Interestingly, both classes of lysins display similar C-terminal choline-binding domain but different N-terminal catalytic domains. Like cocktail phages, in a study, it was evident that a blend of two lysins with diverse peptidoglycan specificities was more applicable in shielding against a disease (Loeffler and Fischetti 2003; Schmelcher et al. 2015). It should be noted that a combination of lysins significantly enhances the effectivity of killing kinetics and also reduces the chance of emergence for enzyme-resistant mutants.
Considering an unusual combination (lysin and antibiotics), pneumococcal lysin Cpl-1 and gentamicin are highly effective in slaying pneumococci, while Cpl-1 and penicillin displayed synergy against an extremely penicillin-resistant pneumococcal variety. In another experiment, staphylococcal phage lysins and antibiotics against methicillin-resistant Staphylococcus aureus (MRSA) also found to be effective in in vitro and in vivo condition (Daniel et al. 2010). So it can be concluded that a correct combination of lysin and antibiotic could not only control antibiotic-resistant bacteria but also reestablish the use of specific antibiotics (against which resistance already reported). The good news regarding lysin is that till now no such bacterial resistance is reported. Experiments have been designed to comprehend the resistance pattern of bacterial cells against lysin. A 10 μl drop of dilute lysin was dropped on a bacterial lawn. After incubation, a clear lytic zone was formed and colonies from the periphery of the clear zone were taken to test the resistance. But in >10 cycles of bacterial contact to diluted concentrations of lysin in liquid media, no resistance was found (Loeffler et al. 2001; Schuch et al. 2002). It is obvious that the lack of resistance development is connected with the evolution of binding domain, which may be designed to avert lysin spill during lysis phenomenon.
3.10 Clinical Approach of Lysin
In 2016, Czaplewski et al. revealed in a review that diverse strategies has been considered earlier to find best substitute against antibiotics which include bacteriophage lysins as therapeutics, antibodies and vaccines as prophylactics, and probiotics as a treatment. In 2018, a company called ContraFect has used lysin (CF-301) against S. aureus bacteremia and endocarditis in phase 2 trial worldwide. Results of that trial against MRSA explored a 42% improvement over antibiotic treatment. Later, follow-up studies also supported the above findings, as humans treated with drug–lysin combination were discharged earlier and had fewer relapses compared to drug-alone-treated patients.
As we are surrounded by multidrug-resistant gram-positive and gram-negative pathogens, phages can be employed in indirect way to isolate and apply lysins against them. The lysins under use or under trial are astonishingly heat stable (up to 60 °C) and can be purified and marketed in large quantities with rapid manner. In the future, protein engineering, domain swapping, and gene shuffling approaches could be handy in coping up against continuously adapting bacterial pathogens.
4 Genetic Engineering of Bacteriophages
Bacteriophage diversity and adaptive potentials definitely make them an ideal candidate for genome engineering. Engineered phages can be applied to different sectors like in synthetic biology (Lemire et al. 2018), material science (Cao et al. 2016), and biomedical fields (Kilcher et al. 2018). There are several techniques to engineer phages.
4.1 Homologous Recombination–Based Techniques
This phenomenon talks about the swap of sequences in between two DNA strands, which share identical sequences. Phage cross is such a classical strategy to produce a mutant phage with a desired phenotype by combining characteristics from two parents (Karam and Drake 1994). Sometimes, host cells were coinfected with two dissimilar phages, and the homologous recombination inside the host cell generates the desired progeny with the desired phenotype. The progenies were chosen by nominating selective markers.
Homologous recombination among the plasmid and phage genome was also developed where plasmid containing the desired mutation flanked by homologous sequences was commenced into the host cells, which is also coinfected with the desired phage to be engineered (Namura et al. 2008). Though the overall rate of frequency of recombination is very low (Tanji et al. 2004), in some cases, a high frequency of recombination is also observed (Oda et al. 2004). So this genetic strategy is complicated and time consuming to find the preferred recombinants unless there is a properly designed selection strategy.
4.2 Bacteriophage Recombineering of Electroporated DNA (BRED)
This is a very popular technique based on the homologous recombination principle. This method exploits a RecE/RecT system of Rac prophage and Red system of lambda phage to enrich the frequency of recombination (Murphy 2012; Nafissi and Slavcev 2014). Red system is a well-coordinated system, composed of exo (α), bet (β), and gam (γ) genes. The target of exo gene is double-stranded DNA (dsDNA) end to transform to single-stranded DNA (ssDNA) substrate, whereas beta performs as a ssDNA-binding protein that anneals ssDNA target and cognate recombination zone in phage genome. Lastly, gamma works as an inhibitor of E. coli RecBCD exonuclease complex (designated to degrade linear dsDNA substrate).
BRED needs co-electroporation of the bacteriophage DNA and donor DNA expressing RecE/T-like proteins into recipient cells via either plasmid or chromosomally inserted so that homologous recombination can be promoted in an efficient manner (Marinelli et al. 2008, 2012; Thomason et al. 2009). It was observed earlier that recombination occurs only after starting of bacteriophage genome replication, and further purification of recombined phage is highly essential apart from initial PCR screening (Marinelli et al. 2008). This technique was first employed on mycobacteriophage and later applied to several other phages by constructing replacements, deletions, and insertions of heterologous genes (Marinelli et al. 2012). This technique is not so useful regarding gram-negative bacteria as they have low transformation efficiency, compared to gram-negative bacteria. So in the case of gram-positive bacteria, instead of transforming both bacteriophage DNA and donor DNA, bacteria that enclose phage-defined recombination system can be transformed with donor DNA only (Pan et al. 2017). The phage mutants can then be generated by infecting bacterial cells with wild-type phage. The main issues with high background of wild-type phage are a matter of concern, and also, it needs screening of recombinants.
4.3 CRISPR–Cas System–Based Engineering
At the dusk of the last century, a novel, highly effective immune system of prokaryotes was discovered. It’s a defense mechanism of prokaryotes against foreign invasion, named as clustered regularly interspaced short palindromic repeats (CRISPR)–Cas system. This system has two main components CRISPR RNA (crRNA) and Cas proteins. CRISPR–Cas system can be classified and can be further diversified into six types. These six types can be brought under two classes (class 1 or 2) based on mechanism and phylogeny (Koonin et al. 2017). It was proved that class 1 systems include types I, III, and IV recruit multiple Cas proteins in association with effector complex, whereas class 2 systems (type II, V, VI) employ single Cas protein in association with effector complexes to nick the target DNAs (Koonin et al. 2017).
The CRISPR–Cas effector complex is target specific and binds their target sequences intervened by the crRNA with a complementary region to the sequence of action. Here Cas proteins cleave the DNA and generate a break into the double-stranded DNA (Shmakov et al. 2017; Knott and Doudna 2018). It is well documented that the CRISPR–Cas system was first applied in genome editing in 2014 to identify a phage mutant with a deletion of a non-essential gene, gene1.7 (Kiro et al. 2014). This study denotes CRISPR–Cas system can be easily used as a screening utility to eradicate wild-type phage from the recombinant population. The plasmid relied type I CRISPR–Cas system was defined to target gene 1.7 and cleave the wild-type T7 phage genome, whereas mutant phages lacking gene 1.7 were protected from Cas9 complex and could propagate normally. Later in 2016, Box et al. used type I CRISPR–Cas system of Vibrio cholerae to engineer V. cholerae lytic phages. In the above study, both the donor DNA and CRISPR–Cas components were assembled in a single plasmid. Those plasmids were then harbored by V. cholerae; as phages attacked the cells, phage genomes were cleaved by CRISPR–Cas but immediately repaired by homologous recombination with the donor DNA and give rise to recombinant phages with deletion or insertion mutation (Box et al. 2016).
Streptococcus pyogenes CRISPR–Cas is the most commonly used type II system, selected for bacteriophage genome engineering (Lemay et al. 2017; Schilling et al. 2018; Shen et al. 2018). It should be noted the first type II CRISPR–Cas system utilized for phage genome modification belonged to Streptococcus thermophilus (Martel and Moineau 2014). Recent works on Listeria monocytogenes also confirmed the existence of CRISPR–Cas system and are found useful in valuable engineering program for Listeria bacteriophages (Hupfeld et al. 2018). Generally, the three components Cas9, crRNA, and transactivating crRNA (tracrRNA) of CRISPR–Cas system were attached or cloned to a single plasmid. It was interesting to observe that crRNA and tracrRNA could be expressed either separately (Lemay et al. 2017) or as a sole fusion RNA (Schilling et al. 2018). Due to a successful transformation into recipient cells, the machineries form CRISPR–Cas9 complex will specifically connect to the phage genome and craft a double-stranded DNA break during phage genome invasion. It was also explored that Cas9 of S. pyogenes is highly capable of cleaving T4 phage genome (highly protected from most of the restriction endonuclease enzymes due to high levels of 5-hydroxy methylation and glucosylation to its cytosine molecules (Tao et al. 2017, 2018b). It has to be mentioned that sometimes cleavage potential of CRISPR–Cas9 complex on its target crRNA (protospacer sequence) is very high, and then, only the recombinant phages can survive. Consequently, all resultant progeny phages are recombinant mutants. If somehow the protospacer sequence is feebly cleaved or an overburden of parental phage was employed that time, it could lead to error-prone repair and also may result in inclusion of random mutations in the protospacer sequence ensuing in dodging from CRISPR–Cas cleavage. This phenomenon is occasionally reported from equally type I and II CRISPR–Cas systems (Barrangou et al. 2007; Fineran et al. 2014).
Next, Staphylococcus epidermidis represents the type III CRISPR–Cas system. This system allows engineering of virulent staphylococcal phages. Type III system comprises of native endogenous CRISPR–Cas10 system but is accompanied with the crRNA transcribed from an exogenous plasmid (Bari et al. 2017). Interestingly, CRISPR–Cas10 system has an excellent cleavage frequency against elevated dose of staphylococcal phage infection.
5 Future Prospects
Bacteriophage till now, evidenced to be a versatile candidate, can be utilized both as prophylaxis and as therapy against an infection. Therefore, it can be applied equally before and after bacterial infection (Debarbieux et al. 2010; Chanishvili 2016; Tao et al. 2019).
5.1 Vaccines
Recent studies ventured the potential of phages in vaccine platform other than prophylactic or therapeutic platform. Bacteriophages are natural virus that display several properties like size, geometry, multivalent display, and ordered and repetitive structure equivalent to a natural mammalian virus (Bachmann and Jennings 2010; Zepp 2010). These characteristics are crucial for stimulating immune response and guidance for vaccine design, and the only difference is that they infect bacteria (Jończyk-Matysiak et al. 2017). In recent past, several efforts have been employed to enhance the use of phage in vaccine platform, such as T4 (Tao et al. 2013), MS2 (Fu and Li 2016), phages λ (Nicastro et al. 2014), and others (Tissot et al. 2010).
The foundation for using phages as antigen carrier vehicles engages assimilation of the viral antigen on phage capsid either in vivo or in vitro to form an original virus-mimicking particle (VMP) through fusion of antigen to a virus capsid protein. This enabled antigen to be displayed by the phages in a repetitive format, which is crucial for innate immune system activation (Shepardson et al. 2017). For small genome phages, fusion of antigen and capsid protein is easy to produce, in comparison to multifaceted phages such as T4. Bacteriophages do contain CpG (a vital ligand for human immune system Toll-like receptor 9), a crucial viral character, so phages can definitely elevate the level of innate immune response (Sartorius et al. 2015). Hence, this nature of phages displaying antigens confirms itself as a highly effective self-adjuvating vaccine delivery system that is qualified of eliciting a long-lasting immune response without any external adjuvants. Indeed, in 2013, Tao et al. proved that antigens embedded on T4 capsid elicited a much higher immune response associated to their soluble antigens. In another experiment, a phage QB VMP enveloped with CpG sensitizes antigen presenting cells in a faster and better way than a simple mixture of antigen (Gomes et al. 2017). Apart from that, it was also explored that phage QB capsid was able to bind natural IgM due to the display of highly ordered and repetitive format antigen. This capsid can also fix complement component 1q and easily deploy follicular dendritic cells (FDCs). Deploying of FDCs is indispensable or the choice of B cell during germinal center reactions (Link et al. 2012). Another huge advantage of phage VMPs and their display of highly restricted epitope density in a particular zone is the facilitated presentation by both class I and class II major histocompatibility complex (MHC). The feature of phage display sets off both CD4+ and CD8+ T cells, helping to generate long-lasting effective memory immune response (Tao et al. 2013, 2018a). A licensed viral vaccine must contain vastly localized epitope density and that is widely disseminated by bacteriophages (Cheng 2016). A promising strategy to enrich vaccine efficacy is by aiming of antigens to immune cells (Kastenmuller et al. 2014; Macri et al. 2016). Dendritic cells (DCs) are one of the crucial immune cells as they play the connecting link between innate and adaptive immune system (Steinman and Banchereau 2007). Although it is clear that some phage may have mammalian tropism, but most of them are not. So phages can be cleverly engineered to target DCs by displaying DC-specific targeting molecules. Sartorius et al. performed an experiment in 2015, where phage fd was modified to display a single-chain variable fragment of antibody against a DC-specific receptor-205 and separate group of phage also engineered to exhibit only ovalbumin through pVIII and pIII capsid protein. These groups were inoculated into the mice and found phages with DC-specific receptor that generated a higher titer of antibody compared to another group.
It must be mentioned that though phages have exclusive advantages, till now, there is no licensed vaccine based on phage platform that has yet been commercialized. Although several phage carrier–based vaccines are under clinical trial (Low et al. 2014; Huang et al. 2017), but most of these are still constrained to basic biological research. The reason behind this is quite clear: (a) A fair percentage of phages are unable to display the antigen in a highly localized or dense manner as an original mammalian viral vaccine, which is an utmost prerequisite to cultivate high titers of antibody. (b) Several pathogens are quite enabled to mutate specific prime amino acids in the epitopes, making vaccines constructed on one or limited epitopes, less to none effective. (c) Bacteria and bacteriophages don’t display posttranslational modification, so phages cannot display antigens that require posttranslational modifications. (d) Like natural protein nanoparticles, bacteriophages are also naturally enabled to elicit immune responses (Dąbrowska et al. 2014); therefore, these significantly decrease their chance of usage when multiple doses of vaccinations are required, although this setback can be fixed, first by epitope engineering. The epitopes that stimulate robust immune response (immunodominant epitope) (Akram and Inman 2012) can be identified, and their expression levels can be reduced by phage engineering. Second, PEGylation (addition of polyethylene glycol) is a process that enables phages for better solubility and diminution in renal clearance, hence increasing their bioavailability in the circulatory system (Suk et al. 2016).
5.2 Clinical Phage Therapy and Phage-Assisted Approaches
Phage is the undisputed winner in the list of alternative treatments against bacterial infection. Because phages follow “survival of the fittest policy,” they evolve with the selection condition and overcome bacterial resistance mechanism. Restriction modification (R-M) system is employed by bacteria to destroy invading DNA, but phages (not all phages) can integrate base modifications to keep their genome protected from bacterial R-M system (Samson et al. 2013). But phages like T4, restructured their genome cytosines by two alterations, first by 5-hydroxymethylation and second by glucosylation, and this enabled T4 phages to be impervious to majority of the restriction endonucleases of E. coli (Bryson et al. 2015). Phages can escape CRISPR–Cas through either expressing anti-CRISPR proteins (Pawluk et al. 2018) mutation through prime nucleotides responsible for CRISPR–Cas complex binding/cleavage (Tao et al. 2018b). Bacteria can make themselves unreachable by modifying their phage receptors, but phages can reclaim the capability of binding to their receptor by modifying the receptor-binding protein to adapt with the evolving bacterial population (Samson et al. 2013). Hence, co-evolution of phages parallel to host bacteria is a never-ending process, and this phenomenon makes bacteria a less protective form of phage therapy than antibiotics. Bacteriophage therapy also faces issue like highly specific and narrow host range. This feature of phages sometimes limits its use against all strain of a particular species. This limitation can be overcome (De Jonge et al. 2019). The host range can be expanded by the help of genetic engineering. Swapping the long tail fiber genes of T2 bacteriophage with those from phage PP01 swung the host of T2 from E. coli K12 to E. coli O157:H7 (Yoichi et al. 2005). Switching cognate receptor–binding protein genes between more indistinct bacteriophages could even empower an orchestrated E. coli phage to attack Klebsiella bacteria and vice versa (Ando et al. 2015). Various phages aiming various strains can be sequestered from the natural environment to target multiple strains. An excellent example will be a recent study of the “San Diego patient,” who was infected with a multidrug-resistant A. baumannii strain and regained their health after multiple intravenous injection of phage blends (Schooley et al. 2017).
Interestingly, phage-associated lysins, depolymerases and endolysin, can be consumed to lyse bacteria (Maciejewska et al. 2018). Here, depolymerases are polysaccharide-degrading enzymes, which are utilized to disintegrate capsular polysaccharides of pathogens, and in that way, phage gets access to cellular receptor on the bacterial cell surface. In preliminary experiments with depolymerase of PHB02 phage, when injected intraperitoneally, a significant surge in the persistence of mice pre-infected with P. multocida has been observed (Chen et al. 2018). Even though some of the phage-derived enzymes also have constricted infective capability, and they are competent to lyse a given bacterial species other than a single strain (Maciejewska et al. 2018).
All the above experiments do convey a message that synergistic approach of bacteriophages can be the most promising and also an emerging way to counter bacterial resistance in an efficient manner. As predicted, the most noticeable combination is antibiotics and phages. When utilized simultaneously, phages and antibiotics have displayed synergistic effects and effectiveness against biofilms (Chaudhry et al. 2017; Akturk et al. 2019), where the distinct treatments had limited success. Some experiments showed recurring medication with phages, which augmented the biofilm assembly but the mutual use of phage and antibiotics occasioned in biofilm eradication (Henriksen et al. 2019).
Synergism in between phages and antibiotics does not work for all phage–antibiotic blends, and a slightly increased dosage of antibiotics can effectively antagonize phage propagation (Dickey and Perrot 2019). This is predominantly evident when applying antibiotics that aims cellular protein synthesis (Akturk et al. 2019). But in rare cases, even though no synergism was observed, antimicrobial activity is displayed, and the combined utilization of phages and antibiotics drastically lowers or even limits the creation of antibiotic- and phage-resistant bacteria (Coulter et al. 2014; Dickey and Perrot 2019).
Phages and enzymes can be co-administered for enhanced result against stubborn infections, such as simultaneous use of depolymerases and phages that doesn’t naturally express them to get better efficiency against biofilms (Gutiérrez et al. 2015). Phages may be utilized combinedly with DNAse enzymes to diminish the DNA elements of the biofilm matrix and increase effectivity of bacteriophage activity (Hughes et al. 2006). Besides, some distinct productive cases of commingled phages include triclosan, chlorhexidine, chlorine (Zhang and Hu 2013), hydrogen peroxide (Agún et al. 2018), cobalt (II) sulfate (Chhibber et al. 2013), xylitol (Chhibber et al. 2015), probiotics (Woo and Ahn 2014), and honey (Oliveira et al. 2017).
So far, we have observed that majority of the phage-orchestrating technology have concentrated on lytic phages, but some experiments do talk about engineering of temperate bacteriophages for phage remedy purposes. The most confident methodology talks on genetically modifying temperate bacteriophages to become utterly virulent or lytic. This was successfully performed by deletion of the genomic segment accountable for the regulation and instituting lysogeny (Zhang and Hu 2013; Kilcher et al. 2018). The transformation of temperate wild types to otherwise virulent mutant phages can simply explore the miscellany of phages available for therapeutic use. A beautiful example of this methodology can be given; a recent study elaborated utility of cocktail phages constituted of one natural wild-type lytic phage and two engineered temperate phages efficiently that treated a 15-year-old patient with cystic fibrosis with a disseminated Mycobacterium abscessus infection (Dedrick et al. 2019).
Therefore, engineering tactics can possibly expand the antimicrobial features of phages and produce pioneering strategies for rebellious bacterial infections. The significances of genetic remodelling of phage genomes must be sensibly countered, but bacteriophage engineering tactics should be sincerely considered as an impending therapeutic alternative. Furthermore, it is noticed that engineered bacteriophages contain more commercial interest, as getting patent for engineered phages is far more easy than natural phages.
6 Conclusion
We have already entered into the post-antibiotic era and the imminent threat of antibiotic defiance, which requires instant action in the form of phage therapy or phage-assisted therapy. From this chapter, it is crystal clear that phage cure is well matched to denote itself a part of the multidimensional stratagems with versatile administration and engineering opportunities to fight against hostile, stubborn pathogens. So the theory says that phage remedy needs to be commenced in our stash of treatment approaches against multidrug-defiant pathogens, and certainly the sooner the better. Moreover, there is no such outstanding efficient approach to clinical use of phage remedy, and in fact, its miscellany and flexibility with the changing environment are among its utmost advantages. Although there are knowledge gaps that must be enlightened before we can take up the practice of phage remedy on a regular basis, this arena is like a never-ending gold mine and rapidly advancing. A better perceptive of bacteriophage pharmacology, genetics, and immunological interaction over the years has also signify bacteriophages as critical therapeutic agents. In conclusion, we analyzed that phage remedy is a rapidly evolving dynamic alternative against antibiotics, and extensive use of phage therapy is an extremely difficult task to be undertaken, but considering its medical, technological, societal, and economic prospects, it is the one and only leading alternative in repertoire of treatment strategies.
Abbreviations
- CRISPR:
-
Clustered regularly interspaced short palindromic repeat
- DS RNA:
-
Double-stranded RNA
- FDC:
-
Follicular dendritic cells
- GP:
-
Glycoprotein
- ICTV:
-
International Committee on Taxonomy of Viruses
- MHC:
-
Major histocompatibility complex
- PRMC:
-
Phage resisting microbial cells
- SS RNA:
-
Single-stranded RNA
- STIV:
-
Sulfolobus turreted icosahedral virus
References
Abrescia NG, Grimes JM, Kivelä HM, Assenberg R, Sutton GC, Butcher SJ, Bamford JK, Bamford DH, Stuart DI (2008) Insights into virus evolution and membrane biogenesis from the structure of the marine lipid-containing bacteriophage PM2. Mol Cell 31:749–761
Ackermann HW (2007) 5500 phages examined in the electron microscope. Arch Virol 152:227–243
Ackermann HW (2012) Bacteriophage electron microscopy. Adv Virus Res 82:1–32
Adriaenssens E, Brister JR (2017) How to name and classify your phage: an informal guide. Viruses 9:70
Agún S, Fernández L, González-Menéndez E, Martínez B, Rodríguez A, García P (2018) Study of the interactions between bacteriophage phiIPLA-RODI and four chemical disinfectants for the elimination of Staphylococcus aureus contamination. Viruses 10:103
Akram A, Inman RD (2012) Immunodominance: a pivotal principle in host response to viral infections. Clin Immunol 143:99–115
Akturk E, Oliveira H, Santos SB, Costa S, Kuyumcu S, Melo LDR, Azeredo J (2019) Synergistic action of phage and antibiotics: parameters to enhance the killing efficacy against mono and dual-species biofilms. Antibiotics 8:103
Alisky J, Iczkowski K, Rapoport A, Troitsky N (1998) Bacteriophages show promise as antimicrobial agents. J Infect 36:5–15
Ando H, Lemire S, Pires DP, Lu TK (2015) Engineering modular viral scaffolds for targeted bacterial population editing. Cell Syst 1:187–196
Appelt S, Fancello L, Le Bailly M, Raoult D, Drancourt M, Desnues C (2014) Viruses in a 14th-century coprolite. Appl Environ Microbiol 80:2648–2655
Bachmann MF, Jennings GT (2010) Vaccine delivery: a matter of size, geometry, kinetics and molecular patterns. Nat Rev Immunol 10:787–796
Baker ML, Jiang W, Rixon FJ, Chiu W (2005) Common ancestry of herpesviruses and tailed DNA bacteriophages. J Virol 79:14967–14970
Bari SMN, Walker FC, Cater K, Aslan B, Hatoum-Aslan A (2017) Strategies for editing virulent staphylococcal phages using CRISPR- Cas10. ACS Synth Biol 6:2316–2325
Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P (2007) CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709–1712
Benson SD, Bamford JK, Bamford DH, Burnett RM (1999) Viral evolution revealed by bacteriophage PRD1 and human adenovirus coat protein structures. Cell 98:825–833
Benson SD, Bamford JKH, Bamford DH, Burnett RM (2004) Does common architecture reveal a viral lineage spanning all three domains of life? Mol Cell 16:673–685
Bernhardt TG, Wang IN, Struck DK, Young R (2001) A protein antibiotic in the phage Q-beta virion: diversity in lysis targets. Science 292:2326–2329
Bertozzi Silva J, Storms Z, Sauvageau D (2016) Host receptors for bacteriophage adsorption. FEMS Microbiol Lett 363:fnw002
Bloch H (1940) Experimental investigation of the relationship between bacteriophage and malignant tumors. Arch Gesamte Virusforsch 1:481–496
Box AM, Mcguffie MJ, O’Hara BJ, Seed KD (2016) Functional analysis of bacteriophage immunity through a type I-E CRISPR-Cas system in Vibrio cholerae and its application in bacteriophage genome engineering. J Bacteriol 198:578–590
Breitbart M, Hewson I, Felts B, Mahaffy JM, Nulton J, Salamon P, Rohwer F (2003) Metagenomic analyses of an uncultured viral community from human feces. J Bacteriol 185:6220–6223
Brum JR, Schenck RO, Sullivan MB (2013) Global morphological analysis of marine viruses shows minimal regional variation and dominance of non-tailed viruses. ISME J 7:1738–1751
Bruttin A, Brüssow H (2005) Human volunteers receiving Escherichia coli phage T4 orally: a safety test of phage therapy. Antimicrob Agents Chemother 49:2874–2878
Bryson AL, Hwang Y, Sherrill-Mix S, Wu GD, Lewis JD, Black L, Clark TA, Bushman FD (2015) Covalent modification of bacteriophage T4 DNA inhibits CRISPR-Cas9. MBio 6:e00648
Cadwell K (2015) The virome in host health and disease. Immunity 42:805–813
Caliendo AM, Gilbert DN, Ginocchio CC, Hanson KE, May L, Quinn TC, Tenover FC, Alland D, Blaschke AJ, Bonomo RA, Carroll KC, Ferraro MJ, Hirschhorn LR, Joseph WP, Karchmer T, MacIntyre AT, Reller LB, Jackson AF, Infectious Diseases Society of America (IDSA) (2013) Better tests, better care: improved diagnostics for infectious diseases. Clin Infect Dis 57:S139–S170
Campbell A (1961) Conditions for the existence of bacteriophage. Evolution 15:153–165
Cao B, Yang M, Mao C (2016) Phage as a genetically modifiable supramacromolecule in chemistry, materials and medicine. Acc Chem Res 49:1111–1120
Chanishvili N (2016) Bacteriophages as therapeutic and prophylactic means: summary of the soviet and post-soviet experiences. Curr Drug Deliv 13:309–323
Chaudhry WN, Concepción-Acevedo J, Park T, Andleeb S, Bull JJ, Levin BR (2017) Synergy and order effects of antibiotics and phages in killing Pseudomonas aeruginosa biofilms. PLoS One 12:e0168615
Chen Y, Sun E, Yang L, Song J, Wu B (2018) Therapeutic application of bacteriophage PHB02 and its putative depolymerase against Pasteurella multocida capsular type a in mice. Front Microbiol 9:1678
Cheng W (2016) The density code for the development of a vaccine? J Pharm Sci 105:3223–3232
Cheng Q, Nelson D, Zhu S, Fischetti VA (2005) Removal of group B streptococci colonizing the vagina and oropharynx of mice with a bacteriophage lytic enzyme. Antimicrob Agents Chemother 49:111–117
Chhibber S, Kaur S, Kumari S (2008) Therapeutic potential of bacterio- phage in treating Klebsiella pneumoniae B5055-mediated lobar pneumonia in mice. J Med Microbiol 57:1508–1513
Chhibber S, Nag D, Bansal S (2013) Inhibiting biofilm formation by Klebsiella pneumoniae B5055 using an iron antagonizing molecule and a bacteriophage. BMC Microbiol 13:174
Chhibber S, Bansal S, Kaur S (2015) Disrupting the mixed-species biofilm of Klebsiella pneumoniae B5055 and Pseudomonas aeruginosa PAO using bacteriophages alone or in combination with xylitol. Microbiology 161:1369–1377
Chipman PR, Agbandje-McKenna M, Renaudin J, Baker TS, McKenna R (1998) Structural analysis of the Spiroplasma virus, SpV4: implications for evolutionary variation to obtain host diversity among the Microviridae. Structure 6:135–145
Cooper CJ, Khan Mirzaei M, Nilsson AS (2016) Adapting drug approval pathways for bacteriophage-based therapeutics. Front Microbiol 7:1209
Coulter LB, McLean RJ, Rohde RE, Aron GM (2014) Effect of bacteriophage infection in combination with tobramycin on the emergence of resistance in Escherichia coli and Pseudomonas aeruginosa biofilms. Viruses 6:3778–3786
Czaplewski L, Bax R, Clokie M, Dawson M, Fairhead H, Fischetti VA, Foster S, Gilmore BF, Hancock RE, Harper D, Henderson IR, Hilpert K, Jones BV, Kadioglu A, Knowles D, Ólafsdóttir S, Payne D, Projan S, Shaunak S, Silverman J, Thomas CM, Trust TJ, Warn P, Rex JH (2016) Alternatives to antibiotics-a pipeline portfolio review. Lancet Infect Dis 16:239–251
Dąbrowska K, Miernikiewicz P, Piotrowicz A, Hodyra K, Owczarek B, Lecion D, Kaźmierczak Z, Letarov A, Górski A (2014) Immunogenicity studies of proteins forming the T4 phage head surface. J Virol 88:12551–12557
Daniel A, Euler C, Collin M, Chahales P, Gorelick KJ, Fischetti VA (2010) Synergism between a novel chimeric lysin and oxacillin protects against infection by methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 54:1603–1612
De Jonge PA, Nobrega FL, Brouns SJJ, Dutilh BE (2019) Molecular and evolutionary determinants of bacteriophage host range. Trends Microbiol 27:51–63
Debarbieux L, Leduc D, Maura D, Morello E, Criscuolo A, Grossi O, Balloy V, Touqui L (2010) Bacteriophages can treat and prevent Pseudomonas aeruginosa lung infections. J Infect Dis 201:1096–1104
Dedrick RM, Guerrero-Bustamante CA, Garlena RA, Russell DA, Ford K, Harris K, Gilmour KC, Soothill J, Jacobs-Sera D, Schooley RT, Hatfull GF, Spencer H (2019) Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat Med 25:730–733
Devoto AE, Santini JM, Olm MR, Anantharaman K, Munk P, Tung J, Archie EA, Turnbaugh PJ, Seed KD, Blekhman R, Aarestrup FM, Thomas BC, Banfield JF (2019) Megaphages infect Prevotella and variants are widespread in gut microbiomes. Nat Microbiol 4:693–700
Dickey J, Perrot V (2019) Adjunct phage treatment enhances the effectiveness of low antibiotic concentration against Staphylococcus aureus biofilms in vitro. PLoS One 14:e0209390
Doron S, Melamed S, Ofir G, Leavitt A, Lopatina A, Keren M, Amitai G, Sorek R (2018) Systematic discovery of antiphage defense systems in in the microbial pangenome. Science 359:eaar4120
Duda RL, Teschke CM (2019) The amazing HK97 fold: versatile results of modest differences. Curr Opin Virol 36:9–16
Duerkop BA, Hooper LV (2013) Resident viruses and their interactions with the immune system. Nat Immunol 14:654–659
Dutilh BE, Cassman N, McNair K, Sanchez SE, Silva GG, Boling L, Barr JJ, Speth DR, Seguritan V, Aziz RK, Felts B, Dinsdale EA, Mokili JL, Edwards RA (2014) A highly abundant bacteriophage discovered in the unknown sequences of human faecal metagenomes. Nat Commun 5:4498
El Omari K, Sutton G, Ravantti JJ, Zhang H, Walter TS, Grimes JM, Bamford DH, Stuart DI, Mancini EJ (2013) Plate tectonics of virus shell assembly and reorganization in phage φ8, a distant relative of mammalian reoviruses. Structure 21:1384–1395
Eriksson F, Tsagozis P, Lundberg K, Parsa R, Mangsbo SM, Persson MA, Harris RA, Pisa P (2009) Tumor-specific bacteriophages induce tumor destruction through activation of tumor-associated macrophages. J Immunol 182:3105–3111
Fineran PC, Gerritzen MJ, Suárez-Diez M, Künne T, Boekhorst J, van Hijum SA, Staals RH, Brouns SJ (2014) Degenerate target sites mediate rapid primed CRISPR adaptation. Proc Natl Acad Sci U S A 111:E1629–E1638
Fish R, Kutter E, Wheat G, Blasdel B, Kutateladze M, Kuhl S (2016) Bacteriophage treatment of intransigent diabetic toe ulcers: a case series. J Wound Care 25(Suppl 7):S27–S33
Fish R, Kutter E, Bryan D, Wheat G, Kuhl S (2018) Resolving digital staphylococcal osteomyelitis using bacteriophage-a case report. Antibiotics (Basel) 7:87
Fraser JS, Yu Z, Maxwell KL, Davidson AR (2006) Ig-like domains on bacteriophages: a tale of promiscuity and deceit. J Mol Biol 359:496–507
Fraser JS, Maxwell KL, Davidson AR (2007) Immunoglobulin-like domains on bacteriophage: weapons of modest damage? Curr Opin Microbiol 10:382–387
Fu Y, Li J (2016) A novel delivery platform based on bacteriophage MS2 virus-like particles. Virus Res 211:9–16
Gomes AC, Flace A, Saudan P, Zabel F, Cabral-Miranda G, Turabi AE et al (2017) Adjusted particle size eliminates the need of linkage of antigen and adjuvants for appropriated T cell responses in virus-like particle-based vaccines. Front Immunol 8:226
Gorski A, Dąbrowska K, Switala-Jeleń K, Nowaczyk M, Weber-Dąbrowska B, Boratynski J, Wietrzyk J, Opolski A (2003) New insights into the possible role of bacteriophages in host defense and disease. Med Immunol 2:1–5
Górski A, Wazna E, Dąbrowska BW, Dąbrowska K, Switała-Jeleń K, Miedzybrodzki R (2006) Bacteriophage translocation. FEMS Immunol Med Microbiol 46:313–319
Górski A, Międzybrodzki R, Borysowski J, Dąbrowska K, Wierzbicki P, Ohams M, Korczak-Kowalska G, Olszowska-Zaremba N, Łusiak-Szelachowska M, Kłak M, Jończyk E, Kaniuga E, Gołaś A, Purchla S, Weber-Dąbrowska B, Letkiewicz S, Fortuna W, Szufnarowski K, Pawełczyk Z, Rogóż P, Kłosowska D (2012) Phage as a modulator of immune responses: practical implications for phage therapy. Adv Virus Res 83:41–71
Górski A, Międzybrodzki R, Weber-Dąbrowska B, Fortuna W, Letkiewicz S, Rogóż P, Jończyk-Matysiak E, Dąbrowska K, Majewska J, Borysowski J (2016) Phage therapy: combating infections with potential for evolving from merely a treatment for complications to targeting diseases. Front Microbiol 7:1515
Górski A, Dąbrowska K, Międzybrodzki R, Weber-Dąbrowska B, Łusiak-Szelachowska M, Jończyk-Matysiak E, Borysowski J (2017) Phages and immunomodulation. Future Microbiol 12:905–914
Górski A, Jończyk-Matysiak E, Łusiak-Szelachowska M, Międzybrodzki R, Weber-Dąbrowska B, Borysowski J, Letkiewicz S, Bagińska N, Sfanos KS (2018) Phage therapy in prostatitis: recent prospects. Front Microbiol 9:1434
Gregory AC, Zayed AA, Conceição-Neto N, Temperton B, Bolduc B, Alberti A, Ardyna M, Arkhipova K, Carmichael M, Cruaud C, Dimier C, Domínguez-Huerta G, Ferland J, Kandels S, Liu Y, Marec C, Pesant S, Picheral M, Pisarev S, Poulain J, Tremblay JÉ, Vik D, Tara Oceans Coordinators, Babin M, Bowler C, Culley AI, de Vargas C, Dutilh BE, Iudicone D, Karp-Boss L, Roux S, Sunagawa S, Wincker P, Sullivan MB (2019) Marine DNA viral macro- and microdiversity from pole to pole. Cell 177:1–15
Gutiérrez D, Briers Y, Rodríguez-Rubio L, Martínez B, Rodríguez A, Lavigne R, García P (2015) Role of the pre-neck appendage protein (Dpo7) from phage vb sepis-phiipla7 as an anti-biofilm agent in staphylococcal species. Front Microbiol 6:1315
Hatfull GF (2008) Bacteriophage genomics. Curr Opin Microbiol 11:447–453
Henriksen K, Rørbo N, Rybtke ML, Martinet MG, Tolker-Nielsen T, Høiby N, Middelboe M, Ciofu O (2019) P. aeruginosa flow-cell biofilms are enhanced by repeated phage treatments but can be eradicated by phage–ciprofloxacin combination monitoring the phage P. aeruginosa biofilms interactions. Pathog Dis 77:ftz011
Hermoso JA, Monterroso B, Albert A, Galan B, Ahrazem O, Garcia P, Martinez-Ripoli M, Garcia JL, Menendez M (2003) Structural basis for selective recognition of pneumococcal cell wall by modular endolysin from phage Cp-1. Structure 11:1239–1249
Hodyra-Stefaniak K, Miernikiewicz P, Drapała J, Drab M, Jończyk-Matysiak E, Lecion D, Kaźmierczak Z, Beta W, Majewska J, Harhala M, Bubak B, Kłopot A, Górski A, Dąbrowska K (2015) Mammalian host-versus-phage immune response determines phage fate in vivo. Sci Rep 5:14802
Hoyle N, Zhvaniya P, Balarjishvili N, Bolkvadze D, Nadareishvili L, Nizharadze D, Wittmann J, Rohde C, Kutateladze M (2018) Phage therapy against Achromobacter xylosoxidans lung infection in a patient with cystic fibrosis: a case report. Res Microbiol 169:540–542
Hua J, Huet A, Lopez CA, Toropova K, Pope WH, Duda RL, Hendrix RW, Conway JF (2017) Capsids and genomes of jumbo-sized bacteriophages reveal the evolutionary reach of the HK97 fold. mBio 8:e01579-17
Huang X, Wang X, Zhang J, Xia N, Zhao Q (2017) Escherichia coli derived virus-like particles in vaccine development. NPJ Vaccines 2:3
Huff WE, Huff GR, Rath NC, Donoghue AM (2010) Immune interference of bacteriophage efficacy when treating colibacillosis in poultry. Poult Sci 89:895–900
Hughes G, Walker JT, Sharp R, Hart A (2006) Bacteriophage for the treatment of bacterial biofilms. US7758856B2
Huiskonen JT, de Haas F, Bubeck D, Bamford DH, Fuller SD, Butcher SJ (2006) Structure of the bacteriophage φ6 nucleocapsid suggests a mechanism for sequential RNA packaging. Structure 14:1039–1048
Hupfeld M, Trasanidou D, Ramazzini L, Klumpp J, Loessner MJ, Kilcher S (2018) A functional type II-A CRISPR-Cas system from listeria enables efficient genome editing of large non-integrating bacteriophage. Nucleic Acids Res 46:6920–6933
Jaiswal A, Koley H, Ghosh A, Palit A, Sarkar B (2013) Efficacy of cocktail phage therapy in treating Vibrio cholerae infection in rabbit model. Microbes Infect 15:152–156
Jaiswal A, Koley H, Mitra S, Saha DR, Sarkar B (2014) Comparative analysis of different oral approaches to treat Vibrio cholerae infection in adult mice. Int J Med Microbiol 304:422–430
Jerne NK (1952) Bacteriophage inactivation by antiphage serum diluted in distilled water. Nature 169:117–118
Jerne NK (1956) The presence in normal serum of specific antibody against bacteriophage T4 and its increase during the earliest stages of immunization. J Immunol 76:209–216
Jończyk-Matysiak E, Weber-Dąbrowska B, Owczarek B, Międzybrodzki R, Łusiak-Szelachowska M, Łodej N, Górski A (2017) Phage-phagocyte interactions and their implications for phage application as therapeutics. Viruses 9:pii: E150
Karam JD, Drake JW (1994) Molecular biology of bacteriophage T4. American Society for Microbiology, Washington, DC
Kastenmuller W, Kastenmuller K, Kurts C, Seder RA (2014) Dendritic cell-targeted vaccines–hope or hype? Nat Rev Immunol 14:705–711
Kauffman KM, Hussain FA, Yang J, Arevalo P, Brown JM, Chang WK, VanInsberghe D, Elsherbini J, Sharma RS, Cutler MB, Kelly L, Polz MF (2018) A major lineage of non-tailed dsDNA viruses as unrecognized killers of marine bacteria. Nature 554:118–122
Kaur S, Harjai K, Chhibber S (2014) Bacteriophage-aided intracellular killing of engulfed methicillin-resistant Staphylococcus aureus (MRSA) by murine macrophages. Appl Microbiol Biotechnol 98:4653–4661
Khayat R, Tang L, Larson ET, Lawrence CM, Young M, Johnson JE (2005) Structure of an archaeal virus capsid protein reveals a common ancestry to eukaryotic and bacterial viruses. Proc Natl Acad Sci U S A 102:18944–18949
Kilcher S, Studer P, Muessner C, Klumpp J, Loessner MJ (2018) Cross- genus rebooting of custom-made, synthetic bacteriophage genomes in L-form bacteria. Proc Natl Acad Sci U S A 115:567–572
Kim MS, Bae JW (2018) Lysogeny is prevalent and widely distributed in the murine gut microbiota. ISME J 12:1127–1141
Kiro R, Shitrit D, Qimron U (2014) Efficient engineering of a bacteriophage genome using the type I-E CRISPR-Cas system. RNA Biol 11:42–44
Knott GJ, Doudna JA (2018) CRISPR-Cas guides the future of genetic engineering. Science 361:866–869
Koonin EV, Makarova KS, Zhang F (2017) Diversity, classification and evolution of CRISPR-Cas systems. Curr Opin Microbiol 37:67–78
Krupovic M, Prangishvili D, Hendrix RW, Bamford DH (2011) Genomics of bacterial and archaeal viruses: dynamics within the Prokaryotic virosphere. Microbiol Mol Biol Rev 75:610–635
Kurzepa A, Da̧browska K, Skaradziński G, Górski A (2009) Bacteriophage interactions with phagocytes and their potential significance in experimental therapy. Clin Exp Med 9:93–100
Labrie SJ, Samson JE, Moineau S (2010) Bacteriophage resistance mechanisms. Nat Rev Microbiol 8:317–327
Langdon A, Crook N, Dantas G (2016) The effects of antibiotics on the microbiome throughout development and alternative approaches for therapeutic modulation. Genome Med 8:39
Lebedev AA, Krause MH, Isidro AL, Vagin AA, Orlova EV, Turner J, Dodson EJ, Tavares P, Antson AA (2007) Structural framework for DNA translocation via the viral portal protein. EMBO J 26:1984–1994
Legendre DP, Muzny CA, Marshall GD, Swiatlo E (2014) Antibiotic hyper- sensitivity reactions and approaches to desensitization. Clin Infect Dis 58:1140–1148
Lemay ML, Tremblay DM, Moineau S (2017) Genome engineering of virulent lactococcal phages using CRISPR-Cas9. ACS Synth Biol 6:1351–1358
Lemire S, Yehl KM, Lu TK (2018) Phage-based applications in synthetic biology. Annu Rev Virol 5:453–476
Letkiewicz S, Miedzybrodzki R, Fortuna W, Weber-Dąbrowska B, Górski A (2009) Eradication of Enterococcus faecalis by phage therapy in chronic bacterial prostatitis - case report. Folia Microbiol (Praha) 54:457–461
Letkiewicz S, Międzybrodzki R, Kłak M, Jończyk E, Weber-Dąbrowska B, Górski A (2010) The perspectives of the application of phage therapy in chronic bacterial prostatitis. FEMS Immunol Med Microbiol 60:99–112
Lim ES, Zhou Y, Zhao G, Bauer IK, Droit L, Ndao IM, Warner BB, Tarr PI, Wang D, Holtz LR (2015) Early life dynamics of the human gut virome and bacterial microbiome in infants. Nat Med 21:1228–1234
Link A, Zabel F, Schnetzler Y, Titz A, Brombacher F, Bachmann MF (2012) Innate immunity mediates follicular transport of particulate but not soluble protein antigen. J Immunol 188:3724–3733
Loeffler JM, Fischetti VA (2003) Synergistic lethal effect of a combination of phage lytic enzymes with different activities on penicillin-sensitive and -resistant Streptococcus pneumoniae strains. Antimicrob Agents Chemother 47:375–377
Loeffler JM, Nelson D, Fischetti VA (2001) Rapid killing of Streptococcus pneumoniae with a bacteriophage cell wall hydrolase. Science 294:2170–2172
Loessner MJ (2005) Bacteriophage endolysins-current state of research and applications. Curr Opin Microbiol 8:480–487
Lood R, Winer BY, Pelzek AJ, Diez-Martinez R, Thandar M, Euler CW, Schuch R, Fischetti VA (2015) Novel phage lysin capable of killing the multidrug-resistant gram-negative bacterium Acinetobacter baumannii in a mouse bacteremia model. Antimicrob Agents Chemother 59:1983–1991
Low JG, Lee LS, Ooi EE, Ethirajulu K, Yeo P, Matter A, Connolly JE, Skibinski DA, Saudan P, Bachmann M, Hanson BJ, Lu Q, Maurer-Stroh S, Lim S, Novotny-Diermayr V (2014) Safety and immunogenicity of a virus-like particle pandemic influenza A (H1N1) 2009 vaccine: results from a double-blinded, randomized phase I clinical trial in healthy Asian volunteers. Vaccine 32:5041–5048
Łusiak-Szelachowska M, Zaczek M, Weber-Dąbrowska B, Międzybrodzki R, Kłak M, Fortuna W, Letkiewicz S, Rogóż P, Szufnarowski K, Jończyk-Matysiak E, Owczarek B, Górski A (2014) Phage neutralization by sera of patients receiving phage therapy. Viral Immunol 27:295–304
Maciejewska B, Olszak T, Drulis-Kawa Z (2018) Applications of bacteriophages versus phage enzymes to combat and cure bacterial infections: an ambitious and also a realistic application? Appl Microbiol Biotechnol 02:2563–2581
Macri C, Dumont C, Johnston AP, Mintern JD (2016) Targeting dendritic cells: a promising strategy to improve vaccine effectiveness. Clin Transl Immunol 5:e66
Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL (2012) Multidrug-resistant, extensively drug-resistant and pandrug- resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 18:268–281
Majewska J, Beta W, Lecion D, Hodyra-Stefaniak K, Kłopot A, Kaźmierczak Z, Miernikiewicz P, Piotrowicz A, Ciekot J, Owczarek B, Kopciuch A, Wojtyna K, Harhala M, Mąkosa M, Dąbrowska K (2015) Oral application of T4 phage induces weak antibody production in the gut and in the blood. Viruses 7:4783–4799
Manrique P, Bolduc B, Walk ST, van der Oost J, de Vos WM, Young MJ (2016) Healthy human gut phageome. Proc Natl Acad Sci U S A 113:10400–10405
Marinelli LJ, Piuri M, Swigonová Z, Balachandran A, Oldfield LM, van Kessel JC, Hatfull GF (2008) BRED: a simple and powerful tool for constructing mutant and recombinant bacteriophage genomes. PLoS One 3:e3957
Marinelli LJ, Hatfull GF, Piuri M (2012) Recombineering: a powerful tool for modification of bacteriophage genomes. Bacteriophage 2:5–14
Martel B, Moineau S (2014) CRISPR-Cas: an efficient tool for genome engineering of virulent bacteriophages. Nucleic Acids Res 42:9504–9513
Maslov S, Sneppen K (2017) Population cycles and species diversity in dynamic kill-the-winner model of microbial ecosystems. Sci Rep 7:39642
Matsuzaki S, Uchiyama J, Takemura-Uchiyama I, Daibata M (2014) Phage therapy: experiments using animal infection models. In: Borysowski J, Międzybrodzki R, Górski A (eds) Phage therapy: current research and applications. Caister Academic Press, Norfork, pp 237–256
McKenna R, Xia D, Willingmann P, Ilag LL, Krishnaswamy S, Rossmann MG, Olson NH, Baker TS, Incardona NL (1992) Atomic structure of single-stranded DNA bacteriophage φX174 and its functional implications. Nature 355:137–143
McMahon SA, Miller JL, Lawton JA, Kerkow DE, Hodes A, Marti-Renom MA, Doulatov S, Narayanan E, Sali A, Miller JF, Ghosh P (2005) The C-type lectin fold as an evolutionary solution for massive sequence variation. Nat Struct Mol Biol 12:886–892
Medhekar B, Miller JF (2007) Diversity-generating retroelements. Curr Opin Microbiol 10:388–395
Merril CR (2008) Bacteriophage ecology. Cambridge University Press, Cambridge
Merril CR, Trigg ME, Geier MR (1973) Fate of bacteriophage Lambda in non-immune germ-free mice. Nature 246:221–223
Merril CR, Biswas B, Carlton R, Jensen NC, Creed GJ, Zullo S, Adhya S (1996) Long-circulating bacteriophage as antibacterial agents. Proc Natl Acad Sci U S A 93:3188–3192
Międzybrodzki R, Kłak M, Jończyk-Matysiak E, Bubak B, Wójcik A, Kaszowska M, Weber-Dąbrowska B, Łobocka M, Górski A (2017) Means to facilitate the overcoming of gastric juice barrier by a therapeutic staphylococcal bacteriophage A5/80. Front Microbiol 8:467
Miernikiewicz P, Dąbrowska K, Piotrowicz A, Owczarek B, Wojas-Turek J, Kicielińska J, Rossowska J, Pajtasz-Piasecka E, Hodyra K, Macegoniuk K, Rzewucka K, Kopciuch A, Majka T, Letarov A, Kulikov E, Maciejewski H, Górski A (2013) T4 phage and its head surface proteins do not stimulate inflammatory mediator production. PLoS One 8:e71036
Minot S, Sinha R, Chen J, Li H, Keilbaugh SA, Wu GD, Lewis JD, Bushman FD (2011) The human gut virome: inter- individual variation and dynamic response to diet. Genome Res 21:1616–1625
Minot S, Grunberg S, Wu GD, Lewis JD, Bushman FD (2012) Hypervariable loci in the human gut virome. Proc Natl Acad Sci 109:3962–3966
Murphy KC (2012) Phage recombinases and their applications. Adv Virus Res 83:367–414
Nafissi N, Slavcev R (2014) Bacteriophage recombination systems and biotechnical applications. Appl Microbiol Biotechnol 98:2841–2851
Namura M, Hijikata T, Miyanaga K, Tanji Y (2008) Detection of Escherichia coli with fluorescent labeled phages that have a broad host range to E. coli in sewage water. Biotechnol Prog 24:481–486
Navarro F, Muniesa M (2017) Phages in the human body. Front Microbiol 8:566
Nelson D, Loomis L, Fischetti VA (2001) Prevention and elimination of upper respiratory colonization of mice by group A streptococci by using a bacteriophage lytic enzyme. Proc Natl Acad Sci U S A 98:4107–4112
Nguyen S, Baker K, Padman BS, Patwa R, Dunstan RA, Weston TA (2017) Bacteriophage transcytosis provides a mechanism to cross epithelial cell layers. MBio 8:e01874–e01817
Nicastro J, Sheldon K, Slavcev RA (2014) Bacteriophage lambda display systems: developments and applications. Appl Microbiol Biotechnol 98:2853–2866
Nigro OD, Jungbluth SP, Lin HT, Hsieh CC, Miranda JA, Schvarcz CR, Rappé MS, Steward GF (2017) Viruses in the oceanic basement. mBio 8:1–15
Nilsson AS (2014) Phage therapy constraints and possibilities. Ups J Med Sci 119:192–198
O’Neill J (2014) Antimicrobial resistance: tackling a crisis for the health and wealth of nations/The Review on Antimicrobial Resistance, chaired by Jim O’Neill. http://www.jpiamr.eu/wp-content/uploads/2014/12/AMR-Review-Paper-Tackling-a-crisis-for-the-health-and-wealth-of-nations_1-2.pdf
Ochs HD, Davis SD, Wedgwood RJ (1971) Immunologic responses to bacteriophage phi-X 174 in immunodeficiency diseases. J Clin Invest 50:2559–2568
Oda M, Morita M, Unno H, Tanji Y (2004) Rapid detection of Escherichia coli O157:H7 by using green fluorescent protein-labeled PP01 bacteriophage. Appl Environ Microbiol 70:527–534
Oliveira A, Ribeiro HG, Silva AC, Silva MD, Sousa JC, Rodrigues CF, Melo LDR, Henriques AF, Sillankorva S (2017) Synergistic antimicrobial interaction between honey and phage against Escherichia coli biofilms. Front Microbiol 8:2407
Paez-Espino D, Eloe-Fadrosh EA, Pavlopoulos GA, Thomas AD, Huntemann M, Mikhailova N, Rubin E, Ivanova NN, Kyrpides NC (2016) Uncovering Earth’s virome. Nature 536:425–430
Pan YJ, Lin TL, Chen CC, Tsai YT, Cheng YH, Chen YY, Hsieh PF, Lin YT, Wang JT (2017) Klebsiella phage PhiK64-1 encodes multiple depolymerases for multiple host capsular types. J Virol 91:pii: e02457-16
Pawluk A, Davidson AR, Maxwell KL (2018) Anti-CRISPR: discovery, mechanism and function. Nat Rev Microbiol 16:12–17
Peralta B, Gil-Carton D, Castaño-Díez D, Bertin A, Boulogne C, Oksanen HM, Bamford DH, Abrescia NG (2013) Mechanism of membranous tunnelling nanotube formation in viral genome delivery. PLoS Biol 11:e1001667
Perepanova TS, Merinov DS, Kazachenko AV, Khazan PL, Malova YA (2020) Bacteriophage therapy of urological infections. Urologiia 5:106–114
Petrovic Fabijan A, Lin RCY, Ho J, Maddocks S, Ben Zakour NL, Iredell JR et al (2020) Safety of bacteriophage therapy in severe Staphylococcus aureus infection. Nat Microbiol 5:465–472
Pietilä MK, Laurinmäki P, Russell DA, Ko CC, Jacobs-Sera D, Hendrix RW, Bamford DH, Butcher SJ (2013) Structure of the archaeal head–tailed virus HSTV-1 completes the HK97 fold story. Proc Natl Acad Sci U S A 110:10604
Pritchard DG, Dong S, Kirk MC, Cartee RT, Baker JR (2007) LambdaSa1 and LambdaSa2 prophage lysins of Streptococcus agalactiae. Appl Environ Microbiol 73:7150–7154
Puig A, Araujo R, Jofre J, Frias-Lopez J (2001) Identification of cell wall proteins of Bacteroides fragilis to which bacteriophage B40-8 binds specifically. Microbiology 147:281–288
Reyes A, Haynes M, Hanson N, Angly FE, Heath AC, Rohwer F, Gordon JI (2010) Viruses in the fecal microbiota of monozygotic twins and their mothers. Nature 466:334–338
Rixon FJ, Schmid MF (2014) Structural similarities in DNA packaging and delivery apparatuses in herpesvirus and dsDNA bacteriophages. Curr Opin Virol 5:105–110
Roach DR, Leung CY, Henry M, Morello E, Singh D, Di Santo JP, Weitz JS, Debarbieux L (2017) Synergy between the host immune system and bacteriophage is essential for successful phage therapy against an acute respiratory pathogen. Cell Host Microbe 22:38–47
Roux S, Krupovic M, Daly RA, Borges AL, Nayfach S, Schulz F, Sharrar A, Matheus Carnevali PB, Cheng JF, Ivanova NN, Bondy-Denomy J, Wrighton KC, Woyke T, Visel A, Kyrpides NC, Eloe-Fadrosh EA (2019) Cryptic inoviruses are pervasive in bacteria and archaea across Earth’s biomes. Nat Microbiol 4:1895–1906
Rubinstein A, Mizrachi Y, Bernstein L, Shliozberg J, Golodner M, Liu GQ, Ochs HD (2000) Progressive specific immune attrition after primary, secondary and tertiary immunizations with bacterio-phage phi X174 in asymptomatic HIV-1 infected patients. AIDS 14:F55–F62
Samson JE, Magadan AH, Sabri M, Moineau S (2013) Revenge of the phages: defeating bacterial defences. Nat Rev Microbiol 11:675–687
Sarker SA, McCallin S, Barretto C, Berger B, Pittet AC (2012) Oral T4-like phage cocktail application to healthy adult volunteers from Bangladesh. Virology 434:222–232
Sartorius R, D’Apice L, Trovato M, Cuccaro F, Costa V, De Leo MG, Marzullo VM, Biondo C, D’Auria S, De Matteis MA, Ciccodicola A, De Berardinis P (2015) Antigen delivery by filamentous bacteriophage fd displaying an anti-DEC-205 single-chain variable fragment confers adjuvanticity by triggering a TLR9-mediated immune response. EMBO Mol Med 7:973–88
Schilling T, Dietrich S, Hoppert M, Hertel R (2018) A CRISPR-Cas9- based toolkit for fast and precise in vivo genetic engineering of Bacillus subtilis phages. Viruses 10:pii: E241
Schmelcher M, Powell AM, Camp MJ, Pohl CS, Donovan DM (2015) Synergistic streptococcal phage λSA2 and B30 endolysins kill streptococci in cow milk and in a mouse model of mastitis. Appl Microbiol Biotechnol 99:8475–8486
Schooley RT, Biswas B, Gill JJ, Hernandez-Morales A, Lancaster J, Lessor L, Barr JJ, Reed SL, Rohwer F, Benler S, Segall AM, Taplitz R, Smith DM, Kerr K, Kumaraswamy M, Nizet V, Lin L, McCauley MD, Strathdee SA, Benson CA, Pope RK, Leroux BM, Picel AC, Mateczun AJ, Cilwa KE, Regeimbal JM, Estrella LA, Wolfe DM, Henry MS, Quinones J, Salka S, Bishop-Lilly KA, Young R, Hamilton T (2017) Development and use of personalized bacteriophage-based therapeutic cocktails to treat a patient with a disseminated resistant Acinetobacter baumannii infection. Antimicrob Agents Chemother 261:e00954–e00917
Schuch R, Nelson D, Fischetti VA (2002) A bacteriolytic agent that detects and kills Bacillus anthracis. Science 418:884–889
Shearer WT, Lugg DJ, Rosenblatt HM, Nickolls PM, Sharp RM, Reuben JM, Ochs HD (2001) Antibody responses to bacteriophage φX-174 in human subjects exposed to the Antarctic winter-over model of spaceflight. J Allergy Clin Immunol 107:160–164
Shen J, Zhou J, Chen GQ, Xiu ZL (2018) Efficient genome engineering of a virulent klebsiella bacteriophage using CRISPR-Cas9. J Virol 92:pii: e00534-18
Shepardson KM, Schwarz B, Larson K, Morton RV, Avera J, McCoy K, Caffrey A, Harmsen A, Douglas T, Rynda-Apple A (2017) Induction of antiviral immune response through recognition of the repeating subunit pattern of viral capsids is toll-like receptor 2 dependent. MBio 8(6):e01356-17
Shmakov S, Smargon A, Scott D, Cox D, Pyzocha N, Yan W, Abudayyeh OO, Gootenberg JS, Makarova KS, Wolf YI, Severinov K, Zhang F, Koonin EV (2017) Diversity and evolution of class 2 CRISPR-Cas systems. Nat Rev Microbiol 15:169–182
Speck P, Smithyman A (2016) Safety and efficacy of phage therapy via the intravenous route. FEMS Microbiol Lett 363:fnv242
Steinman RM, Banchereau J (2007) Taking dendritic cells into medicine. Nature 449:419–426
Suk JS, Xu Q, Kim N, Hanes J, Ensign LM (2016) PEGylation as a strategy for improving nanoparticle-based drug and gene delivery. Adv Drug Deliv Rev 99:28–51
Tanji Y, Furukawa C, Na SH, Hijikata T, Miyanaga K, Unno H (2004) Escherichia coli detection by GFP-labeled lysozyme-inactivated T4 bacteriophage. J Biotechnol 114:11–20
Tao P, Mahalingam M, Kirtley ML, van Lier CJ, Sha J, Yeager LA, Chopra AK, Rao VB (2013) Mutated and bacteriophage T4 nanoparticle arrayed F1-V immunogens from Yersinia pestis as next generation plague vaccines. PLoS Pathog 9:e1003495
Tao P, Wu X, Tang WC, Zhu J, Rao V (2017) Engineering of bacteriophage T4 genome using CRISPR-Cas9. ACS Synth Biol 6:1952–1961
Tao P, Mahalingam M, Zhu J, Moayeri M, Sha J, Lawrence WS, Leppla SH, Chopra AK, Rao VB (2018a) A bacteriophage T4 nanoparticle-based dual vaccine against Anthrax and Plague. MBio pii: e01926-18
Tao P, Wu X, Rao V (2018b) Unexpected evolutionary benefit to phages imparted by bacterial CRISPR-Cas9. Sci Adv 4:eaar4134
Tao P, Zhu J, Mahalingam M, Batra H, Rao VB (2019) Bacteriophage T4 nanoparticles for vaccine delivery against infectious diseases. Adv Drug Deliv Rev 145:57–72
Thandar M, Lood R, Winer BY, Deutsch DR, Euler CW, Fischetti VA (2016) Novel engineered peptides of a phage lysin as effective antimicrobials against multidrug-resistant Acinetobacter baumannii. Antimicrob Agents Chemother 60:2671–2679
Thomason LC, Oppenheim AB, Court DL (2009) Modifying bacteriophage lambda with recombineering. Methods Mol Biol 501:239–251
Tissot AC, Renhofa R, Schmitz N, Cielens I, Meijerink E, Ose V, Jennings GT, Saudan P, Pumpens P, Bachmann MF (2010) Versatile virus-like particle carrier for epitope-based vaccines. PLoS One 5:e9809
Valegard K, Liljas L, Fridborg K, Unge T (1990) The three-dimensional structure of the bacterial virus MS2. Nature 345:36–41
Van Belleghem JD, Clement F, Merabishvili M, Lavigne R, Vaneechoutte M (2017) Pro- and anti-inflammatory responses of peripheral blood mononuclear cells induced by Staphylococcus aureus and Pseudomonas aeruginosa phages. Sci Rep 7:8004
Vidaver AK, Koski RK, Van Etten JL (1973) Bacteriophage φ6: a lipid-containing virus of pseudomonas phaseolicola. J Virol 11:799–805
Wang IN, Smith DL, Young R (2000) Holins: the protein clocks of bacteriophage infections. Annu Rev Microbiol 54:799–825
Wang IN, Deaton J, Young R (2003) Sizing the holin lesion with an endolysin-beta-galactosidase fusion. J Bacteriol 185:779–787
Weber-Dąbrowska B, Mulczyk M, Górski A (2000) Bacteriophage therapy of bacterial infections: an update of our institute’s experience. Arch Immunol Ther Exp 48:547–551
Wenger SL, Turner JH, Petricciani JC (1978) The cytogenetic, proliferative and viability effects of four bacteriophages on human lymphocytes. In Vitro 14:543–549
White DW, Suzanne Beard R, Barton ES (2012) Immune modulation during latent herpesvirus infection. Immunol Rev 245:189–208
Whittard E, Redfern J, Xia G, Millard A, Ragupathy R, Malic S, Enright MC (2021) Phenotypic and genotypic characterization of novel polyvalent bacteriophages with potent in vitro activity against an international collection of genetically diverse Staphylococcus aureus. Front Cell Infect Microbiol 11:698909
Woo J, Ahn J (2014) Assessment of synergistic combination potential of probiotic and bacteriophage against antibiotic-resistant Staphylococcus aureus exposed to simulated intestinal conditions. Arch Microbiol 196:719–727
World Health Organization (2014) Antimicrobial resistance: global report on surveillance 2014. WHO Press, Geneva
World Health Organization (2015) Global action plan on antimicrobial resistance. WHO Press, Geneva
Xu J, Dayan N, Goldbourt A, Xiang Y (2019) Cryo-electron microscopy structure of the filamentous bacteriophage IKe. Proc Natl Acad Sci U S A 116:5493
Yoichi M, Abe M, Miyanaga K, Unno H, Tanji Y (2005) Alteration of tail fiber protein gp38 enables T2 phage to infect Escherichia coli O157:H7. J Biotechnol 115:101–107
Żaczek M, Łusiak-Szelachowska M, Jończyk-Matysiak E, Weber-Dąbrowska B, Międzybrodzki R, Owczarek B, Kopciuch A, Fortuna W, RogóŻ P, Górski A (2016) Antibody production in response to staphylococcal MS-1 phage cocktail in patients undergoing phage therapy. Front Microbiol 7:1681
Zepp F (2010) Principles of vaccine design-lessons from nature. Vaccine 28:C14–C24
Zhang Y, Hu Z (2013) Combined treatment of Pseudomonas aeruginosa biofilms with bacteriophages and chlorine. Biotechnol Bioeng 110:286–295
Zuo T, Lu XJ, Zhang Y, Cheung CP, Lam S, Zhang F, Tang W, Ching JYL, Zhao R, Chan PKS, Sung JJY, Yu J, Chan FKL, Cao Q, Sheng JQ, Ng SC (2019) Gut mucosal virome alterations in ulcerative colitis. Gut 68:1169–1179
Acknowledgments
I wish to acknowledge Dr. Bipransh Kumar Tiwary, Dr. Patha Sarathi Satpathi, and Dr. Nilanjan Chakraborty for encouraging me in writing this chapter. I also thank ICMR-DHR for giving me the platform to carry forward my research activities effectively.
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Ethics declarations
None to declare.
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Jaiswal, A. (2022). Phage Therapy: Genomics to Applications and Future Prospects. In: Saha, T., Deb Adhikari, M., Tiwary, B.K. (eds) Alternatives to Antibiotics. Springer, Singapore. https://doi.org/10.1007/978-981-19-1854-4_5
Download citation
DOI: https://doi.org/10.1007/978-981-19-1854-4_5
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-19-1853-7
Online ISBN: 978-981-19-1854-4
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)