Abstract
Alternaria spp. is a polyphagous necrotrophic pathogen and infects many crops. In tomato, two species of Alternaria, namely A. solani and A. alternata cause infection. Particularly, A. solani infects leaves/stem and causes early blight (EB), which is a major yield-limiting disease of tomato worldwide, while A. alternata only infects fruit and stem leading to canker disease. This virulent pathogen causes severe damages to both fruits and plants of tomato. In the past decades, this disease was managed through an integrated approach using chemicals and bio-fungicides as well as through host-plant resistance. In the era of molecular biology, the ongoing efforts to reduce the pathogenic nature of Alternaria species, integration of omics technologies such as genomics, transcriptomics, proteomics, and metabolomics have recently been an advanced approach for understanding the pathogenesis and defense mechanisms involved in Alternaria and tomato plant interaction. The studies of omics will offer a basis for improving breeding programs through genetic manipulation that will ultimately lead to the possible protection of tomatoes from EB infection. In this chapter, we have described the disease symptoms, epidemiology, and current integrated management practices for EB along with knowledge gaps. In addition, an attempt is made to highlight the current research progress in tomato plant responses against EB stress using omics tools. We also deliberate the break that recent technologies of omics can provide to investigate tomato–EB pathogen interaction to project potential management strategies through crop improvement.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Alternaria spp.
- Solanum lycopersicum
- Chemical and biological control
- Genomics
- Transcriptomics
- Proteomics
- Metabolomics
- Disease control
4.1 Introduction
Worldwide, tomato, Solanum lycopersicum L. is one of the most important vegetables cultivated for its edible fruits, grown for various purposes such as for use as fresh as well as several industrial purposes (Islam et al. 2013). According to a report of FAO (2018), the total world production of tomato was 182 million tones (MT), with China as the largest producer of tomato producing 61.5% MT of tomato annually, followed by India (19.4 MT), USA (12.6 MT), Turkey (12.2 MT), and Egypt (6.6 MT). Tomato is a rich source of 17% of vitamin C of the daily value, contains 4% carbohydrates, and <1% each of protein and fat (FAO 2018). However, the worldwide production of tomatoes is constrained by several biotic and abiotic stresses, which adversely affect the quantity, quality, and profitability (Engindeniz and Ozturk 2013).
In biotic stresses, the diseases caused by fungal pathogens are particularly crucial in terms of production and quality (Sain and Pandey 2016). During the cropping periods, tomato plants are attacked by several roots and foliar fungal diseases. The wilt caused by Rhizoctonia solani and Fusarium oxysporum f. sp. lycopersici and damping-off by Pythium aphanidermatum are the major root rot diseases, while early blight incited by Alternaria solani or A. alternata, Septoria leaf spot by Septoria lycopersici, and late blight by Phytophthora infestans are the major foliar fungal diseases (Agrios 2005).
Among these diseases, early blight (EB) is one of the most severe diseases of tomato, causing 50–90% loss of the total production worldwide under favorable condition (Iqbal et al. 2019). For the management of this disease, growers rely on the use of chemicals (Mizubuti et al. 2007) and biological fungicides. But, the bio-fungicides are slow in their activity, and due to the retention of chemical fungicide residues in the vegetables, their use should be minimized (Stangarlin et al. 2011) and necessitates an alternative for disease management. In addition, small farmers growing tomatoes do not practice protective gears during the application of chemical fungicides and are not aware of the dilution instructions, thus compromising their own safety (Damalas and Koutroubas 2015). Therefore, these requirements have become more severe, especially in the amounts of chemical residues remaining in the fresh vegetables (European Commission 2012).
During recent years, using omics technology for the management of diseases of tomatoes has been found helpful to reduce the fungicidal risks problems (AbuQamar et al. 2016). Understanding the host responses and mechanisms toward a particular disease by deploying omics technologies is essential to improve the defense mechanism of tomato plants through breeding programs or by emerging ad hoc biotechnology strategies. Particularly, there is a great interest to improve tomato crops that could be free from EB, due to its global relevance as fresh and processed produce. Available literature revealed that little work has been done on the role of omics technology such as genomics or transcriptomics, proteomics, and metabolomics in understanding the Alternaria × tomato interaction and the management of EB. This chapter captures the latest significant studies in epidemiology, host range, and current integrated disease management strategies. In addition, we focus on the modern approaches regarding recent omics interventions for the potential management of EB disease along with knowledge gaps to deliver a role for the exploitation of candidate genes of interest and their additional analyses, offering trait-specific markers suitable for the improvement of tomato.
4.2 Disease Symptoms and the Biology of Causal Organism
Different pathogenic species of Alternaria can be distinguished by the symptoms produced on different plant parts. Initially, symptoms appear on the lower leaves as concentric rings in dark brown spots, which is the primary characteristic symptom of this disease (Fig. 4.1a, b). During humid weather, the disease progresses upwards, the areas affected by pathogen merge and form dark brown patches on the whole leaves. Under severe conditions, infected leaves may shrink and fall prematurely, resulting in early defoliation. On fruits, the infection takes place at the stalk end in the form of dark brown spots near the place of attachment with the fruit (Fig. 4.1c).
Worldwide, five different species of Alternaria, namely A. alternata, A. linariae (syn. A. tomatophila), A. solani, A. tenuissima, and A. grandis have been identified as the causal agents of EB of tomato (Bessadat et al. 2017). However, A. solani (Ell. And Mart) and A. alternata (Fr.) are the prevalent species. The mycelium of A. solani consists of branched, septate, light brown hyphae, which with age become darker. Conidiophores are relatively shorter, i.e. 50–90μm with dark color. Alternaria conidia are typically beaked, muriform, dark, and borne single or in chains, with 5–10 transverse septa and some time in each conidium a few longitudinal septa are present (Fig. 4.1d). Alternaria alternata possesses much fluffy margin with off white color colonies, which turn into dusky neutral gray within 96 h. Later these colonies become nearly grayish black.
4.3 Host Range and Pathogen Variability
The Alternaria species has a wide host range. It infects both arable crops such as crucifers, solanaceous crops, leafy vegetables (Loganathan et al. 2016) and plantation crops like tea, coconut, etc. (Rao and Subrahamanyam 1976). Based on pathogenicity tests on tomatoes, both A. alternata and A. solani isolates have been classified under the virulent category (Loganathan et al. 2016). Few species of A. solani (non-pathogenic) have been found to promote growth in chili plants instead of its pathogenic nature (Mauricio-Castillo et al. 2020). It is also reported the A. solani isolated from the different hosts exhibited pronounced variability in their pathogenicity. Also, the growth of isolates was influenced by the type of nutrients provided in the media, and among the different sources of nutrition provided, V8 juice agar supported the sporulation of the fungus (Pasche et al. 2004; Kumar et al. 2008). Several researchers reported the effect of lights such as blue or UV light on the sporulation of A. solani and other species, A. tegetica, A. alternata, and A. kikuchiana (Prasad and Dutt 1974; Cotty 1987; Fourtouni et al. 1998).
4.4 Epidemiology and Disease Development
The Alternaria species infect tomatoes are overwintered in diseased plant debris. It can survive in, or on the soil, atleast one of perhaps several years. The pathogen is seed-borne (Khulbe and Sati 1987; Shahida and Abdul 1995) and can be introduced through the infected seeds. Primary infection takes place first on lower leaves, and conidia are formed in crop debris left in the soil. The conidia developed on the primary spot helps in the secondary spread of the disease. These conidia are blown by wind or water or insects through the neighboring leaves/plants. The infection generally occurs through stomata, but Alternaria spp. are also capable of direct penetration.
The disease severity was reported maximum in crops sown during June–July compared to September–October and January–April planted crops (Data and Mayee 1981). Prevalence of high humidity and soil moisture favors the disease development during July, August, and September months. The optimum temperature required for the growth of Alternaria spp. is 28–30 °C for A. solani and 20–25 °C for A. alternata (Sahi 1990; Singh 1995). Once the infection has occurred, conidial dispersion continues throughout the growing season. Datar and Mayee (1982) reported the maximum dispersal of conidia occurs during the advanced stage of the symptom development and particularly between 9 am and 12 pm.
Among the fungal diseases, EB incited by A. solani or A. alternata is one of the major severe concerns due to substantial yield losses in tomatoes. This ascomycete pathogen usually infects tomato, potato, and eggplant. The disease is promoted by warm temperature with long periods of leaf wetness, dew, rainfall, and dense cropping. During the fruiting period, tomato plants become more susceptible to this pathogen (Cerkauskas 2005; Momel and Pemezny 2006). Although the disease is termed as EB, it may occur at all stages of development. Early blight occurs in three phases, leaf spots, fruit rot, and stem canker. Still, the foliar phase is more destructive and accountable for significant economic losses sustained by tomato producers (Chaerani and Voorrips 2006). The EB fungus can survive for several days on the infected seeds, but it is still speculative that in the next season, whether the seed-borne inoculum serves as a source of primary infection (Datar and Mayee 1982).
4.5 Existing Disease Mitigation Strategies
For the long term management of this disease, integrated disease management (IDM) strategies such as crop rotation, breeding of resistant cultivars of tomato, use of chemical and bio-fungicides have been practiced. Since Alternaria is both seed and soil-borne pathogen, both seed treatment and foliar application are recommended for disease management. Chemical and biological controls are the frequently adopted control measure for EB.
4.5.1 Chemical Fungicides
As far as chemical fungicides are concerned, mancozeb, hexaconazole, and zineb are effective at different concentrations against EB in both in vitro and in vivo conditions (Raza et al. 2016). At present, mancozeb is the most frequently used fungicide against EB (Singh et al. 2020). Majumder et al. (2016) reported that ED50 (effective dose) of nanoformulation of mancozeb against A. solani was in the range of 1.31–2.79 mg/L. In addition, mancozeb has also reduced the disease incidence of EB in the Pusa Ruby variety of tomato (Kumar and Srivastava 2013; Gondal et al. 1993). Besides, mancozeb, hexaconazole (0.05%), and azoxystrobin have also significantly managed the EB (Kumar et al. 2007). However, in the study of Arunkumar (2006) only azoxystrobin at 0.05, 0.1, and 0.15% was found to be more effective against EB than chlorothalonil, pyraclostrobin, and mancozeb. On the contrary, Singh and Singh (2006) reported that hexaconazole was more effective than chlorothalonil, azoxystrobin, mancozeb, propineb, and copper oxychloride. Recently, Farooq et al. (2019) observed that pyraclostrobin was more efficient against EB pathogen at 500 ppm, than that of hexaconazole and carbendazim. The variable range of efficacy reported for the fungicides may be due to the different isolates of the pathogens or active ingredients present in the chemical fungicides.
In addition, the fungicide resistance has also been reported for the EB pathogen, A. solani due to the higher pathogenic and genetic variability among different isolates, isolated from various agro-climatic regions (Pasche et al. 2004) and it could also break down the genetic resistance of the host (van derWaals et al. 2004). Therefore, to reduce the risk of chemical fungicide resistance, fungicides rotation strategies, use of different modes of action of fungicides through mixing should be executed at the regional and national level where fungicide resistance is a severe problem in EB prone areas. The increased use of fungicides to mitigate EB of tomato requires the implementation of alternative disease control practices.
4.5.2 Biological and Botanical Control
In recent years, to minimize the use of chemical fungicides, investigations were carried out to use the microbial biocontrol agents (MBCAs) and botanicals to combat EB where it was severe. There are several formulations of Trichoderma spp. and Pseudomonas spp. available in the markets that can be used against EB, and their efficacy has been confirmed by conducting several investigations. In the late 2000s, Varma et al. (2008) investigated that foliar spray of T. viride reduced EB severity caused by A. solani. Other reports also evidenced that the antagonist’s Bacillus amyloliquefaciens, Pseudomonas fluorescens, and T. harzianum were efficiently controlled EB incidence in tomato (El-Rafai et al. 2003; Camlica and Tozlu 2019).
The antagonistic potential of these MBCAs is attributed to several extracellular enzymes, PAL (phenylalanine ammonia-lyase), defense enzyme and oxidative enzymes (polyphenol oxidase, peroxidase and superoxide dismutase), several antifungal metabolites, presence of several enzymes and secondary metabolites (β-1,3-glucanase) produced by these MBCAs (Montealegre et al. 2010; Chowdappa et al. 2013). However, the slow activity of the MBCAs based fungicides limits their application in EB management. In addition, the application of neem leaf extracts has also been used to control EB incidence in tomato (Raza et al. 2016). In particular, the active ingredient of neem leaf can be used for the formulation of next generation fungicides that will have broad application in IDM as well as to reduce the residue level and fungicide resistance problems.
4.6 Exploitation of Omics Approaches in Understanding Tomato × Alternaria Interactions and for EB Management
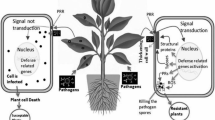
To reduce the losses in tomato due to EB, developing resistant varieties can be an economical and most effective management strategy (Panthee and Chen 2010; Adhikari et al. 2017). For the development of resistant varieties, investigators applied several genetic approaches. In particular, tomato plants show a high degree of similarity in gene sequence with other solanaceous crops (Kumar and Khurana 2014), making the investigation easy to understand the genetic programs based on interspecies knowledge transfer. The recent methodologies have established many efficient omics methods to untangle the molecular mechanisms of tomato plant response to A. solani to improve the detection and diagnosis of the pathogen (Fig. 4.2).
Historically, in an organism the genome is a whole set of chromosomes, which comprises all genes. The entire set of non-coding and coding RNAs is called a transcriptome, while the collected proteins derived from a genome are termed as proteome. Conversely, all metabolites present in the plant system are called the metabolome. However, the defense systems in plants against a particular pathogen cannot be studied uniquely through the genomic or transcriptomic methods, as they involve not only the expression of several defense-related genes, but also the incidence of post-translational modification or metabolites accumulation, affecting the final gene products expression.
The omics tools such as metabolomics and proteomics, enabling the proteins and metabolites interactions downstream of plant gene expression, may be practically pooled with genome and transcriptome. Although these approaches are complex, they can enhance our understanding of plant response mechanisms to fungal pathogen and other associated MBCAs, endophytes, and PGPR in a comprehensive way. Moreover, the methods of metagenomics enable the further understanding of the plant × associated microorganisms, offering an innovative prospect to sustain and manage the production of tomatoes at larger scale, based on microbiomes.
4.6.1 Search for the Resistant Cultivars against EB and Nature of Resistance
The investigations were carried out worldwide to search the resistant/tolerant cultivars of tomato against EB (Adhikari et al. 2017). However, till date there are few EB-resistant tomato genotypes available. Out of 401 tomato genotypes screened by Akhtar et al. (2019), only one genotype, i.e. “21,396” was found resistant against EB. In addition, some investigators found that several wild species (Solanum pimpinellifolium, S. peruvianum, S. chilense, and S. habrochaites) have been identified as potential sources of resistance against EB (Poysa and Tu 1996; Thirthamallappa 2000; Foolad et al. 2000). Thus, these wild species can be exploited in the breeding program.
In addition, HRC-G90.158, HRC90.145, HRC90.159, (Poysa and Tu 1996), and IHR1816 F (Thirthamallappa 2000) have shown resistance toward EB. From India, Lohith et al. (2011) reported four genotypes, such as EC251717, EC251709, EC164295, and LE15 of tomato resistant against EB. In a recent report six genotypes of tomato, such as NCEBR-1, NCEBR-4, Arka Rakshak, Arka Alok, Arka Saurabh, and 8-3-3 have shown EB resistance (Amarnath et al. 2019a, b); however, these genotypes were resistant in lab conditions and need further screening in the field.
Unfortunately, in the germplasm of tomatoes, there are only a few studies describing wide explorations for promising resistance sources to an EB pathogen (Adhikari et al. 2017; Nasr Esfahani 2019). In addition to 401 genotypes, Akhtar et al. (2019) also screened inbred lines and 72 genotypes from ten species of wild Solanum and found that none of the inbred lines was immune, highly resistant, or resistant. However, some genotypes derived from S. galapagense (1), S. peruvianum (1), S. pimpinellifolium (5), S. habrochaites (5 introgression lines), S. pennellii (2 introgression lines), S. lycopersicum E-6203 × S. pimpinellifolium LA1589 (eight RILs) showed moderately resistant reaction. In tomato, the nature of resistance is reported as polygenic in nature. Consequently, some genes present in tomato may confer resistance to the leaf blight, whereas others may contribute stem or fruit rot resistance (Stancheva et al. 1991; Chaerani et al. 2007). However, Barksdale and Stoner (1977) reported that stem lesion resistance of EB was independent of EB resistance on the leaves.
In the past decades, in the genotypes C1943 and 71B2, the EB resistance genes were reported recessive and not allelic (Maiero et al. 1989). However, the F1 hybrids were intermediate when these two resistance genes were crossed with another susceptible genotype, indicating partial dominance or additive genetic control (Maiero et al. 1989). Besides, the recessive genes have also been identified in the genotypes 83,602,029 (Stancheva et al. 1991) and IHR1816 and IHR1939 (Thirthamallappa 2000) derived from S. lycopersicum. In addition to this, in S. pimpinellifolium and S. habrochaites the partial dominant inheritance has been reported (Martin and Hepperly 1987). Another tomato genotype, i.e. 87B187 derived from PI390662 (S. habrochaites), shared common resistance genes with the genotype NCEBR-2 (Maiero et al. 1990a, b), even though this genotype was developed via S. lycopersicum source, C1943. Moreover, Thirthamallappa (2000) investigated independent genes in the genotypes IHR1816 and IHR1939, which were derived from S. pimpinellifolium and S. habrochaites, respectively.
4.6.2 Identification of Quantitative Trait Loci (QTLs) for Resistance to EB
The quantitative trait nature of EB makes selection more problematic as compared to the qualitative traits. In the tomato breeding programs, QTL analysis and development of molecular markers has been carried out in order to cognize the genetic control of EB resistance and to enable its introgression in tomatoes. Foolad et al. (2002) identified ten QTLs for EB through the crossing of resistant (PI126445) genotype derived from S. habrochaites and susceptible genotype (NC84173) of tomato, and each QTL explained total phenotypic variation in the range of 8.4–25.9%, while the collective effect was more than 57%. A list of QTLs identified by Foolad et al. (2002) for EB resistance in tomato is presented in Table 4.1. Later on, by selective genotyping, Zhang et al. (2003) identified QTLs conferring EB resistance in a L. esculentum × L. hirsutum cross. In addition, they also detected seven QTLs for EB resistance in a trait marker analysis (Zhang et al. 2003). However, the success in incorporating resistance in tomato is limited because most of the breeding lines such as NCEBR-4 (Gardner and Shoemaker 1999), NCEBR1, NCEBR-2 (Gardner 1988), and HRC90.303, HRC91.341 (Poysa and Tu 1996) were late maturing, relatively low yielding, and indeterminate. These accessions were derived from L. hirsutum. In 2007, Chaerani et al. (2007) identified three resistant QTLs to stem lesions from F2 and F3 populations derived from a cross between S. peruvianum LA2157 (resistant) and S. lycopersicum cv. Solentos (susceptible), and that explained 35% of the phenotypic variance. These QTLs can be used for the development of markers against EB in tomatoes.
4.6.3 Genomics Studies of Host and Pathogen
In the recent era of molecular biology, gene sequencing-based approaches remain economical, and both the pyro-sequencing and traditional Sanger dideoxy nucleotide have demonstrated their usefulness for confirmatory sequencing (Pareek et al. 2011). The EB pathogen, A. alternata or A. solani has become perfect for dividing the complexity of necrotrophic fungal pathogens and a wide range of pathogenicity of various crops. The pathogen may survive in diverse ecological stresses that promote or inhibit the infections on their host plants such as tomato (Ahlem et al. 2012).
Recently, based on conserved DNA sequences the genus Alternaria has been modernized (Ozkilinc et al. 2017; Woundenberg et al. 2014). It was confirmed that some species of Alternaria, i.e. A. grandis and A. protenta closely related to A. solani (Duarte et al. 2014), can also incite EB in tomato and potato (Ayad et al. 2017; Bessadat et al. 2016). To understand the A. alternata or A. solani–plant interactions in-depth at whole genome level, the whole genome sequence of A. alternata isolated from onion was studied. Its total genome size was 33.12 Mb with 50.9% GC content and 11,701 predicted coding sequences (Bihon et al. 2016). In addtion, A. alternata isolates from sorghum had 27 scaffolds, and the total genome size was 33.5 Mb (Nguyen et al. 2016). However, the partial sequence for A. alternata isolated from tomato is available (Gherbawy et al. 2018). Although, in the past, genomes of many Alternaria species (Hu et al. 2012), including A. solani, have been sequenced (Dang et al. 2015; Woudenberg et al. 2015), but due to analysis based on short-read sequencing, most of these genome assemblies were highly fragmented. Still, in discovering new genes, this information can be useful to clarify the classification and taxonomy of Alternaria species, and they enable comparative genomics.
Therefore, to produce fungal genomes having high-quality assemblies, use of long reads derived from PacBio-SMRT (Pacific Biosciences-single-molecule real-time) sequencing tools is a most prevalent method (Faino et al. 2015). This has been recently explained for the pathogen A. alternata (Nguyen et al. 2016). In particular, the assembly of a contiguous genome for the study of plant pathogenic fungi is essential because the genes coding the disease development effector proteins are often existing in fast-evolving that are challenging to assemble (Thomma et al. 2016). Likewise, understanding about related chromosomes and the gene organization helps in the gene cluster identification that has a major role in the secondary metabolite production, and together the characterization of potential provisionally expendable chromosomes helps in studying the pathogenicity of Alternaria spp. (Thomma et al. 2016).
Recently, Wolters et al. (2018) sequenced the A. solani causing EB in tomato and potato of genome size 33.1 Mb comprises about 99% of the total length of chromosomes. They identified that A. solani has ten chromosomes. Similar results were reported in an earlier study, in which genome sizes of A. solani was in the range of 32.6–32.9 Mb (Dang et al. 2015; Woudenberg et al. 2015). Besides A. solani genome sequenced by Wolters et al. (2018) showed a major advancement than that of the earlier A. solani genome assemblies, which consisted of over 100 separate contigs. Their genome sequencing analysis provides a concrete basis for the performance of comparative genomics, which will help to understand the molecular basis of pathogenicity of A. solani and other Alternaria species.
As far as the host is concerned, the first full genome sequence of tomato was carried out (Tomato Genome Consortium 2012), which describes 35,000 genes on 12 chromosomes. Later on, Li et al. (2018) sequenced genome of 360 varieties of tomato followed by Bolger et al. (2014) who sequenced the genome of S. pennellii, a stress-tolerant tomato wild species. The tomato plant contains 83 SlWRKY genes, which have several roles in the defense responses to both biotic and abiotic stresses (Bai et al. 2018). In the tomato plant, most of the WRKYs genes act as positive regulators of host responses to biotic stresses, whereas a lesser number of genes act as negative regulators.
The releases of sequences of whole genome of A. alternata and A. solani and their hosts will help in tackling the candidate genes responsible for virulence of Alternaria species and the potential target genes in the tomato plant associated with resistance against it. The genome sequences of tomatoes are very useful in understanding the plant defense system against Alternaria species. The sequencing of the genome of both host and pathogen will be also useful for the tomato breeders in developing resistant hybrids through the selection of defense-related genes in host crop or modification in virulent genes of the pathogen. As long as both Alternaria sp. and tomato genomes have been sequenced, the gene expression analysis through whole genome sequencing will tackle the critical factors in the pathogenesis of Alternaria spp. and mechanisms of EB resistance in tomato.
4.6.4 Transcriptomics
The comparative gene expression analyses can be utilized to mine the guiding information through transcriptomic technologies to generate data on biotic stress modulations of gene expression in tomato plants. In the modern era of molecular biology, RNAseq-based approaches are being used to study the transcriptomics in both model and non-model plants or pathogens (Warren et al. 2007). Remarkably, the transcriptome analysis of an organism helps to determine the pathogenesis-related proteins to be efficient to various biotic stress conditions (Ali et al. 2018). For instance, after the infection by a pathogen, plants produce pathogenicity-related (PR) proteins and chitinase in response to chitin, which is a major component of the cell wall of fungi (Adhikari et al. 2017).
The antifungal influence of chitinases and several hydrolytic enzymes has been determined against several foliar fungal pathogens, including A. solani. In addition, the genes accountable for the production of PR proteins have considerable enhanced resistance against pathogens causing EB and other pathogens in several arable crops (McNeece et al. 2019; Upadhyay et al. 2014a, b). When Alternaria infects the tomato plants, it suppresses both photosynthesis and metabolic processes such as glycolysis, electron transport chain, etc. At the same time, the defense-related genes, for instance, that encode chitinase, PR protein (PR2 and 3), and β-1, 3-glucanase showed a higher level in the highly EB-resistant species of tomato (Moghaddama et al. 2019). In addition, the expression of many secondary metabolites and defense-related genes in tomato plants were also upregulated when attacked by Alternaria.
In addition to the PR proteins, WRKY proteins also have a major role in the plant defense against the pathogens (Yang et al. 2018). In this regard, Moghaddam et al. (2019) reported that the expression pattern of antifungal genes 7 PR and 5 SlWRKYs genes in tomato increased 1–50-fold, when infected by A. alternata, and were upregulated among the resistant tomato varieties. In addition, the differential expression patterns of genes SlWRKY1 and SlWRKY11 were consistent with the expression pattern of genes PR7 and PDF1.2, which suggest that these transcription factors have a possible role in the enhancement of expression of PR genes in response to A. alternata infection.
Apart from EB, tomato plants also showed improved resistance to late blight caused by Phytophthora infestans (Cui et al. 2019) and S. arcanum to EB, A. solani (Shinde et al. 2018) due to the overexpression of WRKY1 gene. In an investigation, SlWRKY39 gene present in tomato was significantly upregulated in response to Pseudomonas syringae infection (Bai et al. 2018). In the same line, in response to Botrytis cinerea and A. brassicicola, the expression pattern of AtWRKY70 gene was altered, and changes in activity of AtWRKY70 genes might increase the susceptibility to B. cinerea, Erysiphe cichoracearum, gall formation by Linaria vulgaris, and Macrophomina phaseolina (Ulker et al. 2007; Lawaju et al. 2018; Pandey et al. 2016; Zorića et al. 2019).
In addition, the variable expression pattern of WRKY and PR defense-related genes is controlled not only by salicylic acid and jasmonic acid mediated signal events, but in between the resistant and susceptible genotypes of tomato infected by Alternaria species, the level of gene expression also varied (Pathak et al. 2017; Yang et al. 2015). Conversely, some plants showed resistance at seedling stages while becoming moderately resistant/susceptible at mature stage as has been reported for EB in potatoes incited by A. alternata (Nasr Esfahani et al. 2017). The experiment of Moghaddama et al. (2019) revealed that the tomato variety Esfahan local inoculated with pathogen showed an enhanced expression of defense-related genes and significant resistance at both young and mature stages, while the tomato variety Rio Grande showed resistance only at maturity stage. They also reported that in the inoculated EB-resistant tomato variety (H.a.s 2274) the expression of PR7 was upregulated at transplanting stage, and a strong expression in the inoculated resistant genotypes (Esfahan local, H.a.s 2274 and Rio Grande) was reported at the maturing stage (Moghaddama et al. 2019). Therefore, from the above findings, it is suggested that these are the key genes activating the defense response in host plant to the pathogen.
Besides, in tomato plants PR7 gene encoding 69 endopeptidase (Moghaddama et al. 2019), has been reported as proteases induced by the pathogen (Jorda and Vera 2000), and the fungal activities of PR7 gene is shown by another investigation (Golshani et al. 2015). The PR7 defense gene has also been found to be expressed during in several others interaction of the pathogen with hosts, comprising Pseudomonas syringae (Jorda and Vera 2000) and Phytophthora infestans (Tian et al. 2007) infections. The enzymatic activity of PR2 and PR3 proteins (β-1, 3-glucanase and chitinase) in the enhancement of defenses in tomato against EB has also been studied (Moghaddama et al. 2019), which revealed that both enzymes had a significant contribution to the protection of tomato from EB. Some studies revealed that the release of glucanase and chitinase in the form of hydrolytic products of induced PR genes disturbs the virulence of fungal pathogens and endorses the plant immunity responses (Kumar et al. 2018; Pusztahelyi 2018). Further, an investigation reported that among 32 genes present in the resistant genotype of tomato (EC-520061), 20 genes were upregulated against EB whereas in case of the CO-3, a susceptible genotype, no significant upregulation in fold change was examined (Upadhyay et al. 2016). Thus, these studies showed that these enzymes and genes significantly impact the EB resistance in tomatoes.
These results approve the crosstalk existence at the tomato plant retorts to Alternaria spp., involving several hormone signaling pathways, which alter the rate of photosynthesis, transport of proteins and their synthesis, thereby emphasizing the complexity of cellular signaling networks in tomato plants (AbuQamar et al. 2016). In addition, the incorporation of genomics and transcriptomics data of tomato or EB pathogen, along with proteomics will detect the biomarkers for EB pathogen. These omics data sets (transcriptomics and proteomics data) can build a vigorous model of functional features of biological pathways linking the transcripts and proteins.
4.6.5 Proteomics
In a host plant, the outcome of the incompatible and compatible host-pathogen interaction is determined through proteome analysis and associated metabolites. Independently, proteomic and metabolic profiling, or in the permutation with transcriptome data, provides additional understanding about the mechanisms of host defense response at the molecular level (Sharma et al. 2007; Tenenboim and Brotman 2016; Kumar et al. 2014). As far as EB of tomato is concerned, fewer studies regarding the proteomics analysis of tomato plants infected with EB have been carried out. However, literature is available for the other hosts such as Brassica and other crops infected by Alternaria species. The level of 48 proteins was significantly affected at several points in the tolerant lines of Brassica spp. when infected by A. brassicae, which suggested that the role of ROS (reactive oxygen supply) mediated auxin signaling in pathosystem of Alternaria sp. (Sharma et al. 2007).
Likewise, the level of 210 proteins in the Mentha arvensis leaves affected/altered during infection by A. alternata identified by matrix assisted laser desorption or ionization time of flight-mass spectrometry of them 29% of the proteins was defense-related proteins (Sinha and Chattopadhyay 2011). In another pathosystem of tomato, it was found that there was alternation in 186 proteins in wild-type mature green fruits infected by Botrytis cinerea, which were unaltered in wild-type red ripe fruits (RR). However, less defense-related proteins were altered in mature wild-type green tomato fruits than in RR tomato fruits (Shah et al. 2012). Therefore, further investigations are required to study the proteome analysis of tomato plants infected with EB to understand changes in the protein level. However, as far as tomato-EB interaction is concerned, the proteomic study can be compounded by the existence of pathogen proteins, which can be determined through the accessible full genome sequence of tomato and Alternaria spp.
4.6.6 Metabolomics
Each plant or pathogen contained metabolites, and these are organic compounds classify under the end product of plant metabolism or gene expression. Secondary metabolites present in plants have several roles in defense against pests and pathogens, and any changes in these metabolites affect the plant defense to the pathogens (Yuan et al. 2017). Conversely, secondary metabolites react with particular stress conditions, either biotic or abiotic, for example, ROS scavengers, pathogens, coenzymes, regulatory molecules, and antioxidants. Metabolomic profiling is carried out through NMR (nuclear magnetic resonance spectrometry) or MS (mass spectrometry), such as GC (gas chromatography)-MS and LC (liquid chromatography)-MS (Gathungu et al. 2014; Sharma et al. 2018).
Like other necrotrophic fungi infecting arable crops, the genus Alternaria often produces various phytotoxins and secondary metabolites, as “killing” weapons to the host cells from a wide range of plant species (Encinas-Basurto et al. 2017). Approximately, the 70 phytotoxins sill has been recognized that is produced by the several species of Alternaria, some are host-specific, and some are non-host specific (Johann et al. 2012; Escrivá et al. 2017). The major toxins produced by Alternaria include alternariol monomethyl ether (AME), alternariol (AOH), and altertoxin I and altertoxin II (Jarolim et al. 2017) which have several side effects in humans as well as in plants (Wenderoth et al. 2019). However, these phytotoxins have a phytotoxic minor impact on the host plant. Still, majorly they support in the colonization process of the pathogen inside the host by compensating the response of plant hypersensitive (Touhami et al. 2018). After colonization, they inhibit the enzymatic reactions within the host tissue or lead to death or necrosis of plant cells. In more resistant tomato genotypes, a correlation between the reduction in the production of AOH and a hogA mutant of A. alternata was taken as a sign for the role of Alternaria toxin AOH as a supporting factor in the virulence and colonization (Wojciechowska et al. 2014). However, AOH supports the colonization of the fungus (Wenderoth et al. 2019).
In addition, during interaction of wild tomato × A. solani, a significant modulation in secondary metabolites have been identified. In this regard, Shinde et al. (2017) reported that secondary metabolites (phytoalexins, phenylpropanoids, lignin accumulation) synthesized in a resistant wilt tomato species, namely S. arcanum through steroidal-glycoalkaloid and phenylpropanoid pathways has significant role in protection against EB. The WRKY and MYB in WRKY1 genes had major role in secondary metabolites synthesis pathways, and in resistant plant, the lignin biosynthesis that was regulated by transcription factors was upregulated (Shinde et al. 2017).
During the infection, both host and pathogen release metabolites mediate the resistance response in the host. Some secondary metabolites, such as 3-methyl-2-butenal, dimethyl disulfide, 1-butanol, hexanol, and 2-methyl-1-butanol acetate responsible for resistance in tomato fruits were synthesized on tomato only when infected by A. alternata (Johanson and Thurston 1990). The primary, secondary metabolites correlated to tomato-EB resistance include a higher level of flavonol, tannin, and phenolic compounds in both stems and leaves, as has been reported in EB-resistant cultivars (Bhatia et al. 1972).
The production of these metabolites is associated with several mechanisms. The peroxidase (PO) present in the host plant plays an important role in the production of reactive oxygen (RO), these RO are directly or indirectly toxic to the fungal pathogen infecting plants (Hammond-Kosack and Jones 1996). The phenylalanine lyase (PAL) is also an important enzyme in the secondary molecules synthesis (Mauch-Mani and Slusarenko 1996), which help in the activation of the expression pattern of a variety of pathogenesis-related genes. Moreover, in response to the A. solani, the polyphenol oxidase (PPOs) is systemically upregulated, examined in the upper nodes of leaves, but absent in the lower nodes of tomato leaves (Thipyapong and Steffens 1997). This induction pattern of PR genes in tomato leaves accords with the observation of transient resistance of young leaves of tomato to A. solani infection (Johanson and Thurston 1990). The oxidation of phenols to quinones is catalyzed by PPOs and sensitive molecules that encourage the death of pathogen cell and blocks to the secondary infection in the host plant (Thipyapong and Steffens 1997).
The expression pattern of PR-1B increased when salicylic acid was applied on tomato roots to prevent the infection from EB (Spletzer and Enyedi 1999). After leaf treatment of tomato with arachidonic acid, the PR-1-like protein level increased (Coquoz et al. 1995), and the sequential expression of ST-ACS4 and ST-ACS5, ACC synthase genes also reported in potato plants (Schlagnhaufer et al. 1997). In a recent report, the total phenol contents of tomato were significantly increased as a response to A. solani infection (Attiaa et al. 2020). The remarkable metabolic changes in tomato upon infection with Alternaria spp. cause metabolic perturbations, both in the plant and the fungal pathogen. A recent study reports that, at transplanting stage, the activity of PAL increased 5-fold and TPC 4-fold, when the resistant tomato plant was inoculated by EB pathogen, while 2–3 fold increased in TPC activity and 3-fold in POD was reported at maturity stage (Alizadeh-Moghaddam et al. 2020).
Thus, the above described results suggest the resistant genotypes of tomato can be differentiated from susceptible genotypes through using both genetic and enzymatic diversity to EB.
4.7 Conclusions and Future Remarks
The descriptive information on the EB of tomato unveils a wealth of information which is regarded, for instance, pathogen epidemiology, integrated disease management, and role of omics in EB management. However, investigations in some important areas need further attention. There are few EB-resistant varieties of tomato and QTLs are available, and it will need to be investigated in the future research program. Although few QTLs are available for EB of tomato, however, to avoid integration of large parts of the donor genome along with the resistance gene, fine mapping is needed before these can be used in a marker-assisted breeding program. Also, before QTLs are deployed in a tomato breeding program, their pleiotropic effects on other traits should be investigated in future research projects.
In the era of molecular approaches, there is no doubt that the exploitation of omics in the potential disease mitigation is delivering toward understanding the mechanism at the molecular level of the tomato plant resistance to the Alternaria sp. The breeders are making potential efforts to link the resistant genes with traits to improve the resistance of tomato cultivars and understand the mechanisms of disease resistance. Therefore, to make the sustainable production of tomato, scientists must adopt innovative technologies to develop the high yield and EB-resistant varieties of tomato.
Omics enables the researchers to identify, isolate the desired genes and traits. It helps to interpret the complex interaction among genes and helps in creating tools to enhance crop productivity. This article provides a comprehensive overview of “omics” technologies and its application in agriculture to combat major problems of crops especially related to field pathogens, for example, EB of tomato. Through omics technologies, the consistency and predictability of plant genetic engineering and breeding will be significantly improved by reducing the time and expense for producing EB-resistant tomato crops. There is an urgent need to create an environment where modern tools like omics can be conveniently used and comprehensively regarded as important keys to combat other diseases of tomato crops, including EB. These can still be used with conventional tools of disease diagnostics and management, thus bridging the knowledge gaps and enabling us with a better understanding of plant disease management under conditions like climate change.
References
AbuQamar SF, Moustafa K, Tran L-SP (2016) ‘Omics’ and plant responses to Botrytis cinerea. Front Plant Sci 7:1658
Adhikari P, Oh Y, Panthee DR (2017) Current status of early blight resistance in tomato: an update. Int J Mol Sci 18:1–22
Agrios GN (2005) Plant pathology, 5th edn. Academic Press, Oxford
Ahlem H, Mohammed E, Badoc A, Ahmed L (2012) Effect of pH, temperature and water activity on the inhibition of Botrytis cinerea by Bacillus amyloliquefaciens isolates. Afr J Biotechnol 11:2210–2217
Akhtar KP, Ullah N, Saleem MY, Iqbal Q, Asghar M, Khan AR (2019) Evaluation of tomato genotypes for early blight disease resistance caused by Alternaria solani in Pakistan. J Plant Pathol 101:1159–1170
Ali S, Ganai BA, Kamili AN, Bhat AA, Mir ZA, Bhat JA et al (2018) Pathogenesis-related proteins and peptides as promising tools for engineering plants with multiple stress tolerance. Microbiol Res 212:29–37
Alizadeh-Moghaddam G, Rezayatmand Z, Nasr-Esfahani M, Khozaei M (2020) Bio-genetic analysis of resistance in tomato to early blight disease, Alternaria alternata. Phytochemistry 179:112486
Amarnath M, Ravishankar KV, Satish D, Sriram S, Sadashiva AT (2019a) Screening of tomato genotypes against early blight pathogen Alternaria solani using detached leaf assay method. Pest Manage Horticult Ecosyst 25:93–101
Amarnath M, Ravishankar KV, Satish D, Sriram S, Sadashiva AT (2019b) Screening of tomato genotypes against early blight pathogen Alternaria solani using detached leaf assay method. Pest Manage Hortic Ecosyst 25:93–101
Arunkumar KT (2006) Studies on Alternaria solani (Ellis and Martin) Jones and Grout causing early blight of tomato. M.Sc. thesis, University of Agricultural Science, Bangalore
Attiaa MS, El-Sayyad GE, Abd Elkodousd M (2020) The effective antagonistic potential of plant growth-promoting rhizobacteria against Alternaria solani-causing early blight disease in tomato plant. Sci Hortic 266:109289
Ayad D, Leclerc S, Hamon B, Kedad A, Bouznad Z, Simoneau P (2017) First report of early blight caused by Alternaria protenta on potato in Algeria. Plant Dis 101:836
Bai Y, Sunarti S, Kissoudis C, Visser RGF, van der Linden CG (2018) The role of tomato WRKY genes in plant responses to combined abiotic and biotic stresses. Front Plant Sci 9:801
Barksdale TH, Stoner AK (1977) A study of the inheritance of tomato early blight resistance. Plant Dis Rep 61:63–65
Bessadat N, Hamon B, Henni DE, Simoneau P (2016) First report of tomato early blight caused by Alternaria grandis in Algeria. Plant Dis 100:533
Bessadat N, Berruyer R, Hamon B, Bataille-Simoneau N, Benichou S, Kihal M, Henni DE, Simoneau P (2017) Alternaria species associated with early blight epidemics on tomato and other Solanaceae crops in northwestern Algeria. Eur J Plant Pathol 148:181–197
Bhatia IS, Uppal DS, Bajaj KL (1972) Study of phenolic contents of resistant and susceptible varieties of tomato (Lycopersicum esculentum) in relation to early blight disease. Indian Phytopathol 25:231–235
Bihon W, Cloete M, Gerrano AS, Oelofse D, Adebola P (2016) Draft genome sequence of Alternaria alternata isolated from onion leaves in South Africa. Genome Announc 4:e01022–e01016
Bolger A et al (2014) The genome of the stress-tolerant wild tomato species Solanum pennellii. Nat Genet 46:1034–1038
Camlica E, Tozlu E (2019) Biological control of Alternaria solani in tomato. Fresen Environ Bull 28(10):7092–7100
Cerkauskas R (2005) Early blight, AVRDC, the world vegetable centre, www.avrdc.org
Chaerani R, Voorrips RE (2006) Tomato early blight (Alternaria solani): the pathogen, genetics, and breeding for resistance. J Gen Plant Pathol 72:335–347
Chaerani R, Smulders MJM, Van Der Linden CG, Vosman B, Stam P, Voorrips RE (2007) QTL identification for early blight resistance (Alternaria solani) in a Solanum lycopersicum × S arcanum cross. Theor Appl Genet 114:439–450
Chowdappa P, Kumar SM, Lakshmi MJ, Upreti KK (2013) Growth stimulation and induction of systemic resistance in tomato against early and late blight by Bacillus subtilis OTPB1 or Trichoderma harzianum OTPB3. Biol Control 65:109–117
Coquoz JL, Buchala AJ, Meuwly PH, Métraux JP (1995) Arachidonic acid induces local but not systemic synthesis of salicylic acid and confers systemic resistance in potato plants to Phytophthora infestans and Alternaria solani. Phytopathology 85:1219–1224
Cotty JP (1987) Modulation of sporulation of Alternaria tagetica by carbon dioxide. Mycologia 74:508–513
Cui J, Jiang N, Meng J, Yang G, Liu W, Zhou X, Ma N (2019) LncRNA33732- respiratory burst oxidase module associated with WRKY1 in tomato- Phytophthora infestans interactions. Plant J 97:933–946
Damalas CA, Koutroubas DS (2015) Farmers’ exposure to pesticides: toxicity types and ways of prevention. Toxics 4:1–10
Dang HX, Pryor B, Peever T, Lawrence CB (2015) The Alternaria genomes database: a comprehensive resource for a fungal genus comprised of saprophytes, plant pathogens, and allergenic species. BMC Genomics 16:239
Data VV, Mayee CD (1981) Epidemiology of early blight of tomato foliage to early blight caused by Alternaria solani. Indian Phytopathol 33:83–86
Datar VV, Mayee CD (1982) Conidial dispersal of Alternaria solani in tomato. Indian Phytopathol 35:69–70
Duarte HSS, Zambolim L, Rodrigues FA, Paul PA, Pádua JG, Ribeiro Júnior JI, Júnior AFN, Antonio F, Rosado AW (2014) Field resistance of potato cultivars to foliar early blight and its relationship with foliage maturity and tuber skin types. Trop Plant Pathol 39:294–306
El-Rafai IM, Asswah SMW, Awdalla OA (2003) Biocontrol of some tomato diseases using some antagonistic microorganisms. Pak J Biol Sci 6:399–406
Encinas-Basurto D, Valenzuela-Quintanar MI, Sánchez-Estrada T-HM, Rodríguez-Félix A, Troncoso-Rojas R (2017) Alterations in volatile metabolites profile of fresh tomatoes in response to Alternaria alternata (Fr) Keissl infection. Chilean J Agric Res 77:194
Engindeniz S, Ozturk GC (2013) An economic comparison of pesticide application for processing and table tomatoes, a case study for Turkey. J Plant Prot Res 53:230–237
Escrivá L, O’ueslate S, Font G, Manyes L (2017) Alternaria mycotoxins in food and feed. An overview. J Food Quality 2017:1569748
European Commission (2012) Amending Annex I to Regulation (EC) No 669/2009 implementing Regulation (EC) No 882/2004 of the European Parliament and of the Council as regards the increased level of official controls on imports of certain feed and food of non-animal origin. Off J Eur Union 54:45–50
Faino L, Seidl MF, Datema E, van den Berg GCM, Janssen A, Wittenberg AHJ, Thomma BPHJ (2015) Single-molecule real-time sequencing combined with optical mapping yields completely finished fungal genome. M Bio 6:e00936–e00915
FAO (2018) Tomato production in 2018, crops/regions/world list/production quantity (pick lists). UN Food and Agriculture Organization, Corporate Statistical Database (FAOSTAT), Rome
Farooq S, Jat RR, Gupta A, Singh R, Majeed M, Nabi SU et al (2019) Evaluation of different fungicides against Alternaria solani (Ellis & Martin) Sorauer cause of early blight of tomato under laboratory conditions. Pharma Innov J 8:140–142
Foolad MR, Ntahimpera N, Christ BJ, Lin GY (2000) Comparison between field, greenhouse, and detached-leaflet evaluations of tomato germplasm for early blight resistance. Plant Dis 84:967–972
Foolad MR, Zhang LP, Khan AA, Niño-Liu D, Lin GY (2002) Identification of QTLs for early blight (Alternaria solani) resistance in tomato using backcross populations of a Lycopersicon esculentum X Lycopersicon hirsutum cross. Theor Appl Genet 104:945–958
Fourtouni A, Menetas Y, Christias C (1998) Effect of UV-B radiation on growth, pigmentation and spore production in phytopatho-genic fungus Alternaria solani. Can J Bot 76:2093–2099
Gardner RG (1988) NC EBR-1 and NC EBR-2 early blight resistant tomato breeding lines. Hort Sci 23:779–781
Gardner RG, Shoemaker PB (1999) “Mountain supreme” early blight resistant hybrid tomato and its parents, NC EBR-3 and NC EBR-4. Hort Sci 34:745–746
Gathungu RM, Bird SS, Sheldon DP, Kautz R, Vouros P, Matson WR et al (2014) Identification of metabolites from LC-EC profiling: GC-MS and re- fractionation provide essential information orthogonal to LCMS/micro NMR. Anal Biochem 454:23–32
Gherbawy Y, Hussein MA, Runge F, Spring O (2018) Molecular characterization of Alternaria alternata population isolated from upper Egyptian tomato fruits. J Phytopathol 166:709–721
Golshani F, Fakheri BA, Behshad E, Mohammadpour Vashvaei R (2015) PRs proteins and their mechanism in plants. BFAIJ 7:477–495
Gondal AS, Ijaz M, Riaz K, Khan AR (1993) Compendium of tomato diseases. American Phytopathological Society, St. Paul, pp 28–29
Hammond-Kosack KE, Jones JDG (1996) Resistance gene-dependent plant defense responses. Plant Cell 8:1773–1791
Hu J, Chen C, Peever T, Dang H, Lawrence C, Mitchell T (2012) Genomic characterization of the conditionally dispensable chromosome in Alternaria arborescens provides evidence for horizontal gene transfer. BMC Genomics 13:171
Iqbal T, Altaf S, Dar JA (2019) Status of early blight [Alternaria solani (Ellis and Martin) jones and grout] disease of tomato in Kashmir. J Pharmacog Phytoch 8:2152–2154
Islam RMD, Mondal C, Hossain I, Meah MB (2013) Organic management: an alternative to control late blight of potato and tomato caused by Phytophthora infestans. Int J Theor Appl Sci 5:32–42
Jarolim K, Del Favero G, Pahlke G, Dostal V, Zimmermann K, Heiss E et al (2017) Activation of the Nrf2-ARE pathway by the Alternaria alternata mycotoxins altertoxin I and II. Arch Toxicol 91:203–216
Johanson A, Thurston H (1990) The effect of cultivar maturity on the resistance of potatoes to early blight caused by Alternaria solani. Am Potato J 67:615–623
Johann S, Rosa LH, Rosa CA, Perez P, Cisalpino PS, Zani CL, Cota BB (2012) Antifungal activity of altenusin isolated from the endophytic fungus Alternaria sp. against the pathogenic fungus Paracoccidioides brasiliensis. Rev Iberoam Micol 29:205–209
Jorda L, Vera P (2000) Local and systemic induction of two defense-related Subtilisin-like protease promoters in transgenic Arabidopsis plants, Lucifer in induction of PR gene expression. Plant Physiol 124:1049–1057
Khulbe RD, Sati MC (1987) Seed transmission of Alternaria solani in tomato. Indian Phytopathol 40:106–107
Kumar R, Khurana A (2014) Functional genomics of tomato: opportunities and challenges in post-genome NGS era. J Biosci 39:917–929
Kumar S, Srivastava K (2013) Screening of tomato genotypes against early blight Alternaria solani under field condition. Bioscan 8:189–193
Kumar V, Gupta RC, Singh PC, Pandey KK, Kumar R, Rai AS et al (2007) Management of early blight disease of tomato cv. Ksahi Amrit through fungicides, bioagents and cultural practices in India. Veg Sci 34:206–207
Kumar V, Haldar S, Pandey KK, Singh RP, Singh AK, Singh PC (2008) Cultural, morphological, pathogenic and molecular variability amongst tomato isolates of Alternaria solani in India. World J Microbiol Biotechnol 24:1003–1009
Kumar A, Bimolata W, Kannan M, Kirti PB, Qureshi IA, Ghazi IA (2014) Comparative proteomics reveals differential induction of both biotic and abiotic stress response associated proteins in rice during Xanthomonas oryzae pv. oryzae infection. Funct Integr Genomics 15(4):425–437
Kumar M, Brar A, Yadav M, Chawade A, Vivekanand V, Pareek N (2018) Chitinases-potential candidates for enhanced plant resistance towards fungal pathogens. Agri 8:88. https://doi.org/10.3390/agriculture8070088
Lawaju BR, Lawrence KS, Lawrence GW, Klink VP (2018) Harpin-inducible defense signaling components impair infection by the ascomycete Macrophomina phaseolina. Plant Physiol Biochem 129:331–348
Li T, Zhang XY, Huang Y, Xu ZS, Wang F, Xiong AS (2018) An R2R3-MYB transcription factor, SlMYB28, involved in the regulation of TYLCV infection in tomato. Sci Hortic 237:192–200
Loganathan M, Venkataravanappa V, Saha S, Rai AB, Tripathi S, Rai RK, Chowdappa P (2016) Morphological, pathogenic and molecular characterizations of Alternaria species causing early blight of tomato in northern India. Proc Nat Acad Sci, Ind Sec B: Biol Sci 86:25–330
Lohith MR, Chandrashekar Reddy K, Venkata Ramana C, Venkata Rao P, Ravendra Reddy K, Lokanadha Reddy D (2011) Screening of tomato genotypes against early blight (Alternaria solani) by detached leaf method. Acta Hortic 914:465–468
Maiero M, Ng TJ, Barksdale TH (1989) Combining ability estimates for early blight resistance in tomato. J Am Soc Hort Sci 114:118–121
Maiero M, Ng TJ, Barksdale TH (1990a) Genetic resistance to early blight in tomato breeding lines. Hort Sci 25:344–346
Maiero M, Ng TJ, Barksdale TH (1990b) Inheritance of collar rot resistance in the tomato breeding lines C1943 and NC EBR-2. Phytopathology 80:1365–1368
Majumder S, Shakil NA, Kumar J, Banerjee T, Sinha P, Singh BB, Garg P (2016) Eco-friendly PEG-based controlled release nano-formulations of mancozeb: synthesis and bioefficacy evaluation against phytopathogenic fungi Alternaria Solani and Sclerotium Rolfsii. J Environ Sci Health B 51:873–887
Martin FW, Hepperly P (1987) Sources of resistance to early blight, Alternaria solani, and transfer to tomato, Lycopersicon esculentum. J Agric Univ Puerto Rico 71:85–95
Mauch-Mani B, Slusarenko AJ (1996) Production of salicylic acid precursors is a major function of phenylalanine ammonium lyase enzymes in the resistance of Arabidopsis to Peronospora parasitica. Plant Cell 8:203–212
Mauricio-Castillo JA, Salas-Muñoz S, Reveles-Torres LR, Salas-Luevano MA, Salazar-Badillo FB (2020) Could Alternaria solani IA300 be a plant growth-promoting fungus? Eur J Plant Pathol 157:413–419
McNeece BT, Sharma K, Lawrence GW, Lawrence KS, Klink VP (2019) The mitogen activated protein kinase (MAPK) gene family functions as a cohort during the Glycine max defense response to Heterodera glycines. Plant Physiol Biochem 137:25–41
Mizubuti GSE, Junior VL, Forbes GA (2007) Management of late blight with alternative products. Pest Technol 1:106–116
Moghaddama GA, Rezayatmanda Z, Esfahania MN, Khozaeic M (2019) Genetic defense analysis of tomatoes in response to early blight disease Alternaria alternata. Plant Physiol Biochem 142:500–509
Momel TM, Pemezny KL (2006) Florida plant disease management guide: tomato. Florida Cooperation Extensive Service, Institute of Food and Agriculture Sciences, Florida
Montealegre J, Valderrama L, Sánchez S, Herrera R, Besoain X, Pérez LM (2010) Biological control of Rhizoctonia solani in tomatoes with Trichoderma harzianum mutants. Electron J Biotechnol 13:2
Nasr Esfahani M (2019) An IPM plan for early blight disease of potato Alternaria solani sorauer and A. alternata (Fries.) Keissler. Arch Phytopathol Plant Protect. https://doi.org/10.1080/03235408.2018.1489600
Nasr Esfahani M, Alizadeh Moghaddam G, Karimkhah MA (2017) The relation of leaf micro-morphological components with early blight resistant potatoes varieties. Iran Plant Prot Sci 39:51–64
Nguyen HD, Lewis CT, Lévesque CA, Gräfenhan T (2016) Draft genome sequence of Alternaria alternata ATCC 34957. Genome Announc 4:e01554–e01515
Ozkilinc H, Sarpkaya K, Kurt S, Can C, Polatbilek H, Yasar A, Sevinc U, Uysal A, Konukoglu F (2017) Pathogenicity, morpho-species and mating types of Alternaria spp. causing Alternaria blight in Pistacia spp. in Turkey. Phytoparasitica 45:719–728
Pandey D, Kunnumal Rajendran SRC, Gaur M, Sajeesh PK, Kumar A (2016) Plant defense signaling and responses against necrotrophic fungal pathogens. J Plant Growth Regul 35:1159–1174
Panthee DR, Chen F (2010) Genomics of fungal disease resistance in tomato. Curr Genomics 11:30–39
Pareek CS, Smoczynski R, Tretyn A (2011) Sequencing technologies and genome sequencing. J Appl Genet 52:413–435
Pasche JS, Wharam CM, Gudmestad NC (2004) Shift in sensitivity of Alternaria solani in response to QoI fungicides. Plant Dis 88:181–187
Pathak RK, Baunthiyal M, Shukla R, Pandey D, Taj G, Kumar K (2017) In silico identification of mimicking molecules as defense inducers triggering jasmonic acid mediated immunity against Alternaria blight disease in Brassica species. Front Plant Sci 8:609
Poysa V, Tu JC (1996) Response of cultivars and breeding lines of Lycopersicon spp. to Alternaria solani. Can Plant Dis Surv 76:5–8
Prasad B, Dutt BL (1974) Inducing sporulation of Alternaria solani II. Effect of light. Mycopathol Mycol Appl 49:141–146
Pusztahelyi T (2018) Chitin and chitin-related compounds in plant–fungal interactions. Mycology 9:189–201
Rao AP, Subrahamanyam K (1976) A new record of Alternaria on coconut leaf [India]. FAO, Rome
Raza W, Ghazanfar MU, Iftikhar Y, Ahmed KS, Haider N, Rasheed MH (2016) Management of early blight of tomato through the use of plant extracts. Int J Zool Stud 1:1–4
Sahi HPS (1990) Epidemiology and management of Alternaria leaf spots of tomato in Himachal Pradesh. Ph.D thesis, Dr. Y.S. Parmar University of Horticulture and Forestry, Solan, p 97
Sain SK, Pandey AK (2016) Biological spectrum of Trichoderma harzianum Rifai isolates to control fungal diseases of tomato (Solanum lycopersicon L.). Arch Phytopath Plant Prot 49:507–521
Schlagnhaufer CD, Arteca RN, Pell EJ (1997) Sequential expression of two 1-aminocyclopropane-1-carboxylate synthase genes in response to biotic and abiotic stresses in potato (Solanum tuberosum L.) leaves. Plant Mol Biol 35:683–688
Shah P, Powell ALT, Orlando R, Bergmann C, Gutierrez-Sanchez G (2012) Proteomic analysis of ripening tomato fruit infected by Botrytis cinerea. J Proteome Res 11:2178–2192
Shahida P, Abdul G (1995) Seed borne mycoflora of tomato. Pakistan J Bot 27:201–208
Sharma N, Rahman MH, Strelkov S, Thiagarajah M, Bansal VK (2007) Proteome-level changes in two Brassica napus lines exhibiting differential responses to the fungal pathogen Alternaria brassicae. Plant Sci 172:95–110
Sharma K, Sarma S, Bohra A, Mitra A, Sharma NK, Kumar A (2018) Plant metabolomics: an emerging technology for crop improvement. In: Çelik O (ed) New visions in plant science. IntechOpen, London, pp 65–79
Shinde BA, Dholakia BB, Hussain K, Panda S, Meir S, Rogachev I, Aharoni A et al (2017) Dynamic metabolic reprogramming of steroidal glycol-alkaloid and phenylpropanoid biosynthesis may impart early blight resistance in wild tomato (Solanum arcanum Peralta). Plant Mol Biol 95:411–423
Shinde BA, Dholakia B, Hussain KH, Aharoni A, Giri AP, Kamble AC (2018) WRKY1 acts as a key component improving resistance against Alternaria solani in wild tomato, Solanum arcanum Peralta. Plant Biotechnol J 16:1502–1513
Singh RS (1995) Diseases of vegetable crops, 3rd edn. Oxford and IBH Publishers, New Delhi, pp 107–109
Singh PC, Singh D (2006) In vitro evaluation of fungicides against Alternaria alternata. Ann Plant Prot Sci 14:500–502
Singh RN, Kushal R, Chugh RK, Sangwan R (2020) Evaluation of plant extracts, fungicides and bio-agents against early blight disease of tomato incited by Alternaria solani under in vitro and field conditions. Int J Curr Microbiol App Sci 9:1914–1920
Sinha R, Chattopadhyay S (2011) Changes in the leaf proteome profile of Mentha arvensis in response to Alternaria alternata infection. J Proteomics 74:327–336
Spletzer ME, Enyedi AJ (1999) Salicylic acid induces resistance to Alternaria solani in hydroponically grown tomato. Phytopathology 89:722–727
Stancheva I, Lozanov I, Achkova Z (1991) Sources of resistance to Alternaria solani in the tomato in wild growing species of the genus Lycopersicon. Genetika-i-Selektsiya 24:126–130
Stangarlin JR, Kuhn OJ, Assi L, Schwan-Estrada KRF (2011) Control of plant diseases using extracts from medicinal plants and fungi. Formatex 2011:1033–1042
Tenenboim H, Brotman Y (2016) Omic relief for the biotically stressed: metabolomics of plant biotic interactions. Trends Plant Sci 21:781–791
The Tomato Genome Consortium (2012) The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485:635–641
Thipyapong P, Steffens JC (1997) Tomato polyphenol oxidase: differential response of the polyphenol oxidase F promoter to injuries and wound signals. Plant Physiol 115:409–418
Thirthamallappa, Lohithaswa HC (2000) Genetics of resistance to early blight (Alternaria solani Sorauer) in tomato (Lycopersicum esculentum L.). Euphytica 113:187–193
Thomma BPHJ, Seidl MF, Shi-Kunne X, Cook DE, Bolton MD, van Kan JAL, Faino L (2016) Mind the gap; seven reasons to close fragmented genome assemblies. Fungal Genet Biol 90:24–30
Tian M, Win J, Song J, Van der Hoorn R, Van der Knaap E, Kamoun SA (2007) Phytophthora infestans cystatin-like protein targets a novel tomato papain-like apoplastic protease. Plant Physiol 143:364–377
Touhami N, Soukup ST, Schmidt-Heydt M, Kulling SE, Geisen R (2018) Citrinin as an accessory establishment factor of P. expansum for the colonization of apples. Int J Food Microbiol 266:224–233
Ulker B, Shahid M, Somssich IE (2007) The WRKY70 transcription factor of Arabidopsis influences both the plant senescence and defense signaling pathways. Planta 226:125–137
Upadhyay P, Rai A, Kumar R, Singh M, Sinha B (2014a) Differential expression of pathogenesis related protein genes in tomato during inoculation with A Solani. J Plant Pathol Microb 5:217
Upadhyay P, Rai A, Kumar R, Singh M, Sinha B (2014b) Differential expression of pathogenesis related protein genes in tomato during inoculation with A solani. J Plant Pathol Microbiol 5:217
Upadhyay P, Ganie SH, Rai A, Singh M, Sinha B (2016) Identification of transcription factors in tomato, potentially related to early blight resistance at invasion inhost tissue, using microarray expression profiling. S Afr J Bot 106:165–173
van derWaals JE, Korsten L, Slippers B (2004) Genetic diversity among Alternaria solani isolates from potatoes in South Africa. Plant Dis 88:959–964
Varma PK, Gandhi SK, Singh S (2008) Biological control of Alternaria solani, the causal agent of early blight of tomato. J Biol Contr 22:67–72
Warren RL, Sutton GG, Jones SJM, Robert RA (2007) Assembling millions of short DNA sequences using SSAKE. Bioinformatics 23:500–501
Wenderoth M, Garganese F, Schmidt-Heydt F, Soukup ST, Ippolito A, Sanzani SM, Fischer R (2019) Alternariol as virulence and colonization factor of Alternaria alternata during plant infection. Mol Microbiol 112:131–146
Wojciechowska E, Weinert CH, Egert B, Trierweiler B, Schmidt-Heydt M, Horneburg B et al (2014) Chlorogenic acid, a metabolite identified by untargeted metabolome analysis in resistant tomatoes, inhibits the colonization by Alternaria alternata by inhibiting alternariol biosynthesis. Eur J Plant Pathol 139:735–747
Wolters PJ, Faino L, van den Bosch TBM, Evenhuis B, Visser RGF, Seidl MS et al (2018) Gapless genome assembly of the potato and tomato early blight pathogen Alternaria solani. Resour Announce 31:7
Woudenberg JHC, Seidl MF, Groenewald JZ, de Vries M, Stielow JB, Thomma BP, Crous PW (2015) Alternaria section Alternaria: species, formae speciales or pathotypes? Stud Mycol 82:1–21
Woundenberg JHC, Truter M, Groenewald JZ et al (2014) Large-spored Alternaria pathogens in section Porri disentangled. Stud Mycol 79:1–47
Yang X, Hu H, Yu D, Sun Z, He X, Zhang J, Chen Q (2015) Candidate resistant genes of sand pear (Pyrus pyrifolia Nakai) to Alternaria alternata revealed by transcriptome sequencing. PLoS One 10:e0135046
Yang X, Li H, Yang Y, Wang Y, Mo Y, Zhang R, Zhang Y, Ma J, Wei C, Zhang X (2018) Identification and expression analyses of WRKY genes reveal their involvement in growth and abiotic stress response in watermelon (Citrullus lanatus). PLoS One 16:e0191308
Yuan Y et al (2017) Metabolomic analyses of banana during postharvest senescence by 1H-high resolution-NMR. Food Chem 218:406–412
Zhang LP, Lin GY, Nino-Liu D, Foolad MR (2003) Mapping QTLs conferring early blight (Alternaria solani) resistance in a Lycopersicon esculentum × L hirsutum cross by selective genotyping. Mol Breed 12:3–19
Zorića AS, Morina F, Toševskic I, Tostie T, Jovićd J, Krstićd O, Veljović-Jovanović S (2019) Resource allocation in response to herbivore and gall formation in Linaria vulgaris. Plant Physiol Biochem 135:224–232
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Pandey, A.K., Savani, A.K., Singh, P. (2021). The Early Blight of Tomato: Omics Interventions Toward Controlling Disease Spread and Development. In: Kumar, A., Kumar, R., Shukla, P., Pandey, M.K. (eds) Omics Technologies for Sustainable Agriculture and Global Food Security Volume 1. Springer, Singapore. https://doi.org/10.1007/978-981-16-0831-5_4
Download citation
DOI: https://doi.org/10.1007/978-981-16-0831-5_4
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-16-0830-8
Online ISBN: 978-981-16-0831-5
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)