Abstract
The Magnaporthaceae family includes fungal species that cause devastating diseases on cereals and grasses. The causal agent of take-all disease of wheat Gaeumannomyces graminis, the rice blast fungus Magnaporthe oryzae, and Magnaporthe poae which causes the grey leaf spot on turfgrasses, belong to this family. M. poae and G. graminis are considered root pathogens, whereas M. oryzae is found on aerial plant tissues. Remarkably, M. oryzae can also infect roots and distinct mechanisms control its root infection ability compared to leaf colonisation. Since G. graminis and M. poae are genetically intractable, M. oryzae underground infection process can be used to dissect genetic pathways and molecular mechanisms underlying root infection in other members of Magnaporthaceae. Interestingly, M. oryzae root infection process also shares similarities with ancient mycorrhizal associations. Here, we highlight the latest advances on the mechanisms regulating pathogenicity in these economically significant plant pathogens.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
4.1 Taxonomy of the Magnaporthaceae Family
The Magnaporthaceae family (P.F. Cannon) has a complex taxonomic history. Gaeumannomyces and Magnaporthe species were grouped in the Magnaporthaceae based on the common morphology of their teleomorphs and similarities in host range (Cannon 1994). The Magnaporthaceae family is included within the class Sordariomycetes (Cannon and Kirk 2007). Traditionally, the genus Gaeumannomyces belonged to the order Diaporthales. Since the Magnaporthaceae family expanded to comprise fungal species that are not limited to Diaporthales fungi (Berbee 2001; Castlebury et al. 2002), enough evidence has been found to classify these fungal species at a new order level, and currently the order Magnaporthales has been proposed (Thongkantha et al. 2009). The Magnaporthaceae is a small family that comprises 17 genera and nearly a 100 species (Kirk et al. 2008; Thongkantha et al. 2009). The genera Buergenerula, Ceratosphaerella, Clasterosphaeria, Clasterosporium, Gaeumannomyces, Gibellina, Harpophora, Herbampulla, Magnaporthe, Muraeriata, Mycoleptodiscus, Nakataea, Omnidemptus, Ophioceras, Pseudohalonectria, Pyricularia and Yukonia belong to this family (Thongkantha et al. 2009). It is interesting to highlight that marine fungal species are included within this family such as Buergenerula spartinae, Gaeumannomyces medullaris (anamorph Trichocladium medullare) and Pseudohalonectria halophila (Jones et al. 2009).
Some Phialophora species have been reported as anamorphs of several Gaeumannomyces species including the take-all fungus Gaeumannomyces. graminis. The Gaeumannomyces species together with their Phialophora anamorphs and other root colonisers of non-pathogenic Phialophora species form the Gaeumannomyces–Phialophora complex (Bateman et al. 1992; Bryan et al. 1995; Henson 1992; Ulrich et al. 2000). Within the genus Gaeumannomyces, taxonomists have identified up to nine different fungal species (Table 4.1).
Phylogenetic analyses using partial sequences of the 18S and 28S ribosomal genes of fungal isolates from different Magnaporthaceae genera suggested a monophyletic origin of this family (Thongkantha et al. 2009). However, more recent studies based on a six-gene phylogeny strongly support that both genera Magnaporthe and Gaeumannomyces are polyphyletic (Zhang et al. 2011c), meaning that they share a number of morphological signatures, but their origin probably is not from a common ancestor (convergent evolution). Therefore, the classification and evolution of Magnaporthe/Gaeumannomyces species needs further analyses by additional multigene phylogenies and whole genome comparison approaches (see Sect. 4.5).
The first original report describing the fungus Pyricularia grisea as the causal agent of grey leaf spot on the grass Digitaria sanguinalis appeared in 1880 (Saccardo 1880). A few years later, in 1892, Cavara published a report naming Pyricularia oryzae as the causal agent of rice blast disease (Cavara 1892). Subsequently, the name P. oryzae was applied for rice-infecting isolates; the isolates from other cereals and grasses kept the name P. grisea. However, P. oryzae was considered as a synonym of P. grisea based on morphological commonalities and interfertility between P. oryzae strains from rice and P. grisea strains from different grass hosts. Then, it was corrected to name rice-infecting isolates as P. grisea (Rossman et al. 1990). Concomitantly, the teleomorph of P. grisea was identified as Magnaporthe grisea (T.T. Hebert) M.E. Barr (Hebert 1971), and taxonomically it was more correct to name the sexual stage of the fungus (see further information on this subject below). As a result, scientists working on blast disease have been using four different names to refer to rice-infecting blast isolates (P. grisea, P. oryzae, M. grisea and M. oryzae).
To gain clarification on the taxonomy of Magnaporthe species, rice and all the other grass isolates are currently included within the M. grisea species complex (Couch and Kohn 2002). Globally, M. grisea species complex can infect a wide range of plant hosts, although one strain infects only one or few host species. Frequently one strain is susceptible only to a specific cultivar of the host (Borromeo et al. 1993; Valent and Chumley 1991). Phylogenetic analysis has inferred the presence of two monophyletic intersterile groups within the M. grisea species complex based on three unrelated gene sequences (actin, β-tubulin and calmodulin) and mating compatibility tests (Couch and Kohn 2002). The lineage M. grisea has been kept for fungal isolates associated with the host grass Digitaria spp. The second lineage contains rice and related fungal isolates and has been renamed as a new species, M. oryzae, although no morphological differences exist between isolates of these two groups.
The teleomorph is the sexual form (reproductive stage) of a fungus, while the anamorph is its asexual form. It has been a common practice to name differently the anamorph and teleomorph of a particular fungus. Based on the Article 59 of the International Code of Botanical Nomenclature, a particular fungal species with both reproductive stages, the teleomorph name takes prevalence over the anamorph name (Hawksworth 2011). However, molecular phylogenetic approaches and whole genome sequences have revolutionised taxonomy, and this dual nomenclature rule based on morphological features is going to disappear. The rule “one fungus, one name” was approved at the Melbourne International Botanical Congress in 2011 and will be applied from January 2013 onwards (Hawksworth 2011). This has originated an important debate about the maintenance of the genus Magnaporthe or Pyricularia for the rice blast fungus among community members (http://www.magnaporthe.blogspot.com.es/).
4.2 The Take-All Fungus Gaeumannomyces graminis
Take-all is an extremely damaging disease of cereals and grasses caused by G. graminis, a homothallic soilborne fungus that colonises preferentially belowground plant tissues (Asher and Shipton 1981; Freeman and Ward 2004; Hornby 1998). Isolates of G. graminis are classified into four varieties based on morphological traits (ascospore size or hyphopodial structure) and pathogenicity features (host range and aggressiveness):
-
1.
Isolates of G. graminis var. tritici infect mainly wheat (Triticum aestivum L.) but also invade barley (Hordeum vulgare L.) and rye (Secale cereale L.; Walker 1972). G. graminis var. tritici isolates can be further classified as R or N isolates based on their ability [R] or inability [N] to infect rye (Bryan et al. 1995).
-
2.
Isolates of G. graminis var. avenae [Gaeumannomyces cariceti] infect oats (Avena spp.) although they can also infect rice and wheat (Dennis 1960).
-
3.
Isolates of G. graminis var. graminis are less aggressive and are normally found on maize (Zea mays L.), rice and other grasses such as Bermuda grass (Cynodon dactylon L.; Arx and Olivier 1952).
-
4.
Isolates of G. graminis var. maydis are found in maize and can infect Sorghum species (Yao et al. 1992).
It is noteworthy to mention that G. graminis var. tritici and avenae strains are more closely related to each other than to G. graminis var. graminis isolates (Bryan et al. 1995). Little is known about the molecular mechanisms underlying the interaction of G. graminis with cereal roots due to the difficulty of generating stable transformants in this fungal species, although genetic crosses and gene disruption approaches have been successfully achieved in the past with Gaeumannomyces strains (Bowyer et al. 1995; Frederick et al. 1999). In addition, it is a real challenge to identify and introgress take-all resistance genes in polyploid hosts such as wheat and oat.
The take-all caused by G. graminis var. tritici on wheat is one of the major agronomical problems in this crop. The most characteristic symptom is the blackening of the root due to extended necrotic lesions preceded by a complete disruption of the root architecture in severely affected crops (Fig. 4.1a). It is possible to observe black mycelia at the stem base on diseased plants. As a consequence of the collapse of the root, diseased plants tiller poorly and do not fill their heads, which become white (“whiteheads”). In the field, these symptoms are observed as round white patches (Fig. 4.1b). Disease symptoms can be present at early stages on seedlings (Fig. 4.1c). Penetration of roots by G. graminis is mediated by simple or lobed hyphopodia (Fig. 4.1d). Lobed hyphopodia can be melanised, but the role of the melanin in hyphopodia-mediated penetration is ambiguous in Gaeumannomyces species. Melanins are dark pigmented secondary metabolites produced by fungi and other organisms and play an important role in protecting these organisms against environmental stresses (Henson et al. 1999). In some phytopathogenic fungi, such as M. oryzae, melanin is required to keep the osmotic pressure that exerts the force for appressorium-mediated leaf penetration (De Jong et al. 1997; Howard and Valent 1996). M. oryzae melanin-deficient mutants are non-pathogenic. However, the involvement of melanin in pathogenicity among Gaeumannomyces species varies (Henson et al. 1999). Melanin-deficient mutants of G. graminis var. graminis are as virulent as the wild-type strain on rice roots. By contrast, melanin is important for pathogenesis on G. graminis var. tritici isolates although the corresponding Phialophora anamorphs which are heavily melanised are non-pathogenic (Henson et al. 1999). This diverse role of melanin in root penetration and colonisation might indicate a prevalence of the mechanisms by which G. graminis varieties penetrate roots, i.e. turgor generation versus cell wall-degrading enzymes. Several laccases, possibly involved in the melanin biosynthesis pathway, have been biochemically characterised in G. graminis var. tritici. However, their participation in fungal virulence remains unclear due to their functional redundancy (Litvintseva and Henson 2002).
G. graminis var. tritici disease symptoms. (a) Take-all symptoms on roots of an adult wheat plant, courtesy of Kansas State University. (b) Visible white heads and stunted plants on a wheat field infected with take-all, courtesy of Richard Gutteridge (Rothamsted Research, UK). (c) Necrotic lesions of a 14 days-old wheat seedling infected with G. graminis. (d) Lobed hyphopodia developed by G. graminis strains, with permission of Fungal Genetics and Biology. (e) Typical circular patches of yellow-brown colour of summer patch disease (M. poae) on turfgrass, courtesy of Dr. Lane Tredway, North Carolina State University
Plant cell wall represents the first barrier that any invader has to overcome to colonise the plant host. Fungal plant pathogens have developed combined strategies to cross plant cell walls. One of them is the secretion of cell wall-degrading enzymes during host invasion. Cellulose is the major polysaccharide polymer of plant cell walls (Fry 2004). It is composed of linear β(1 → 4)-linked d-glucose monomers. G. graminis var. tritici secretes endoglucanases and β-glucosidases during in vitro and in planta growth (Dori et al. 1995). In G. graminis, these enzymes have been grouped based on their acidic (4.0–5.6) and basic (≥9.3) isoelectric point. They are supposed to play an important role in cell wall degradation during G. graminis var. tritici growth on root tissues, but genetic approaches are required to confirm this hypothesis (Dori et al. 1995).
Preformed antimicrobial compounds produced by plants play an important role in plant immunity acting as first barriers to prevent pathogen attack (Field et al. 2006). The saponin avenacin is a triterpene metabolite present in the epidermal layer of oat root tips. Avenacins are a mixture of four glycosylated compounds (avenacins A1, A2, B1 and B2), and avenacin A1 is the most abundant isoform in oats (Crombie et al. 1984). Wheat roots cannot synthesise these triterpenes. While strains of G. graminis var. avenae infect oats due to their ability to synthesise avenacinase and also infect wheat, G. graminis var. tritici isolates only infect wheat roots (but not oat roots) and lack the avenacinase enzyme (Crombie et al. 1986). The fungal avenacinase detoxifies avenacin A1 into other less harmful compounds that do not affect G. graminis var. avenae growth. Taking in account that isolates of G. graminis var. tritici can infect a diploid oat species (Avena longiglumis L.) that lacks avenacin (Osbourn et al. 1994), a correlation exists between the presence of avenacins and the resistance of oats to non-host G. graminis fungal species. This was further confirmed by generating avenacinase-deficient mutants of G. graminis var. avenae, which no longer infected oat roots (Bowyer et al. 1995). Therefore, a single gene confers to G. graminis var. avenae the ability to detoxify avenacin and to control its host range.
4.3 Magnaporthe poae: A Root-Infecting Fungus of Turfgrasses
Magnaporthe poae affects roots and crowns of turfgrasses of the genera Poa, Festuca and Agrostis that are widely used in golf and other sport courses, parks and residential gardens (Landschoot and Jackson 1989a). Consequently, it is a commercially significant root-infecting fungal pathogen. The disease caused by M. poae is called summer patch due to the emergence of symptoms during the hot season in circular patches, which can increase up to 1 m in diameter (Fig. 4.1e). Temperature and high relative humidity favour fungal root penetration. Once inside the host, the fungus can progress through the vascular tissue to the aerial parts of the plant leading to subsequent foliar necrosis. Two new Magnaporthe species affecting warm-season turfgrasses have been recently described in Australia whose symptoms look similar to those produced by M. poae (Wong et al. 2012). These are Magnaporthe garrettii [P. T. W. Wong and M. L. Dickinson sp. nov.] found on couch (Cynodon dactylon) and Magnaporthe griffinii [P. T. W. Wong and A. M. Stirling sp. nov.] associated with a disease complex (“summer decline”) of hybrid couch (C. dactylon × C. transvaalensis). These Magnaporthe species can be accurately identified in infected roots by PCR, providing a reliable method for early detection and disease management of summer patch (Zhao et al. 2012).
There is very limited information about mechanisms regulating M. poae infection process or plant resistance genes against summer patch disease (Tredway 2006). Serine protease activity has been observed during M. poae root colonisation (Sreedhar et al. 1999), suggesting an important role for this enzyme during fungal infection. Interestingly, the genome sequences of the M. poae strain ATCC 64411 and the Ggt isolate R3-111a-1 have been released since May 2010. The comparative analysis of the currently available genomes of Magnaporthaceae strains is still pending publication (Magnaporthe comparative Sequencing Project, Broad Institute of Harvard and MIT; http://www.broadinstitute.org/). A link may exist between their genetic intractability and their ability to colonise roots, where they have to subsist with other living organisms in the rhizosphere. Undoubtedly, the analysis of their genomes will provide many insights that will help to understand the molecular basis of ecological niche adaptation and pathogenicity in these fungal species.
4.4 Rice Blast Disease: An Important Constraint to Rice Production
Rice (Oryza sativa L.) is one of the most important cereal crops and staple diet of more than three billion people. Fungal blast is considered a major threat to rice crops and costs farmers a loss of nearly $5 billion a year (Skamnioti and Gurr 2009). Not surprisingly, it accounts for the world’s largest fungicide market. The Japanese market alone for blast fungicides is estimated at US$400 million per year (Skamnioti and Gurr 2009). Rice blast is caused by the fungus Magnaporthe oryzae (Couch and Kohn 2002), and this fungal species can also cause diseases in other staple food crops including finger millet, maize and wheat, representing a serious risk for food security globally and a significant challenge in developing countries (FAO 2009). The damage produced by blast in rice crops oscillates between 10 and 30 % every year. Under disease-conducive conditions, the fungus can destroy the entire crop (Thinlay et al. 2000). Rice blast is present in all rice-growing areas worldwide, including Western Australia where rice-growing areas were free of this disease until last year (You et al. 2012).
Rice blast is a polycyclic disease since M. oryzae can undergo multiple infection cycles during a rice-growing season. However, disease progression highly relies on favourable weather conditions, increasing the difficulty to effectively control blast. High humidity or long periods of rain followed by relatively warm temperatures favour spore germination and fungal penetration (Ou 1985). Wind-dispersed or water-splashed conidia are the main source of inoculum in the field (Ou 1985). However, M. oryzae can overwinter on alternative weed hosts and infested plant debris for almost 3 years, playing possibly an important role in the epidemiology of the disease (Harmon and Latin 2005). This fungus can form resting structures on roots and plant debris such as microsclerotia and vesicles, which can germinate even after 4 years of dormancy (Gangopadhyay and Row 1986; Sesma and Osbourn 2004) (Fig. 4.2a, b). M. oryzae can penetrate rice roots and spread through the vascular system to the aerial parts of the plant to produce blast disease symptoms (Fig. 4.2c, d), although the relevance of the underground infection process under field conditions is not proven yet (Besi et al. 2009; Sesma and Osbourn 2004). Domestic travellers and the transport of infected material (souvenirs made with seeds, weeds or rice straw) probably also contribute to the dissemination of the disease (You et al. 2012). PCR-based methods have been developed for detection of the fungus, offering a quick method to control the dissemination of infected material (Harmon et al. 2003).
Rice blast disease symptoms. (a) Fungal vesicles and (b) microsclerotia produced on root surfaces. (c) Cross section of a barley root infected with a GFP-tagged M. oryzae strain showing heavy colonisation of the vascular system. (d) Necrotic blast lesions of a 15-day-old rice seedling infected with M. oryzae. (e) Leaf blast symptoms. (f) Panicle blast in the field. (g) Neck blast symptoms. Images f and g courtesy of M. Pau Bretó (IVIA, Spain)
In the field, rice blast disease symptoms are visible at any growth stage and at any part of the aerial plant tissue: leaf, collar, nodes, panicle neck and panicles (Fig. 4.2e–g). The shape, colour and size of the lesions largely depend on the rice cultivar, the age of the lesion and environmental conditions (Ou 1985). On leaves, blast lesions are eyespot shaped with white to grey colour and surrounded by a dark red-brown margin. Lesion size varies but commonly ranges between 1–1.5 cm long and 0.3–0.5 cm wide. The collar rot appears on the junction between the leaf blade and leaf sheath affecting the entire leaf. The neck rot is the most damaging symptom in the field. Typically a necrotic or rotten neck is visible at the base of the panicle often affecting the entire panicle, which becomes white and partially filled or completely unfilled. The blast symptoms in the panicle or nodes are brown or black. On roots, blast lesions show brown necrotic areas, and root architecture is maintained suggesting less aggressive damage compared to take-all symptoms caused by G. graminis.
4.5 From Genome Sequences into Underlying Mechanisms Regulating Fungal Pathogenicity
Due to the genomic resources available for both the rice host and the fungus, the genetic tractability of M. oryzae and the economic relevance of blast disease, the rice–M. oryzae interaction has become a leading pathosystem for studying fungal pathogenicity and plant immunity in crops (Dean et al. 2012). The laboratory strain 70–15 was the first M. oryzae rice-infecting strain whose genome sequence was made available to the research community (Dean et al. 2005). It also represented the first genome publication of a fungal plant pathogen. The genome of M. oryzae is approximately 41 Mb in size (eight annotation, Magnaporthe comparative Sequencing Project, Broad Institute of Harvard and MIT; http://www.broadinstitute.org). Gene prediction programmes estimate the presence of 12,827 protein-coding genes, which are distributed in seven chromosomes. Optical mapping has allowed an accurate DNA alignment of the seven chromosomes.
The genome sequence of M. oryzae has revealed several pathogenicity-associated features. Predicted secreted proteins, which likely act as potential effectors modulating plant physiology and reducing basal host immune response, are more abundant in M. oryzae (~1,600) compared to Neurospora crassa (~800) or Aspergillus nidulans (~900). In addition, these non-pathogenic saprophytic fungi contain up to 10 genes encoding chitin-binding proteins, while M. oryzae genome has undergone an expansion on this protein family (~40 genes), indicating the complexity of chitin metabolism in M. oryzae. The rice blast fungus also presents an increase in seven transmembrane integral proteins, normally involved in activation of signalling pathways that help the fungus to adapt to specific external stimuli. A subgroup of these type of receptors contain CFEMs (conserved fungal-specific extracellular motif), which include an extracellular cysteine-rich EGF-like domain present exclusively in fungi (Kulkarni et al. 2005). One of the CFEM protein members, PTH11, has been shown to be involved in appressorium development and fungal virulence in M. oryzae (DeZwaan et al. 1999).
Different large-scale gene functional studies have been carried out since the release of M. oryzae genome sequence, including large-scale insertional mutagenesis (Betts et al. 2007; Jeon et al. 2007) and gene silencing (Nguyen et al. 2008). Transcriptomic approaches have also revealed global gene expression profiles during nitrogen starvation (Donofrio et al. 2006), appressorium development (Oh et al. 2008; Soanes et al. 2012) and plant infection (Mosquera et al. 2009). From the host perspective, at least 85 resistance gene loci (Pi genes), nine major QTLs defined by molecular markers and additional 350 QTLs have been identified on different rice germplasms to date (Ballini et al. 2008; Chen and Ronald 2011; Liu et al. 2010a). Furthermore, 17 resistance genes and two QTLs have been cloned since the release of the rice genome in 2002 (Table 4.2; Goff et al. 2002; Yu et al. 2002).
In 2010, and as mentioned in Sect. 4.3, two additional genomes of the Magnaporthaceae family have been made available to the scientific community (Magnaporthe comparative Sequencing Project, Broad Institute of Harvard and MIT; http://www.broadinstitute.org). These include the sequence drafts assemblies of the G. graminis var. tritici strain R3-111a-1 and the M. poae strain ATCC 64411. Although G. graminis var. tritici R3-111a-1 and M. poae ATCC 64411 genomes have not been assembled as well as M. oryzae genome, little syntenic regions exist among these three strains as shown by dot plot analysis (http://www.broadinstitute.org/annotation/genome/magnaporthe_comparative/Dotplot.html), in accordance with the polyphyletic origin of Magnaporthe and Gaeumannomyces genera found by multigene phylogeny (Zhang et al. 2011c).
More recently, the genomes of M. oryzae rice-infecting field isolates Y34 and P131 have been sequenced and compared against the genome reference of the laboratory strain 70–15. This genomic comparison has pointed out some relevant features of the field isolates (Xue et al. 2012). Y34 and P131 strains contain several 100 unique genes and have undergone unique DNA duplication events and expansions of pathogenicity-associated gene families. Thousands of transposon-like elements are present on the field isolates, although their genomic locations are poorly conserved among them. This suggests that transposition events might play an important role in genome variation in the rice blast fungus, which can explain the rapid adaptation of M. oryzae isolates to new resistant rice varieties (Kang et al. 2001; Zeigler 1998).
4.6 Evolutionary Implications of M. oryzae Reproduction
M. oryzae is a haploid and heterothallic ascomycetous fungus. Blast isolates with opposite mating types MAT-1.1 and MAT-1.2 (compatible strains) can conjugate and enter into an heterokaryotic stage where mycelia contain unfused nuclei (Valent et al. 1991). Subsequently, this heterokaryotic mycelium enters into a sexual cycle by fusing both nuclei. Within 3 weeks, sexual fruiting bodies or perithecia are formed. The perithecium is filled with asci, each of which contains eight ascospores (sexual spores). The dissection of ascospores is used for classical genetic studies to determine the genetic basis of phenotypic traits looking at the segregation of genetic markers (Talbot 2003; Valent and Chumley 1991; Valent et al. 1991). Blast strains isolated from finger millet (Eleusine coracana) or weeping lovegrass (Eragrostis curvula) are normally hermaphrodites and have been used to conduct early genetic studies (Valent et al. 1991). By contrast, rice blast isolates from the same geographic location reproduce mainly asexually since the same mating type is found normally among local populations and are female sterile (Couch et al. 2005; Zeigler 1998). A few examples of fertile rice isolates have been recovered from the field such as the strain Guy11 (Leung et al. 1988; Valent et al. 1991). Transposable elements or mutations in the mating alleles are directly involved in this lack of fertility (Zeigler 1998). Heterokaryosis and parasexual cycle have been reported for rice blast field isolates (Noguchi et al. 2006). The presence in M. oryzae of repeat-induced point mutation (RIP)-like processes, which only occur in the sexual phase of a fungal life cycle, suggests that sexual reproduction in the rice blast fungus exists or existed in nature (Ikeda et al. 2002). Under laboratory conditions, it is relatively easy to produce sexual crosses between M. oryzae isolates from different grasses. This ability has been used to identify several important gene loci and to generate fertile rice-infecting laboratory strains such as 70–15.
The relevance of sexual reproduction in the field, with the advantage of increasing pathogen fitness, has been addressed in the blast fungus (Saleh et al. 2012). A direct evidence of contemporary sexual reproduction is the identification of sexual structures (perithecia) in nature. However, their visualisation is challenging since M. oryzae perithecia have small size and may be constrained to limited areas or time periods. Molecular tools have been developed to identify recombination events in field population samples such as linkage disequilibrium (LD) associations and diversity of molecular markers (genotyping; Arnaud-Haond et al. 2007). In populations where recombination occurs, high genotypic diversities exist, and the non-random association of alleles at two or more loci (i.e. linkage disequilibrium) is low or not significant.
As mentioned before, a similar mating type is normally found in field populations of the rice blast fungus. Strikingly, ancestral populations of M. oryzae from south and east of Asia, the geographical location where this fungus emerged, show clear signatures of sexual reproduction (Couch et al. 2005; Kumar et al. 1999; Saleh et al. 2012; Zeigler 1998). Molecular evidences such as genotypic richness and linkage disequilibrium data support these findings (Saleh et al. 2012). Female-fertile M. oryzae strains still can be recovered from these locations and can complete the sexual cycle in vitro. This is the only region in the world so far where evidences for sexual reproduction of M. oryzae have been found, confirming the loss of sexual reproduction outside its original location of emergence. In terms of evolution, this geographical area may represent an initial point where M. oryzae isolates have evolved by adaptive selection against new rice cultivars and different hosts (Saleh et al. 2012).
4.7 The M. oryzae Leaf Infection Process
Under high relative humidity conditions, a succession of developmental events initiates the M. oryzae aerial infection (Fig. 4.3; Tucker and Talbot 2001), which begins when a wind-dispersed or water-splashed conidium lands on the leaf surface. Immediately after landing, a preformed adhesive material is secreted from the conidial tip to attach itself to the highly hydrophobic surface. One hour later a short germ tube develops from the apical cell of the conidium. Within a few hours, the apex of the germ tube swells, and a specialised dome-shaped penetration structure known as appressorium is formed. The appressorium is heavily melanised and a tremendous turgor pressure is generated within this structure (De Jong et al. 1997; Howard et al. 1991). A penetration peg emerges at the base of the appressorium and crosses the plant epidermal cell by combining physical force and secretion of cell wall-degrading enzymes (Skamnioti and Gurr 2007). Subsequently, M. oryzae initiates a new morphogenetic programme to colonise the plant epidermal cells. Five to six days after the initial penetration of the fungus, conidiophores emerged on the leaf surface to initiate the last step of the infection with the reproduction of the fungus.
Rice blast disease infection cycle. Right panel: M. oryzae leaf cycle modified from Ribot et al 2008. M. oryzae leaf infection cycle starts when a conidium lands on a leaf and attaches to the surface. Shortly after, the conidium produces a small germ tube, which differentiates into a melanised appressorium. A penetration peg formed at the base of the appressorium crosses the plant cell wall initiating fungal invasion. Invasive growth is different compared to fungal growth on surfaces. The invasive hypha moves beyond the first infected cell during a few days. Finally, conidiophores emerge and the fungus initiates sporulation between 6 and 15 days, releasing thousands of conidia to the environment. Left panel: M. oryzae root infection cycle potentially begins from infected plant debris or dormant structures present in the soil. These resting structures can germinate and penetrate into the plant roots. Fungal hypha colonises the vascular system of the root spreading systemically. The fungus moves to the upper parts of the plant producing typical blast lesions from which conidia are formed. These spores are dispersed to other plants by wind or water, propagating the disease
The rice blast research community has built a large amount of information on each of the steps of the blast disease cycle. Here, a description and latest findings of the M. oryzae leaf infection biology follows.
4.7.1 An Extracellular Matrix Mediates Fungal Adhesion and Differentiation
Spores of M. oryzae get attached immediately to the highly hydrophobic leaf cuticle by secreting a preformed mucilaginous extracellular matrix (ECM). This adhesive material is passively released from the conidial apex upon hydration, meaning that there are no metabolic costs involved in this process (Hamer et al. 1988; Tucker and Talbot 2001). This attachment is required for conidial anchoring and recognition of the surface, steps that precede the subsequent infection-related development. M. oryzae mutants with altered ECM show reduced virulence (Ahn et al. 2004). Initial studies identified components of M. oryzae adhesive material such as glycoproteins and lipids which help to retain moisture, essential for appressorium-mediated penetration (Hamer et al. 1988; Howard 1997). α-Mannosyl and α-glucosyl residues are highly abundant in the ECM based on lectin labelling and protease digestions (Hamer et al. 1988; Xiao et al. 1994). Additional components of M. oryzae ECM have been identified by immunological techniques using antibodies against animal cell adhesion factors (collagen VI, vitronectin, fibronectin, laminin) and integrin α3 (Bae et al. 2007; Inoue et al. 2007). Particularly, collagen (as a major component), vitronectin (as cementing compound), laminin, fibronectin and integrins are present in M. oryzae adhesive material (Bae et al. 2007; Inoue et al. 2007). Evidences suggest that ECM components are synthesised at two different stages of the infection cycle (Inoue et al. 2007). Collagen, vitronectin and integrins seem to be formed earlier than fibronectin and laminin components.
Integrins are transmembrane glycoproteins located at the plasma membrane and act as cell surface receptors modulating the cellular response to environmental stimuli (Kim et al. 2011a; Shattil et al. 2010). Fibronectin and collagen are extracellular ligands of integrin receptors (Kim et al. 2011a; Shattil et al. 2010). Externally applied peptides containing Arg-Gly-Asp amino acids and antibodies against fibronectin reduce conidial adhesion and appressorium development indicating that these processes are modulated by integrin-like proteins in M. oryzae. These defects are restored by manipulating the cAMP response pathway with exogenously applied chemicals (cAMP, cutin monomers and IBMX, a cAMP phosphodiesterase inhibitor; Bae et al. 2007). These results suggest that integrin-like proteins and their cognate extracellular ligands (fibronectin, collagen) activate the cAMP-dependent pathway and possibly other signalling pathways. This activation regulates the subsequent infection-related morphogenesis (Bae et al. 2007; Tucker and Talbot 2001). Integrins are also detected in conidial cell walls (Inoue et al. 2007), suggesting that these transmembrane receptors may play a role in the recognition of substrates by M. oryzae spores at earlier stages, immediately after landing.
4.7.2 Recognition of the Surface Precedes Appressorium Differentiation
Upon hydration, the first germ tube emerges, usually from the apical compartment of the conidium. If the fungus perceives that the surface is not adequate, the germ tube will arrest, blocking any further differentiation. Alternatively, it can develop a second germ tube from the opposite end of the conidium. Two germ tubes are often seen in germinating conidia on artificial substrates (Tucker et al. 2010). It is very unusual to see spores germinating from the middle compartment. The germ tube appears near the adhesion site of the spore in the apical cell and grows in direct contact with the surface of the plant for a short distance. Then, it swells and starts to change direction. This process known as “hooking” takes place before the appressorium development, and it is believed to be an important recognition step (Fig. 4.4a; Bourett and Howard 1990). During germ tube elongation, other processes such as secretion of plant cell wall-degrading enzymes, mobilisation of the metabolic reserves (trehalose) and synthesis of fungal cell wall occur (Tucker and Talbot 2001).
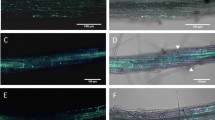
M. oryzae development on artificial and root surfaces. (a) Scanning electron micrograph of a germinating conidium (Co) forming an appressorium (Ap) on hydrophobic coverslips. (b) A two-septate conidium expressing a GFP-tagged nuclear protein; septa (Se) are indicated. (c) Sickle-shaped microconidia (Mi). (d) Conidiophore (Cf)-producing conidia in the stem of a rice seedling. (e) Conidia on roots developing hyphopodia (Hy). (f) Differentiated hyphopodia from fungal hyphae producing infection pegs (Ip) to penetrate rice roots. Scale bar numbers indicate micrometres
Concomitantly with the germination process, M. oryzae secretes additional compounds that contribute to the adhesion of the germ tube and perception of plant physical signals. Among them, hydrophobins have been shown to play a significant role at the early stages of fungal infection (Kim et al. 2005; Linder et al. 2005; Talbot et al. 1996). These specialised proteins are secreted at the interface between the hyphae and a hydrophobic surface and are involved in fungal development and environmental sensing (Linder et al. 2005). In M. oryzae, two hydrophobins play a role during infection, Mpg1 and Mhp1. Mpg1 has been widely characterised in the rice blast fungus (Beckerman and Ebbole 1996; Kershaw et al. 1998; Lau and Hamer 1996; Soanes et al. 2002; Talbot et al. 1993; Talbot et al. 1996). Mpg1 is a class I hydrophobin highly expressed during conidiogenesis, appressorium development and carbon and nitrogen starvation. The Δmpg1 mutants show defects in conidiation and appressoria development, and consequently are less virulent. Mhp1 is a class II hydrophobin and mutants lacking this hydrophobin show pleiotropic effects. Similarly to Mpg1, the hydrophobin Mhp1 is required for fungal morphogenesis, including appressorium development and invasive growth (Kim et al. 2005).
Cutinases and other methyl esterases are relevant enzymes secreted by fungal plant pathogens during early stages of infection (Kolattukudy 1985). Sixteen methyl esterase-encoding genes are present in M. oryzae genome (Dean et al. 2005), and some of them can be components of M. oryzae adhesive material. It is difficult to define their roles in M. oryzae infection biology since they may have redundant functions. Two cutinases have been characterised in the rice blast fungus. The CUT1 gene is dispensable for M. oryzae plant infection (Sweigard et al. 1992). Among all the M. oryzae methyl esterases, CUT2 was selected for further analysis because it is highly induced at 12 h after inoculation on barley leaves (Skamnioti and Gurr 2007). In M. oryzae, Cut2 acts as a surface sensor activating the cAMP/PKA and DAG/PKC signalling cascades which regulate appressorium-mediated penetration. The Δcut2 mutants show that Cut2 is required for appressorium differentiation and full disease symptoms production, but have no defects in adhesion, indicating a specific role for a cutinase in signalling and fungal development (Skamnioti and Gurr 2007).
4.7.3 Orchestrated Cellular Processes Govern Early Stages of Plant Infection
Two important stages take place during the process of appressorium differentiation in M. oryzae. During the recognition phase, the apex of the germ tube begins to hook, and vesicles located in the apical area move towards the surface of the plant (Bourett and Howard 1990; Tucker and Talbot 2001). Then, the tip of the germ tube swells and the appressorium is formed (Fig. 4.4a). This differentiation process is highly orchestrated and is activated in response to starvation stress and physical cues such as hardness and hydrophobicity (Dean 1997; Talbot et al. 1997; Tucker and Talbot 2001). Several interconnected cellular processes regulate these early stages of infection: cell cycle progression followed by cytokinesis and appressorium differentiation (Saunders et al. 2010a, b), programmed cell death (Veneault-Fourrey et al. 2006) and mobilisation of metabolic resources to generate high concentrations of glycerol for turgor-mediated penetration (Howard et al. 1991; Thines et al. 2000).
During germ tube elongation, the nucleus of the germinating cell moves towards the middle of the germ tube. Subsequently, the nucleus undergoes mitosis and one of the daughter nuclei moves towards appressoria, whereas the second nucleus returns to the conidium (Veneault-Fourrey et al. 2006). Concomitantly, storage products are transported towards the appressorium during this first nuclear division, and a septum is formed separating the appressorium from the germ tube (Saunders et al. 2010a). Then, appressorial melanisation begins. Melanin is deposited in the space between cell wall and plasma membrane to maintain appressorium integrity. High levels of glycerol derived from the degradation of stored lipids and glycogen within the appressorium build the osmotic force required for penetration of the cuticle (Thines et al. 2000). Finally, the conidium and germ tube collapse using an autophagic mechanism which is vital for pathogenicity (Talbot and Kershaw 2009; Veneault-Fourrey et al. 2006).
Appressorium morphogenesis, autophagic cell death and mobilisation of carbohydrate and lipid reserves to the appressorium are processes regulated by the mitogen-activated protein kinase (MAPK) Pmk1 pathway (Thines et al. 2000; Veneault-Fourrey et al. 2006; Xu and Hamer 1996). In eukaryotes, MAPKs are involved in the activation of cellular processes in response to environmental cues that help to adapt the cell to the exterior (Zhao et al. 2007). In Saccharomyces cerevisiae, five MAPK pathways exist and have been characterised in detail (Zhao et al. 2007). In M. oryzae, the MAPK Pmk1 (pathogenicity MAP kinase1) has been identified as the homologue of S. cerevisiae FUS3/KSS1 MAPK cascades, which regulate mating and filamentous growth (Xu and Hamer 1996). The Δpmk1 mutants are unable to produce appressorium and are non-pathogenic. However, they can recognise hydrophobic surfaces and react to exogenously applied cAMP. In M. oryzae, Pmk1 is also required for invasive hyphae growth (Xu and Hamer 1996). Homologues of PMK1 are required for pathogenicity in all fungal plant pathogens (biotrophs or necrotrophs) of monocot and dicot plants studied to date, indicating that this MAPK pathway is widely conserved (Zhao et al. 2007). This pathway is under extensive analysis, and genes acting upstream and downstream of Pmk1 have been identified. These include the MAPK kinases Mst7 and Mst11 (Zhao et al. 2005); the PAK kinase Chm1 (Li et al. 2004); the Rho-GTPase MgRac1 (Chen et al. 2008); the scaffold protein Mst50 that interacts with Ras1, Ras2, Ccd42 and the Gβ subunit Mgb1 (Park et al. 2006); the membrane receptors MoMsb2 and MoSho1 (Liu et al. 2011); several transcription factors including Mst12 (Park et al. 2002), Mig1 (Mehrabi et al. 2008), MoSLF1 (Li et al. 2011) and MoMcm1 (Zhou et al. 2011); and two novel Pmk1-interacting proteins Pic1 and Pic5 (Zhang et al. 2011b).
Two other MAPK signalling pathways have been described in M. oryzae. The Mps1-dependent MAPK pathway is implicated in appressoria penetration and cell wall integrity (Xu et al. 1998), whereas the MAPK Osm1 is involved in the cellular response to osmotic stresses and is not required for plant infection (Dixon et al. 1999).
4.7.4 Signalling and Cytoskeletal Dynamics Regulate Fungal Plant Penetration
At the base of the appressorium, a pore ring is formed and the fungus initiates the turgor-driven penetration into plant tissues by developing a specialised hypha or penetration peg (Talbot 2003). The penetration peg enables the fungus to cross the plant cell wall and extend to the epidermal lumen of the plant. This structure is enriched in actin filaments and lacks organelles in its cytoplasm (Bourett and Howard 1992). Particularly two important signalling pathways regulate this step, the Mps1 MAPK cascade (Xu et al. 1998) and the cAMP response pathway (Xu et al. 1997).
The cAMP-dependent cascade acts cooperatively with the PMK1 pathway during M. oryzae plant penetration (Xu and Hamer 1996). The cAMP cascade is required for surface recognition and penetration peg emergence but not appressorium differentiation (Xu et al. 1997). The generation of glycerol and high turgor pressure within the appressorium requires a rapid degradation of lipid and glycogen reserves which is under the control of the cAMP-activated protein kinase A (PKA) pathway (Thines et al. 2000). Several key components of this signalling pathway have been studied such as the adenylate cyclase Mac1 (Choi and Dean 1997), the catalytic subunit of cAMP-dependent PKA CpkA (Xu et al. 1997), the phosphodiesterases PdeL and PdeH (Zhang et al. 2011a) and the Mac1-interating protein Cap1 (Zhou et al. 2012), which regulates the crosstalk between PMK1- and cAMP-dependent pathways through its interaction with Ras2.
Additional genetic determinants have been found to play a role in M. oryzae penetration including the tetraspanin PLS1 (Clergeot et al. 2001; Lambou et al. 2008), the Pmk1-regulated genes GAS1 and GAS2 encoding unknown proteins conserved in filamentous fungi (Xue et al. 2002), the aminophospholipid translocase PDE1 (Balhadere and Talbot 2001) and the isocitrate lyase gene ICL1 of the glyoxylate cycle (Wang et al. 2003).
An actin network organised at the base of the appressorium forces the emergence of a penetration peg (Bourett and Howard 1992). In M. oryzae, this process is regulated by septins (Dagdas et al. 2012). Septins are highly conserved GTPases present in fungi and animals that participate in cytoskeletal-dependent cellular processes such as cytokinesis, polarity and secretion (Gladfelter 2006; Mostowy and Cossart 2012). Septins also act as diffusion barriers. M. oryzae genome contain five septin genes, four of which are core septins present in budding yeast. M. oryzae septins form a dynamic septin ring that contributes to the formation of a toroidal filamentous actin network surrounding the appressorial pore, where the penetration peg differentiates (Dagdas et al. 2012).
4.7.5 Insights into M. oryzae Invasive Growth
Within the host cell, the fungus develops several types of biotrophic invasive hyphae (IH; Kankanala et al. 2007), which contain distinct morphological features compared to the filamentous hyphae produced on the leaf surface or in vitro. Soon after the M. oryzae penetration peg has crossed the epidermal cell wall, it differentiates into a short and thin filamentous hypha known as primary IH. This primary IH precedes the formation of a thicker intracellular pseudohypha called bulbous IH. The bulbous IH grows within the cytoplasm and moves beyond the first invaded cell by crossing with constricted infection pegs at regions of the plasma membrane where plasmodesmata aggregate, also known as pit fields (Bell and Oparka 2011). Thereafter, the bulbous IH differentiates into filamentous IH in the new invaded cell and subsequent fungal invasion continues into neighbouring cells (Kankanala et al. 2007). Importantly, bulbous and filamentous IH are not in direct contact with the plant cell cytoplasm since a plant-derived plasma membrane called extra-invasive hyphal membrane (EIHM) surrounds them. There is no well-established matrix between IH and EIHM, and IH grows in close contact with the EIHM (Kankanala et al. 2007). Secreted fungal proteins and other compounds are retained inside this space such as Slp1 and Bas4 effectors or can be translocated into the plant cytoplasm as it is the case for Pwl2 (described later). An additional morphological feature of M. oryzae invasive growth is the formation of biotrophic interfacial complexes (BICs) where effector proteins accumulate (Khang et al. 2010). When M. oryzae penetrates the first cell, a BIC is formed at the tip of the first bulbous IH, and is left behind, remaining as a discrete structure while the bulbous IH continues growing. New BICs are observed at the tip of each IH growing inside the plant cell. Five to six days after the initial fungal infection, conidiophores start to emerge in the leaf surface, and spores are produced massively during 2 weeks.
The elucidation of the molecular mechanisms involved in M. oryzae invasive growth has been largely overlooked because many of the mutants characterised in this organism are penetration defective. There is an extensive coupling between penetration and invasive growth processes since cell wall degradation and mechanical pressure are also involved during fungal growth inside the host cells (Heath et al. 1992; Xu et al. 1997). As an example, Δmst12 fails to penetrate onion epidermal cells and to infect wounded leaves although it differentiates melanised appressoria, indicating that Mst12 is required for both penetration peg formation and invasive growth differentiation (Park et al. 2002). Very few genes have been found to play specifically a critical role during M. oryzae plant invasion. MIG1 is involved in the late stages of M. oryzae infection since the Δmig1 mutants form normal appressoria, penetrate host cells and develop primary IH but fail to infect wounded leaves. Mig1 is one of the two MADS-box transcription factors present in M. oryzae and a downstream target of the MAPK Mps1 (Mehrabi et al. 2008). The MIR1 gene specifically expressed in M. oryzae IH encodes a protein of unknown function, which is present only in the M. grisea species complex. Despite the fact that MIR1 expression is exclusively found in IH, Δmir1 mutants have no defects in appressorial penetration and are fully pathogenic (Li et al. 2007).
4.7.6 Fungal Metabolism and Plant Infection
M. oryzae has to adapt to the changing nutritional environment during host invasion, and consequently metabolism plays an essential role during M. oryzae invasive growth. M. oryzae is considered a hemibiotrophic fungus based on its nutritional mode during host invasion. However, genes regulating the switch in life style and acquisition of nutrients during plant infection are largely unknown (Fernandez and Wilson 2012; Kankanala et al. 2007). The duration of the biotrophic versus the necrotrophic phase in M. oryzae is also unknown. During early stages of rice colonisation, M. oryzae grows and fulfils its nutritional needs from the plant tissue without killing the host cells due to its ability to manipulate rice physiology as many other biotrophs do (Mendgen and Hahn 2002; Mengiste 2012). During this biotrophic stage, limited amounts of cell wall-degrading enzymes are produced and toxin production is absent according to a biotrophic life style. By contrast, extensive degradation of plant cell walls is observed at later time points of infection and in heavily invaded tissues, both stages associated with the necrotrophic phase of the fungus (Kankanala et al. 2007; Rodrigues et al. 2003). Typically, necrotrophs produce phytotoxic compounds and cell wall-degrading enzymes to kill the cells and cause leakage of nutrients (Mengiste 2012). Plant cell walls nearby M. oryzae hyphae show strong enzymatic digestion, correlating with M. oryzae necrotrophic phase (Kankanala et al. 2007).
Possibly one of the cues that trigger the switch from biotrophic to necrotrophic hyphae in M. oryzae is the lack of carbon sources within the host cell. It is known that nutrient starvation also can act as an environmental cue for infection-related differentiation (Talbot et al. 1997). M. oryzae has to limit the acquisition of nutrients during its biotrophic phase to maintain host cell integrity. Consequently, the use of nutrients must be highly regulated during M. oryzae biotrophic growth in order to respond appropriately to nutrient availability. Several interconnected pathways regulate M. oryzae growth in response to nutrients during plant invasion. These include the target of rapamycin (TOR) signalling cascade, carbon catabolite repression (CCR), nitrogen metabolite repression (NMR) and the integration of carbon and nitrogen metabolism by trehalose-6-phosphate synthase 1 (Tps1).
The TOR signalling cascade is an intracellular regulatory network used by eukaryotic cells to regulate growth according to nutrient availability. The 14-3-3 proteins are involved in key cellular processes and integrate environmental cues through the regulation of signalling pathways, including TOR. The TOR signalling pathway is regulated by the RNA-binding protein Rbp35 (Franceschetti et al. 2011). Rbp35 is a component of the polyadenylation machinery, and it is required for alternative 3' end processing of pre-mRNAs. One of the RBP35 targets is the 14-3-3 pre-mRNA, and this could explain the defects that Δrbp35 shows on TOR signalling and plant infection.
NMR is a highly regulated process in which preferred nitrogen sources, such as ammonia, glutamine and glutamate, are used preferentially. Ammonia is the preferred nitrogen source for M. oryzae. The NMR in M. oryzae occurs through the transcriptional activator Nut1, the M. oryzae AreA/Nit2 orthologue (Froeliger and Carpenter 1996). The expression of a large number of genes encoding enzymes that are involved in the utilisation of various secondary nitrogen sources—nitrate, purines or amino acids—is subject to nitrogen metabolic repression and is positively regulated by Nut1. The Δnut1 mutant can grow on ammonia, which does not require an active Nut1, but Δnut1 is unable to grow on certain alternative nitrogen sources such as nitrate. M. oryzae mutants in genes involved in nitrate assimilation and whose expression is regulated by Nut1 such as NIA1 and NIR1 are fully pathogenic on rice leaves (Lau and Hamer 1996; Wilson et al. 2010). This suggests that NMR is not involved in M. oryzae leaf colonisation and consequently the fungus can assimilate preferred sources of nitrogen (ammonia, glutamine or glutamate) from aerial host tissues. However, genes involved in response to nitrogen availability are important for infection. Two M. oryzae nitrogen-regulatory genes of unknown identity, NPR1 and NPR2, are required for growth on a wide range of secondary nitrogen sources, including nitrate, and do not develop lesions on barley (Lau and Hamer 1996). Therefore, nitrate is not required for M. oryzae leaf infection, but secondary nitrogen sources assimilated via NPR1 or NPR2 are necessary for development of full disease symptoms. Additionally, several studies have shown that nitrogen-limiting conditions result in the expression of genes required for fungal pathogenicity such as the genes encoding the hydrophobin MPG1 and the vacuolar subtilisin-like protease SPM1 (Donofrio et al. 2006; Saitoh et al. 2009; Soanes et al. 2002). We require further studies to understand the molecular mechanisms underlying NMR and their involvement in nitrogen assimilation during M. oryzae plant infection.
CCR is a genetic mechanism that ensures the preferential use of glucose over other, less-preferred carbon sources, and it is also present in M. oryzae (Fernandez et al. 2012). M. oryzae has the ability to use a wide range of mono- and disaccharides as sole carbon source but has a strong preference for glucose (Fernandez and Wilson 2012; Tanzer et al. 2003; Wilson et al. 2007). In A. nidulans, CCR is mediated at DNA level by the global transcriptional repressor CreA. A putative orthologue of CreA (MGG_ 11201) is present in M. oryzae, and its role in fungal pathogenicity has yet to be elucidated.
An interesting interconnection of NMR and CCR is mediated by the sugar sensor trehalose-6-phosphate synthase (Tps1) and trehalose-6-phosphate (Fernandez et al. 2012). Tps1 is one of the three mediators of CCR identified in M. oryzae. The other two mediators are the Nmr1/2/3 inhibitor proteins and Mdt1, a multidrug and toxin extrusion (MATE)–family pump. Tps1 is a metabolic enzyme that synthesises trehalose-6-phosphate (T6P, a trehalose intermediate) from UDP-glucose and glucose-6-phosphate (G6P). Tps1 has two roles, as a biosynthetic enzyme and as signalling component of G6P. The sensing of G6P by Tps1 results in activation of the activity of the enzyme glucose-6-phosphate dehydrogenase (G6PDH), which converts NADP to NADPH using G6P in the pentose phosphate pathway. Therefore, Tps1 controls intracellular levels of NADPH (depending on the concentration of G6P) and subsequent activation of NADPH-dependent signalling cascades that interconnect carbon and nitrogen metabolism. When NADPH levels increase in a Tps1-dependent manner, three NADP-dependent inhibitor proteins (Nmr1 to Nmr3) are inactivated. As a result of inactivation of Nmr proteins, at least three GATA transcription factors become active, one of which is the white collar-2 homologue involved in light sensing (Pas1). The other GATA factor is essential for appressorium formation (Asd4), and the third GATA factor is Nut1 (Wilson et al. 2010). The modulation of GATA factor activity in the NADPH-dependent signalling pathway results in Tps1-dependent expression of at least three known virulence factors: the melanin enzyme Alb1, the seven transmembrane receptor Pth11 and the hydrophobin Mpg1 (Wilson et al. 2007). Accordingly, Δtps1 mutants are non-pathogenic. Tps1 regulation of Nut1 results in similar but not identical growth phenotype of Δtps1 and Δnut1 strains on a wide range of nitrogen sources. An additional regulator of the CCR signal transduction pathway in M. oryzae has been identified during a forward suppressor screening in Δnut1 background (Fernandez et al. 2012). Mdt1 is a member of the MATE protein family required for sporulation and plant infection but not appressorium differentiation. Mdt1 regulates carbon metabolism via extrusion of citrate during infection and growth contributing to M. oryzae in planta nutrient adaptation.
In summary, NADPH signalling, CCR, NMR and TOR are mechanisms by which M. oryzae can sense and adapt its metabolic status to nutrient availability during in planta growth. Future research will determine the interplay among these regulatory pathways that play a pivotal role in the establishment of plant disease.
4.7.7 Secretion Systems: Effectors, Toxins and ABC Transporters
Plant recognition of conserved microbial features (pathogen- or microbial-associated molecular patterns, PAMPs or MAMPs) such as chitin or flagellin (Howard et al. 1991) is mediated by pattern recognition receptors (PRRs; Zipfel 2008). During the coevolution of plants and associated pathogens, plants have developed two levels of immune responses (Jones and Dangl 2006), the PAMP-triggered immunity (PTI) and effector-triggered immunity (ETI). In general, PAMPs are conserved among species of pathogens and play an essential role in pathogenicity. Therefore, PTI represents the first level of immune response in a host. The second type of plant innate immunity, the ETI, is activated upon recognition of highly diverse molecules secreted by the pathogens known as effectors. Fungal effectors play an essential role during invasion (Hogenhout et al. 2009; Stergiopoulos and de Wit 2009). Successful pathogens have managed to produce effectors that overcome PTI. Conversely, some plant resistance genes have evolved to recognise such type of effectors blocking their effect (ETI). Then, plant pathogens no longer can infect their host and become non-pathogenic or avirulent (Jones and Dangl 2006).
M. oryzae contains ~1,600 predicted secreted proteins that may play a role during rice infection (Dean et al. 2005; Soanes et al. 2008). It is not easy to assign a role in pathogenicity to an effector protein by gene disruption due to the large amount of secreted proteins, possibly with functional redundancy, present in M. oryzae (Saitoh et al. 2012). Two effector proteins with virulence functions have been characterised in M. oryzae, MC69 (Saitoh et al. 2012) and Slp1 (Mentlak et al. 2012). MC69 is a single secreted protein that is indispensable for virulence in fungi pathogenic on both monocots and dicots. When MC69 is absent, M. oryzae pathogenicity is severely reduced after penetration into the host cells. However, there are no clear evidences supporting how MC69 contributes to pathogenicity or virulence. The Secreted LysM Protein1 (Slp1) has two LysM domains involved in carbohydrate recognition and is secreted into apoplastic space during initial invasive growth in M. oryzae. This protein is only expressed during the biotrophic phase of M. oryzae (Mentlak et al. 2012). Slp1 can be glycosylated and can form oligomers (Mentlak et al. 2012). The Δslp1 mutants show reduced disease symptoms due to their defects in invasive growth (Mentlak et al. 2012). The Slp1 effector competes with the plant chitin receptor CEBiP to attenuate the rice immune response, the PTI, activated by the presence of M. oryzae chitin oligosaccharides.
To date, the majority of the effectors identified in M. oryzae act as avirulence (AVR) proteins triggering effector-mediated cell death (or ETI) and blocking subsequent pathogen invasion. However, their mode of action is still largely unknown at the molecular level. De novo sequencing of the Japanese rice isolate Ina168 genome and its comparison with the reference genome 70–15 has allowed the identification of a genomic region present only in Ina168 that contained three AVR genes (AVR-Pia, AVR-Pii and AVR- Pik/km/kp) (Table 4.2; Yoshida et al. 2009). An additional effector gene identified by map-based cloning is AVRPiz-t (Li et al. 2009). Knockout mutants in all these genes fail to show virulence phenotypes except in their specific cultivars containing the matching resistance genes. AvrPiz-t is able to suppress BAX-mediated programmed cell death in tobacco leaves in transient expression experiments, providing evidence that this effector may have a role in suppression of plant immunity. An interesting case of a protein with AVR effector function is M. oryzae Ace1. Ace1 is a polyketide synthase–nonribosomal peptide synthetase (PKS-NRPS) located within a gene cluster involved in the biosynthesis of secondary metabolite(s). The metabolite synthesised by the ACE1 gene product represents the only secondary metabolite found in M. oryzae so far with an avirulence role (Collemare et al. 2008; Fudal et al. 2007). M. oryzae isolates containing the ACE1 gene are unable to infect rice cultivars containing the resistance gene Pi33 (Berruyer et al. 2003). The ACE1 gene is exclusively expressed in planta, making it difficult to identify the Ace1-dependent natural product. ACE1 expression is tightly coupled to the onset of appressorium-mediated penetration of the host cuticle.
Effectors are also involved in determining M. oryzae host species specificity. The M. oryzae AVR effector Pwl2 (pathogenicity towards weeping lovegrass 2) prevents M. oryzae isolates from infecting weeping lovegrass (Sweigard et al. 1995). The PWL gene family consists of four genes PWL1, PWL2, PWL3 and PWL4. Pwl1 is a functional AVR effector and has 78 % nucleotide identity with Pwl2. Pwl2 accumulates in the BICs, and this property correlates with its translocation across the plasma membrane into the rice cytoplasm. There are no evidences of avirulence roles for Pwl3 (63 % nucleotide identity) and Pwl4 (65 % nucleotide identity; Kang et al. 1995). Additional AVR genes identified in M. oryzae field isolates are AVR1-CO39, which is broadly present in M. oryzae populations adapted to other host species, and AVR-Pita1 (Valent et al. 1991). AVR-Pita1 is a subtelomeric effector gene which has been extensively studied to understand AVR gene evolution among field isolates in order to generate valuable information for the deployment of resistance genes in field crops (Chuma et al. 2011; Jia et al. 2000).
Four additional biotrophy-associated secreted (Bas1 to Bas4) protein effectors are expressed during biotrophic invasion but not in vitro (Khang et al. 2010). Bas1 is translocated into the rice cell cytoplasm and shows preferential accumulation in BICs, like Pwl2. M. oryzae translocated effectors moved ahead of the fungus and can be seen in the absence of invasive hyphae within the cells, suggesting that these effectors prepare host cells prior to fungal invasion (Khang et al. 2010). It is not clear how M. oryzae delivers effector proteins during its biotrophic phase into the host cells. The MgAPT2-dependent polarised exocytotic processes might contribute to the secretion of effectors during M. oryzae plant colonisation (Gilbert et al. 2006). Bas2 and Bas3 are found in BICs, but they also localise in cell walls of invasive hyphae. Bas4 is a potential matrix protein that preferentially accumulates between the EIHM and the M. oryzae cell wall. The knockout mutants in the BAS genes show no particular phenotype, indicating the functional redundancy of the fungal secretome. Some of these Bas proteins might be involved in altering plant components required for biotrophic invasion, but no clear evidences have been reported (Khang et al. 2010).
In addition to effector proteins, M. oryzae also secretes phytotoxins although this is a largely unexplored area. Pyriculol, tenuazonic acid and pyrichalasin H have been isolated from culture filtrates of M. grisea isolates (Tsurushima et al. 2005). Pyriculol induces necrosis and it is widely distributed among Magnaporthe species. Tenuazonic acid is also present in Alternaria species. Pyrichalasin H is a cytochalasin that prevents polymerisation of actin filaments and is able to inhibit rice seed development although it is not required for leaf disease symptoms. Pyrichalasin H is exclusively produced by blast isolates that infect Digitaria plants, and possibly it represents a host specific toxin (Tsurushima et al. 2005). Lately, the Mag-toxin has been purified from M. oryzae isolates infecting Avena species. Mag-toxin is a derivative of linoleic acid and only causes chlorosis in the presence of light. This toxin is able to induce mitochondria-associated ROS production and cell death (Tsurushima et al. 2010).
Plants secrete toxic compounds to defend themselves from pathogens. The ATP-binding cassette (ABC) transporters play an essential role in fungal survival allowing to efflux plant antimicrobial substances to the cell exterior (Coleman and Mylonakis 2009). M. oryzae has about 50 ABC transporters (Coleman and Mylonakis 2009). Four ABC transporters have been characterised in M. oryzae. Abc1, Abc3 and Abc4 are required for pathogenicity but are dispensable for appressorium differentiation (Gupta and Chattoo 2008; Sun et al. 2006; Urban et al. 1999). Mutants in these genes differentiate normal appressoria and are either unable to penetrate or die shortly after penetrating the host cell. The best characterised ABC transporter is Abc3, which localises in the plasma membrane of appressoria (Sun et al. 2006), and pumps out a plant-derived steroidal glycoside (Patkar et al. 2012).
4.7.8 Conidiation and Light Regulation
The sporulation process is an essential step for fungal reproduction and dispersal and influences largely the disease progression in the field. M. oryzae can produce two types of spores. Some M. oryzae/grisea isolates produce single-celled microconidia (Chuma et al. 2009; Kato et al. 1994) (Fig. 4.4b). Microconidia have thin cell walls and lack nucleoli. They have been identified in other fungi—N. crassa, Botrytis cinerea or Podospora anserina—where they play a role as spermatia during sexual reproduction (Fukumori et al. 2004). Mature microconidia show lower metabolic activity compared to germ tubes, indicating that they may be quiescent or dormant. The M. oryzae MADS-box transcription factor MoMcm1 regulates microconidia production and is also involved in male fertility, supporting the role of microconidia as spermatia during the sexual cycle of M. oryzae (Zhou et al. 2011).
Macroconidia (also named conidia or asexual spores) represent the main dispersal forms of the blast fungus. M. oryzae conidia are pyriform (pear shaped) and bisepted (occasionally 1 or 3 septa can be seen). These two septa generate three distinct cellular compartments in the conidium, each of them enclosing a nucleus (Fig. 4.4c). Conidia size ranges between 19–27 μm long and 8–10 μm wide. Normally, conidia present a basal appendage at the point of attachment to the conidiophore. Conidiophores are specialised hyphae up to 130 × 3–4 μm in size, and conidia are formed in their apex (Fig. 4.4d). A mature M. oryzae conidiophore rarely branches and can form between three and five conidia sympodially arranged. Conidiophores emerge to the plant cell surface and release conidia into the environment.
Molecular mechanisms governing conidiation have been characterised in exquisite detail for the model organisms A. nidulans and N. crassa (Etxebeste et al. 2010; Park and Yu 2012). Conidiation-defective genes and genetic loci have also been identified in the rice blast fungus such as the CON mutants (Shi et al. 1998), ACR1 (Lau and Hamer 1998), COS1 (Zhou et al. 2009), SMO (Hamer et al. 1989), CDC15 (Goh et al. 2011) and COM1 (Yang et al. 2010). A genome-wide expression profile using spores from the rice isolate KJ201 has identified several hundred genes to be up- or downregulated during M. oryzae conidiation, approximately 4.5 % of its total gene content (Kim and Lee 2012). A further comparative transcriptome analysis between the wild-type strain and the Δmohox2 mutant under sporulation conditions has identified a subset of conidiation-related genes regulated by the homeobox transcription factor MoHox2/Htf1 (Kim et al. 2009; Liu et al. 2010b). ΔMohox2 mutants fail to produce conidia indicating that this transcriptional regulator plays an essential role in M. oryzae conidiation process. Not surprisingly, expression of M. oryzae genes MoCON6, ACR1, MoBRLA and MoFLBC is significantly upregulated during conidiation in the wild type but not in Δmohox2. These genes are also highly expressed during sporulation in other fungal species (Adams et al. 1988; Etxebeste et al. 2010; Kwon et al. 2010; Springer and Yanofsky 1992). By contrast, the expression of M. oryzae MoFLBA and MoVOSA (the A. nidulans flbA and vosA orthologues, respectively) is significantly downregulated or unaltered in the wild type while is highly upregulated during conidiation in A. nidulans. The M. oryzae ΔvosA mutant has no defects in conidiation although the A. nidulans VosA is a key regulator of the sporulation process. This may suggest that gene pathways regulating conidiation differ between fungal species because they derive from new mechanisms of gene regulation, rather than biochemical function. Further investigation is necessary to define the genetic pathway and molecular mechanisms controlling conidiation in M. oryzae.
The light is an environmental factor that influences several biological processes in M. oryzae such as conidiation. It is necessary to grow M. oryzae under light/dark conditions to get good sporulation rates (Lee et al. 2006). Asexual development and light regulation are interconnected processes in A. nidulans and N. crassa (Olmedo et al. 2010a, b; Ruger-Herreros et al. 2011). The light during asexual development affects mainly aerial hyphae and conidiophore differentiation. Conidiation in M. oryzae is suppressed by blue light during light/dark cycling and the release of conidia is controlled by both blue and red light (Lee et al. 2006). Therefore, M. oryzae senses the light-to-dark transition, and this environmental cue triggers asexual differentiation and spore release. It is clear that environmental light also influences M. oryzae interaction with rice. It seems that a dark phase applied immediately after pathogen–host contact plays a critical role for disease development (Kim et al. 2011b). Significant light-dependent disease suppression is observed in rice plants infected with M. oryzae when plants are exposed to light (instead of darkness) directly after inoculation (Kim et al. 2011b). In nature, it is difficult to establish the contribution of a particular environmental factor to disease progression since environmental factors are interdependent and can affect the host physiology (plant), the pathogen physiology (fungus) and/or the interaction between both organisms. A partial “blind” strain of M. oryzae required for darkness sensing (a knockout strain in MgWC-1, the blue light photoreceptor gene) has allowed to dissect the effect of light in the fungus during disease development. MgWc-1 is required for light-dependent disease suppression during the dark phase (disease-conducive light condition) after pathogen–host contact. In other words, a full disease progression requires a light/dark cycle after pathogen–host contact and light-to-dark transition sensed by photoreceptors. However, appressorium differentiation and penetration is not regulated by light, and therefore, light does not affect early stages of M. oryzae plant infection. Plants are subject to an overall greater pathogen challenge during the night. Possibly the fungus recognises darkness to mobilise fungal effectors (and also possibly metabolic reserves) during invasive growth, as has been suggested for Cryptococcus neoformans, as a mechanism to avoid the light-regulated increased defence responses in plants (Griebel and Zeier 2008; Idnurm and Heitman 2005). Light-to-dark transitions must be taken in account to understand the crosstalk between plant and associated fungal pathogens, considering that both organisms have an active circadian clock.
4.8 The Dark Phase of Blast: M. oryzae Root Infection Biology
Similar to its close relatives, M. oryzae infects roots (under laboratory conditions) and undertakes a set of developmental programmes typical of root-infecting pathogens (Sesma and Osbourn 2004; Tucker et al. 2010). Several key differences have been found between the mode of penetration of leaves and roots. In contrast to the melanised appressoria observed on leaves, M. oryzae produces hyphal swellings to penetrate roots, resembling the simple hyphopodium seen in root-infecting fungi of the G. graminis–Phialophora complex (Fig. 4.4e). M. oryzae hyphopodia are not melanised and M. oryzae melanin-deficient mutants are able to produce hyphopodia and infect roots (Sesma and Osbourn 2004). The PKA regulates the high turgor pressure within appressoria generated by the degradation of lipid and glycogen reserves (Thines et al. 2000). The M. oryzae Δcpka mutant produces hyphopodia and penetrates roots, indicating that root colonisation is not dependent on CPKA (Sesma and Osbourn 2004). Consequently, M. oryzae penetrates the epidermal root cells through a melanin-independent mechanism and the mechanical entry of the hard leaf surface by osmotic force is not operational during hyphopodia-mediated root penetration. From the host perspective, defence-related gene transcripts of rice showed a different temporal induction pattern during M. oryzae infection of leaves or roots (Marcel et al. 2010), which correlate with the different invasion mechanisms that the rice blast fungus undertakes for colonisation of leaves and roots.
Pre-invasive hyphae (pre-IH) are another type of fungal development observed on root surfaces that also mediates direct penetration of epidermal root cells (Tucker et al. 2010). M. oryzae pre-IH is developed from hyphopodia or germ tubes and penetrates roots directly. The pre-IH can be followed by differential labelling with concanavalin A and wheat germ agglutinin, which indicates that cell wall changes accompanied to this morphogenetic programme. Artificial surfaces such as hydrophilic polystyrene (PHIL-PS) can induce hyphopodia-like structures and pre-IH. The mutant Δpmk1 is non-pathogenic on roots (Dufresne and Osbourn 2001), and this mutant is unable to develop pre-IH on roots and PHIL-PS. Consequently, this fungal differentiation is regulated by the MAPK Pmk1 cascade. Other structures typical of root-infecting fungi seen during M. oryzae root colonisation include microsclerotia and resting structures such as vesicles and swollen cells (Gangopadhyay and Row 1986; Lee et al. 2000; Sesma and Osbourn 2004).
Several lines of evidence have led to the hypothesis that the hyphopodium is an intermediate step before appressorium penetration. It is possible that the primitive hyphopodia evolved by acquisition of melanin and generation of high turgor pressure into a more sophisticated penetration structure, the appressorium (Tucker et al. 2010). The screening of M. oryzae insertional library of 2,885T-DNA transformants looking for altered pre-IH differentiation mutants on PHIL-PS has identified 20 transformants that show reduced virulence or are non-pathogenic on leaves and/or roots (Tucker et al. 2010). Further analysis of these mutants has revealed that appressorium, hyphopodium and pre-IH formation are highly coupled developmental processes, and very few mutants show an organ-specific involvement for infection (Tucker et al. 2010). This indicates that a significant set of common genes are necessary for fungal infection on both plant organs. Out of the 20 mutants, M1373 shows a root-specific infection-deficient phenotype (Table 4.3). This mutant lacks the M. oryzae orthologue of exportin-5/Msn5p (EXP5). The defects of the Δexp5 mutant on disease symptoms production are more evident on roots than on leaves. M. oryzae EXP5 presents a steady-state nuclear localisation under all the conditions tested. Δexp5 mutants show a reduction in conidia production (ca. 40 times lower) and altered preinvasive growth on PHIL-PS. The perimeters of the leaf lesions produced by Δexp5 are smaller, which suggests deficiencies in invasive growth. Pathogenesis-related proteins and/or RNAs transported by this nucleocytoplasmic receptor play a crucial role during M. oryzae infection-associated development.
Exp5 may be involved in the nucleocytoplasmic transport of proteins implicated in nitrogen assimilation. Differences have been found in the role played by nitrogen-related genes during M. oryzae leaf and root colonisation. The assimilation of nitrogen by M. oryzae from underground plant tissues is regulated by the global nitrogen regulator Nut1 (Froeliger and Carpenter 1996). The Δnut1 mutant is non-pathogenic on roots but infects leaves as well as the wild-type strain (Dufresne and Osbourn 2001). Consequently, M. oryzae absorbs nitrogen from less preferred sources in root tissues, and therefore, the NMR plays a crucial role during root infection. The mutants Δnpr1 and Δnpr2 are non-pathogenic on leaves and show opposite phenotypes on roots (Table 4.3), representing an additional evidence of the different roles that nitrogen-related genes play during M. oryzae colonisation of leaves and roots.
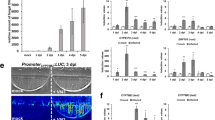
4.8.1 Rice Blast Underground Infection and Arbuscular Mycorrhizal Symbiosis
There are similarities between M. oryzae and the ancient mycorrhizal associations. A global transcriptome profile carried out with the arbuscular mycorrhizal fungus Glomus intraradices and two different root-infecting fungal pathogens (M. oryzae and Fusarium moniliforme) during root infection has demonstrated the presence of common rice genes equally expressed in all three associations. This indicates a common response of rice to fungal invasion (Guimil et al. 2005). A larger set of different genes are shared between the symbiont G. intraradices and M. oryzae than between the G. intraradices and the necrotroph F. moniliforme, as expected for the biotrophic nature of M. oryzae. From the fungal perspective, there are also common protein domains shared by both M. oryzae and the symbiont G. intraradices implicated in root colonisation, suggesting a conservation and expansion of protein families with root colonisation-related functions (Heupel et al. 2010). This is the case for the ERA-like GTPase Erl1 of M. oryzae and the Gin1 protein from the symbiont G. intraradices. The root disease symptoms defects of M. oryzae Δerl1 mutant are restored by reintroduction of the G. intraradices GIN1 gene in Δerl1. Interestingly, the expression of the G. intraradices symbiotic-related gene SP7 into M. oryzae can decrease its necrotic behaviour on rice roots, indicating the general ability of G. intraradices Sp7 protein to contribute to the development of the biotrophic status of G. intraradices and M. oryzae (Kloppholz et al. 2011).
Mitochondrial respiratory activity of symbiotic fungi is stimulated by root exudates (Tamasloukht et al. 2003). Similarly, evidences suggest that mitochondrial respiration is also important for root colonisation of fungal pathogens. The MgFOW1 gene plays an important role during M. oryzae invasion of root cortical cells, but it is dispensable for leaf infection (Sesma and Osbourn 2004). Fow1 was initially identified in the fungal pathogen Fusarium oxysporum as a protein required for colonisation of vascular tissues (Inoue et al. 2002). Fow1 is a mitochondrial carrier protein that shares close sequence similarity with the yeast protein YHM2p required for tricarboxylic acid transport. M. oryzae Δmgfow1 mutants, like Δfow1 mutants in Fusarium oxysporum, are unimpaired in their ability to utilise glycerol as a carbon source in contrast to yeast Δyhm2 “petite” mutants, and deletion of MgFOW1 gene has no effect in fungal growth on a range of rich and minimal media and conidiation, indicating that MgFow1 is dispensable for saprophytic growth. YHM2p associates with mtDNA in vivo and is implicated in replication and segregation of yeast mitochondrial genomes. Maintenance of mtDNA during cell division is essential for progeny to be respiratory competent. In addition, mitochondrial status is sensed by eukaryotic cells through retrograde signalling, a pathway of communication from mitochondria to the nucleus under normal and pathophysiological conditions that regulate changes in nuclear gene expression (Galluzzi et al. 2012). These changes lead to a reconfiguration of metabolism to adapt cells to defects in mitochondria. The function of Fow1-like proteins in phytopathogenic fungi is not known. MgFow1 has the potential to act as a bifunctional protein (mitochondrial carrier and mtDNA-binding protein). Elucidation of Mgfow1 function will represent an important step towards understanding invasion mechanisms of roots and vascular tissues in M. oryzae. The relationship between senescence and mitochondrial respiratory activity is found in ascomycetes (P. anserina, N. crassa), and further investigation in this area may help to clarify the function of the MgFow1 protein during M. oryzae underground infection.
4.9 Concluding Remarks
In the past years, exquisite molecular and cellular approaches have been developed to understand critical processes underlying M. oryzae pathogenicity. However, M. oryzae shows a rapid evolution of host specificity by diverse mutational events, and achieving durable blast resistance represents a challenge. Climate change is likely to alter the geographical range of fungal pathogens, and cereal infection may become more widespread and unpredictable. A clear example of this is the emerging blast disease on wheat in South America (Cruz et al. 2012). As a result of climate change, Europe may become a viable environment for M. oryzae on wheat, our staple cereal crop. Preventing methods and improving protection of staple cereal crops will become vital during the following years. Undoubtedly, a better understanding of M. oryzae plant colonisation will have positive implications for the food security and economic stability of rice- and wheat-dependent populations worldwide. It will also have important implications for the development of new strategies for plant breeding and durable disease control. Fungal root infection processes are poorly understood within the Magnaporthaceae family due to the genetic intractability of root-infecting strains of G. graminis (take-all fungus) and M. poae species. Certainly, the dissection of M. oryzae root infection process will contribute to understand root infection mechanisms undertaken by fungal species of the Magnaporthaceae family.
4.10 Fungal Databases
-
Magnaporthe grisea genome database (Broad Institute): http://www.broadinstitute.org/annotation/genome/magnaporthe_grisea/MultiHome.html
-
Magnaporthe comparative database (Broad Institute): http://www.broadinstitute.org/annotation/genome/magnaporthe_comparative/MultiHome.html
-
Ensembl fungi: http://www.fungi.ensembl.org/Magnaporthe_oryzae/Info/Index
-
FungiDB, an integrated functional genomics database for fungi: http://www.fungidb.org/fungidb/
-
M. oryzae EST database (NIAS): http://www.mg.dna.affrc.go.jp/
-
COGEME EST Database: http://www.ri.imb.nrc.ca/cogeme/index.html
-
M. grisea MPSS database (Massively Parallel Signature Sequencing): http://www.mpss.udel.edu/mg/
-
Orygenes DB: an interactive tool for rice reverse genetics http://www.orygenesdb.cirad.fr/
-
Oryzabase: Integrated Rice Science Database http://www.shigen.nig.ac.jp/rice/oryzabase/top/top.jsp
-
MGOS, Magnaporthe grisea—Oryza sativa interaction database: http://www.mgosdb.org/
-
PHI base (Pathogen–Host Interaction database) offers molecular and biological information on genes involved in host–pathogen interactions. http://www.phi-base.org/
-
Fungal Secretome Database: http://www.fsd.riceblast.snu.ac.kr/index.php?a=view
-
Comparative fungal genomics platform: http://www.cfgp.riceblast.snu.ac.kr/main.php
-
M. oryzae T-DNA analysis platform: http://www.atmt.snu.ac.kr/ and http://www.tdna.snu.ac.kr/
-
Fungal transcription factor database: http://www.ftfd.snu.ac.kr/index.php?a=view
-
Fungal Nomenclature databases: http://www.indexfungorum.org/ and http://www.mycobank.org/
References
Adams TH, Boylan MT, Timberlake WE (1988) BrlA is necessary and sufficient to direct conidiophore development in Aspergillus nidulans. Cell 54:353–362
Ahn N, Kim S, Choi W, Im KH, Lee YH (2004) Extracellular matrix protein gene, EMP1, is required for appressorium formation and pathogenicity of the rice blast fungus, Magnaporthe grisea. Mol Cells 17:166–173
Arnaud-Haond S, Duarte CM, Alberto F, Serrao EA (2007) Standardizing methods to address clonality in population studies. Mol Ecol 16:5115–5139
Arx JA, Olivier DL (1952) The taxonomy of Ophiobolus graminis Sacc. Trans Br Mycol Soc 35:29–33
Asher MC, Shipton PJ (1981) Biology and control of take-all. Academic, London
Ashikawa I, Hayashi N, Yamane H, Kanamori H, Wu JZ, Matsumoto T, Ono K, Yano M (2008) Two adjacent nucleotide-binding site-leucine-rich repeat class genes are required to confer pikm-specific rice blast resistance. Genetics 180:2267–2276
Bae CY, Kim S, Choi WB, Lee YH (2007) Involvement of extracellular matrix and integrin-like proteins on conidial adhesion and appressorium differentiation in Magnaporthe oryzae. J Microbiol Biotechnol 17:1198–1203
Balhadere PV, Talbot NJ (2001) PDE1 encodes a P-Type ATPase involved in appressorium-mediated plant infection by the rice blast fungus Magnaporthe grisea. Plant Cell 13:1987–2004
Balhadere PV, Foster AJ, Talbot NJ (1999) Identification of pathogenicity mutants of the rice blast fungus Magnaporthe grisea by insertional mutagenesis. Mol Plant Microbe Interact 12:129–142
Ballini E, Morel J-B, Droc G, Price A, Courtois B, Nottéghem J-L, Tharreau D (2008) A genome-wide meta-analysis of rice blast resistance genes and quantitative trait loci provides new insights into partial and complete resistance. Mol Plant Microbe Interact 21:859–868
Bateman GL, Ward E, Antoniw JF (1992) Identification of Gaeumannomyces graminis var. tritici and G. graminis var. avenae using a DNA probe and nonmolecular methods. Mycol Res 96:737–742
Beckerman JL, Ebbole DJ (1996) MPG1, a gene encoding a fungal hydrophobin of Magnaporthe grisea, is involved in surface recognition. Mol Plant Microbe Interact 9:450–456
Bell K, Oparka K (2011) Imaging plasmodesmata. Protoplasma 248:9–25
Berbee ML (2001) The phylogeny of plant and animal pathogens in the Ascomycota. Physiol Mol Plant Pathol 59:165–188
Berruyer R, Adreit H, Milazzo J, Gaillard S, Berger A, Dioh W, Lebrun MH, Tharreau D (2003) Identification and fine mapping of Pi33, the rice resistance gene corresponding to the Magnaporthe grisea avirulence gene ACE1. Theor Appl Genet 107:1139–1147
Besi M, Tucker SL, Sesma A (2009) Magnaporthe and its relatives. Encyclopedia of life sciences. Wiley, Chichester
Betts MF, Tucker SL, Galadima N et al (2007) Development of a high throughput transformation system for insertional mutagenesis in Magnaporthe oryzae. Fungal Genet Biol 44:1035–1049
Borromeo ES, Nelson RJ, Bonman JM, Leung H (1993) Genetic differentiation among isolates of Pyricularia infecting rice and weed hosts. Phytopathology 83:393
Bourett TM, Howard RJ (1990) In vitro development of penetration structures in the rice blast fungus Magnaporthe grisea. Can J Bot 68:329–342
Bourett TM, Howard RJ (1992) Actin in penetration pegs of the fungal rice blast pathogen, Magnaporthe grisea. Protoplasma 168:20–26
Bowyer P, Clarke BR, Lunness P, Daniels MJ, Osbourn AE (1995) Host-range of a plant-pathogenic fungus determined by a saponin detoxifying enzyme. Science 267:371–374
Bryan GT, Daniels MJ, Osbourn AE (1995) Comparison of fungi within the Gaeumannomyces-Phialophora complex by analysis of ribosomal DNA sequences. Appl Environ Microbiol 61:681
Bryan GT, Wu K-S, Farrall L et al (2000) A single amino acid difference distinguishes resistant and susceptible alleles of the rice blast resistance gene Pi-ta. Plant Cell 12:2033–2046
Bussaban B, Lumyong S, Lumyong P, Hyde KD, McKenzie EHC (2001) Two new species of endophytes (ascomycetes) from Zingiberaceae sporulating in culture. Nova Hedwigia 73:487–493
Cannon PF (1994) The newly recognized family Magnaporthaceae and its interrelationships. Syst Ascomycetum 13:25–42
Cannon PF, Kirk PM (2007) Fungal families of the world. CABI, Wallingford
Castlebury LA, Rossman AY, Jaklitsch WJ, Vasilyeva LN (2002) A preliminary overview of the Diaporthales based on large subunit nuclear ribosomal DNA sequences. Mycologia 94:1017–1031
Cavara F (1892) Pyricularia oryzae. Fungi Longobardiae exsiccati 2, 49
Chen X, Ronald PC (2011) Innate immunity in rice. Trends Plant Sci 16:451–459
Chen X, Shang J, Chen D et al (2006) A B-lectin receptor kinase gene conferring rice blast resistance. Plant J 46:794–804
Chen J, Zheng W, Zheng S, Zhang D, Sang W, Chen X, Li G, Lu G, Wang Z (2008) Rac1 is required for pathogenicity and Chm1-dependent conidiogenesis in rice fungal pathogen Magnaporthe grisea. PLoS Pathog 4:e1000202
Choi W, Dean RA (1997) The adenylate cyclase gene MAC1 of Magnaporthe grisea controls appressorium formation and other aspects of growth and development. Plant Cell 9:1973–1983
Chuma I, Shinogi T, Hosogi N, Ikeda K, Nakayashiki H, Park P, Tosa Y (2009) Cytological characteristics of microconidia of Magnaporthe oryzae. J Gen Plant Pathol 75:353–358
Chuma I, Isobe C, Hotta Y et al (2011) Multiple translocation of the AVR-Pita effector gene among chromosomes of the rice blast fungus Magnaporthe oryzae and related species. PLoS Pathog 7(7):e1002147
Chumley FG, Valent B (1990) Genetic analysis of melanin deficient, nonpathogenic mutants of Magnaporthe grisea. Mol Plant Microbe Interact 3:135–143
Clergeot PH, Gourgues M, Cots J, Laurans F, Latorse MP, Pepin R, Tharreau D, Notteghem JL, Lebrun MH (2001) PLS1, a gene encoding a tetraspanin-like protein, is required for penetration of rice leaf by the fungal pathogen Magnaporthe grisea. Proc Natl Acad Sci USA 98:6963–6968
Coleman JJ, Mylonakis E (2009) Efflux in fungi: la piece de resistance. PLoS Pathog 5
Collemare J, Pianfetti M, Houlle AE et al (2008) Magnaporthe grisea avirulence gene ACE1 belongs to an infection-specific gene cluster involved in secondary metabolism. New Phytol 179:196–208
Couch BC, Kohn LM (2002) A multilocus gene genealogy concordant with host preference indicates segregation of a new species, Magnaporthe oryzae, from M. grisea. Mycologia 94:683–693
Couch BC, Fudal I, Lebrun MH, Tharreau D, Valent B, van Kim P, Notteghem JL, Kohn LM (2005) Origins of host-specific populations of the blast pathogen Magnaporthe oryzae in crop domestication with subsequent expansion of pandemic clones on rice and weeds of rice. Genetics 170:613–630
Crombie L, Crombie WML, Whiting DA (1984) Isolation of avenacins A-1, A-2, B-1, B-2 from oat roots: structures of their “aglycones”, the avenestergenins. J Chem Soc Chem Comm 4:244–246
Crombie WML, Crombie L, Green JB, Lucas JA (1986) Pathogenicity of “take-all” fungus to oats: its relationship to the concentration and detoxification of the four avenacins. Phytochemistry 9:2075–2083
Cruz CD, Bockus WW, Stack JP, Tang XY, Valent B, Pedley KF, Peterson GL (2012) Preliminary assessment of resistance among U.S. Wheat cultivars to the triticum pathotype of Magnaporthe oryzae. Plant Dis 96:1501–1505
Dagdas YF, Yoshino K, Dagdas G, Ryder LS, Bielska E, Steinberg G, Talbot NJ (2012) Septin-mediated plant cell invasion by the rice blast fungus, Magnaporthe oryzae. Science 336:1590–1595
De Jong JC, McCormack BJ, Smirnoff N, Talbot NJ (1997) Glycerol generates turgor in rice blast. Nature 389:244–245
Dean RA (1997) Signal pathways and appressorium morphogenesis. Annu Rev Phytopathol 35:211–234
Dean RA, Talbot NJ, Ebbole DJ et al (2005) The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 434:980–986
Dean R, Van Kan JAL, Pretorius ZA et al (2012) The top 10 fungal pathogens in molecular plant pathology. Mol Plant Pathol 13:414–430
Dennis RWG (1960) British cup fungi and their allies: an introduction to the Ascomycetes. Ray Society, London
DeZwaan TM, Carroll AM, Valent B, Sweigard JA (1999) Magnaporthe grisea Pth11p is a novel plasma membrane protein that mediates appressorium differentiation in response to inductive substrate cues. Plant Cell 11:2013–2030
Dixon KP, Xu JR, Smirnoff N, Talbot NJ (1999) Independent signaling pathways regulate cellular turgor during hyperosmotic stress and appressorium-mediated plant infection by Magnaporthe grisea. Plant Cell 11:2045–2058
Donofrio NM, Oh Y, Lundy R, Pan H, Brown DE, Jeong JS, Coughlan S, Mitchell TK, Dean RA (2006) Global gene expression during nitrogen starvation in the rice blast fungus, Magnaporthe grisea. Fungal Genet Biol 43:605–617
Dori S, Solel Z, Barash I (1995) Cell wall-degrading enzymes produced by Gaeumannomyces graminis var. tritici in vitro and in vivo. Physiol Mol Plant Pathol 46:189–198
Dufresne M, Osbourn AE (2001) Definition of tissue-specific and general requirements for plant infection in a phytopathogenic fungus. Mol Plant Microbe Interact 14:300–307
Etxebeste O, Garzia A, Espeso EA, Ugalde U (2010) Aspergillus nidulans asexual development: making the most of cellular modules. Trends Microbiol 18:569–576
Fang EGC, Dean RA (2000) Site-directed mutagenesis of the magB gene affects growth and development in Magnaporthe grisea. Mol Plant Microbe Interact 13:1214–1227
FAO (Food and Agriculture Organization) of the United Nations (2009) The state of food insecurity in the world economic crises – impacts and lessons learned. http://www.fao.org/docrep/012/i0876e/i0876e00.htm
Fernandez J, Wilson RA (2012) Why no feeding frenzy? mechanisms of nutrient acquisition and utilization during infection by the rice blast fungus Magnaporthe oryzae. Mol Plant Microbe Interact 25:1286–1293
Fernandez J, Wright JD, Hartline D, Quispe CF, Madayiputhiya N, Wilson RA (2012) Principles of carbon catabolite repression in the rice blast fungus: Tps, 1, Nmr1–3, and a MATE-family pump regulate glucose metabolism during infection. PLoS Genet 8:e1002673
Field B, Jordan F, Osbourn A (2006) First encounters – deployment of defence-related natural products by plants. New Phytol 172:193–207
Franceschetti M, Bueno E, Wilson RA, Tucker SL, Gómez-Mena C, Calder G, Sesma A (2011) Fungal virulence and development is regulated by alternative pre-mRNA 3′ end processing in Magnaporthe oryzae. PLoS Pathog 7:e1002441
Frederick BA, Caesar-Tonthat TC, Wheeler MH, Sheehan KB, Edens WA, Henson JM (1999) Isolation and characterisation of Gaeumannomyces graminis var. graminis melanin mutants. Mycol Res 103:99–110
Freeman J, Ward E (2004) Gaeumannomyces graminis, the take-all fungus and its relatives. Mol Plant Pathol 5:235–252
Froeliger EH, Carpenter BE (1996) NUT1, a major nitrogen regulatory gene in Magnaporthe grisea, is dispensable for pathogenicity. Mol Gen Genet 251:647–656
Frohlich J, Hyde KD (1999) Biodiversity of palm fungi in the tropics: are global fungal diversity estimates realistic? Biodivers Conserv 8:977–1004
Fry SC (2004) Primary cell wall metabolism: tracking the careers of wall polymers in living plant cells. New Phytol 161:641–675
Fudal I, Collemare J, Bohnert HU, Melayah D, Lebrun MH (2007) Expression of Magnaporthe grisea avirulence gene ACE1 is connected to the initiation of appressorium-mediated penetration. Eukaryot Cell 6:546–554
Fukumori Y, Nakajima M, Akutsu K (2004) Microconidia act the role as spermatia in the sexual reproduction of Botrytis cinerea. J Gen Plant Pathol 70:256–260
Fukuoka S, Saka N, Koga H et al (2009) Loss of function of a proline-containing protein confers durable disease resistance in rice. Science 325:998–1001
Galluzzi L, Kepp O, Kroemer G (2012) Mitochondria: master regulators of danger signalling. Nat Rev Mol Cell Biol 13:780–788
Gangopadhyay S, Row KVK (1986) Perennation of Pyricularia oryzae briosi et cav. in sclerotial state. Int J Trop Plant Dis 4:187–192
Gilbert MJ, Thornton CR, Wakley GE, Talbot NJ (2006) A P-type ATPase required for rice blast disease and induction of host resistance. Nature 440:535–539
Gladfelter AS (2006) Control of filamentous fungal cell shape by septins and formins. Nat Rev Microbiol 4:223–229
Goff SA, Ricke D, Lan TH et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296:92–100
Goh J, Kim KS, Park J, Jeon J, Park SY, Lee YH (2011) The cell cycle gene MoCDC15 regulates hyphal growth, asexual development and plant infection in the rice blast pathogen Magnaporthe oryzae. Fungal Genet Biol 48:784–792
Griebel T, Zeier J (2008) Light regulation and daytime dependency of inducible plant defenses in arabidopsis: phytochrome signaling controls systemic acquired resistance rather than local defense. Plant Physiol 147:790–801
Guimil S, Chang HS, Zhu T et al (2005) Comparative transcriptomics of rice reveals an ancient pattern of response to microbial colonization. Proc Natl Acad Sci USA 102:8066–8070
Gupta A, Chattoo BB (2008) Functional analysis of a novel ABC transporter ABC4 from Magnaporthe grisea. FEMS Microbiol Lett 278:22–28
Hamer JE, Howard RJ, Chumley F, Valent B (1988) A mechanism for surface attachment in spores of a plant pathogenic fungus. Science 239:288–290
Hamer JE, Valent B, Chumley FG (1989) Mutations at the SMO genetic locus affect the shape of diverse cell types in the rice blast fungus. Genetics 122:351–361
Harmon PF, Latin R (2005) Winter survival of the perennial ryegrass pathogen Magnaporthe oryzae in north central Indiana. Plant Dis 89:412–418
Harmon PF, Dunkle LD, Latin R (2003) A rapid PCR-based method for the detection of Magnaporthe oryzae from infected perennial ryegrass. Plant Dis 87:1072–1076
Hawksworth DL (2011) A new dawn for the naming of fungi: impacts of decisions made in Melbourne in July 2011 on the future publication and regulation of fungal names. MycoKeys 1:7–20
Hayashi K, Yoshida H (2009) Refunctionalization of the ancient rice blast disease resistance gene Pit by the recruitment of a retrotransposon as a promoter. Plant J 57:413–425
Hayashi N, Inoue H, Kato T et al (2010) Durable panicle blast-resistance gene Pb1 encodes an atypical CC-NBS-LRR protein and was generated by acquiring a promoter through local genome duplication. Plant J 64:498–510
Heath MC, Valent B, Howard RJ, Chumley FG (1992) Ultrastructural interactions of one strain of Magnaporthe grisea with goosegrass and weeping lovegrass. Can J Bot 70:779–787
Hebert TT (1971) Perfect stage of Pyricularia grisea. Phytopathology 61:83–87
Henson JM (1992) DNA hybridization and polymerase chain-reaction (Pcr) tests for identification of Gaeumannomyces, Phialophora and Magnaporthe isolates. Mycol Res 96:629–636
Henson JM, Butler MJ, Day AW (1999) The dark side of the mycelium: melanins of phytopathogenic fungi. Annu Rev Phytopathol 37:447–471
Heupel S, Roser B, Kuhn H, Lebrun MH, Villalba F, Requena N (2010) Erl1, a novel era-like GTPase from Magnaporthe oryzae, is required for full root virulence and is conserved in the mutualistic symbiont glomus intraradices. Mol Plant Microbe Interact 23:67–81
Hogenhout SA, Van der Hoorn RA, Terauchi R, Kamoun S (2009) Emerging concepts in effector biology of plant-associated organisms. Mol Plant Microbe Interact 22:115–122
Hornby D (1998) Take-all disease of cereals: a regional perspective. CAB International, Wallingford
Hornby D, Slope DB, Gutteridge RJ, Sivanesan A (1977) Gaeumannomyces-cylindrosporus, a new ascomycete from cereal roots. Trans Br Mycol Soc 69:21–25
Howard RJ (1997) Breaching the outer barrier – cuticle and cell wall penetration. In: Carroll PTGC (ed) The Mycota V plant relationships, Part A. Springer, Berlin, pp 43–60
Howard RJ, Valent B (1996) Breaking and entering: host penetration by the fungal rice blast pathogen Magnaporthe grisea. Annu Rev Microbiol 50:491–512
Howard R, Ferrari M, Roach D, Money N (1991) Penetration of hard substrates by a fungus employing enormous turgor pressures. Proc Natl Acad Sci USA 88:11281–11284
Hua LX, Wu JZ, Chen CX et al (2012) The isolation of Pi1, an allele at the Pik locus which confers broad spectrum resistance to rice blast. Theor Appl Genet 125:1047–1055
Idnurm A, Heitman J (2005) Light controls growth and development via a conserved pathway in the fungal kingdom. PLoS Biol 3:615–626
Ikeda KI, Nakayashiki H, Kataoka T, Tamba H, Hashimoto Y, Tosa Y, Mayama S (2002) Repeat-induced point mutation (RIP) in Magnaporthe grisea: implications for its sexual cycle in the natural field context. Mol Microbiol 45:1355–1364
Inoue I, Namiki F, Tsuge T (2002) Plant colonization by the vascular wilt fungus Fusarium oxysporum requires FOW1, a gene encoding a mitochondrial protein. Plant Cell 14:1869–1883
Inoue K, Suzuki T, Ikeda K, Jiang S, Hosogi N, Hyong G-S, Hida S, Yamada T, Park P (2007) Extracellular matrix of Magnaporthe oryzae may have a role in host adhesion during fungal penetration and is digested by matrix metalloproteinases. J Gen Plant Pathol 73:388–398
Jeon J, Park SY, Chi MH, Choi J, Park J, Rho HS, Kim S, Goh J, Yoo S (2007) Genome-wide functional analysis of pathogenicity genes in the rice blast fungus. Nat Genet 39:561–565
Jia Y, McAdams SA, Bryan GT, Hershey HP, Valent B (2000) Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J 19:4004–4014
Jones JDG, Dangl JL (2006) The plant immune system. Nature 444:323–329
Jones EBG, Sakayaroj J, Suetrong S, Somrithipol S, Pang KL (2009) Classification of marine Ascomycota, anamorphic taxa and Basidiomycota. Fungal Divers 35:1–187
Kang S, Sweigard JA, Valent B (1995) The PWL host specificity gene family in the blast fungus Magnaporthe grisea. Mol Plant Microbe Interact 8:939–948
Kang S, Lebrun MH, Farrall L, Valent B (2001) Gain of virulence caused by insertion of a Pot3 transposon in a Magnaporthe grisea avirulence gene. Mol Plant Microbe Interact 14:671–674
Kankanala P, Czymmek K, Valent B (2007) Roles for rice membrane dynamics and plasmodesmata during biotrophic invasion by the blast fungus. Plant Cell 19:706–724
Kato H, Mayama S, Sekine R, Kanazawa E, Izutani Y, Urashima A, Kunoh H (1994) Microconidium formation in Magnaporthe grisea. Ann Phytopathol Soc Jpn 60:175–185
Kershaw MJ, Wakley G, Talbot NJ (1998) Complementation of the Mpg1 mutant phenotype in Magnaporthe grisea reveals functional relationships between fungal hydrophobins. EMBO J 17:3838–3849
Khang CH, Berruyer R, Giraldo MC, Kankanala P, Park SY, Czymmek K, Kang S, Valent B (2010) Translocation of Magnaporthe oryzae effectors into rice cells and their subsequent cell-to-cell movement. Plant Cell 22:1388–1403
Kim KS, Lee YH (2012) Gene expression profiling during conidiation in the rice blast pathogen Magnaporthe oryzae. PLoS One 7
Kim S, Ahn IP, Rho HS, Lee YH (2005) MHP1, a Magnaporthe grisea hydrophobin gene, is required for fungal development and plant colonization. Mol Microbiol 57:1224–1237
Kim S, Park SY, Kim KS et al (2009) Homeobox transcription factors are required for conidiation and appressorium development in the rice blast fungus Magnaporthe oryzae. PLoS Genet 5(12):e1000757
Kim C, Ye F, Ginsberg MH (2011a) Regulation of integrin activation. Annu Rev Cell Dev Biol 27:321–345
Kim S, Singh P, Park J, Park S, Friedman A, Zheng T, Lee YH, Lee K (2011b) Genetic and molecular characterization of a blue light photoreceptor MGWC-1 in Magnaporthe oryzae. Fungal Genet Biol 48:400–407
Kirk PM, Cannon PF, Minter DW, Stalpers JA (2008) Ainsworth and bisby’s dictionary of the fungi, 9th edn. CABI, Wallingford
Kloppholz S, Kuhn H, Requena N (2011) A secreted fungal effector of Glomus intraradices promotes symbiotic biotrophy. Curr Biol 21:1204–1209
Kohlmeyer J, Volkmannkohlmeyer B (1995) Fungi on Juncus roemerianus.1. Trichocladium medullare sp. nov. Mycotaxon 53:349–353
Kolattukudy PE (1985) Enzymatic penetration of the plant cuticle by fungal pathogens. Annu Rev Phytopathol 23:223–250
Kulkarni RD, Thon MR, Pan H, Dean RA (2005) Novel G-protein-coupled receptor-like proteins in the plant pathogenic fungus Magnaporthe grisea. Genome Biol 6:R24
Kumar J, Nelson RJ, Zeigler RS (1999) Population structure and dynamics of Magnaporthe grisea in the Indian Himalayas. Genetics 152:971–984
Kwon NJ, Garzia A, Espeso EA, Ugalde U, Yu JH (2010) FlbC is a putative nuclear C2H2 transcription factor regulating development in Aspergillus nidulans. Mol Microbiol 77:1203–1219
Lambou K, Malagnac F, Barbisan C, Tharreau D, Lebrun M-H, Silar P (2008) The crucial role of the Pls1 tetraspanin during ascospore germination in Podospora anserina provides an example of the convergent evolution of morphogenetic processes in fungal plant pathogens and saprobes. Eukaryot Cell 7:1809–1818
Landschoot PJ, Jackson N (1989a) Magnaporthe poae sp. nov., a hyphopodiate fungus with a Phialophora anamorph from grass roots in the United States. Mycol Res 93:59–62
Landschoot PJ, Jackson N (1989b) Gaeumannomyces incrustans sp. nov., a root-infecting hyphopodiate fungus from grass roots in the United States. Mycol Res 93:55–58
Lau G, Hamer JE (1996) Regulatory genes controlling MPG1 expression and pathogenicity in the rice blast fungus Magnaporthe grisea. Plant Cell 8:771–781
Lau GW, Hamer JE (1998) Acropetal: a genetic locus required for conidiophore architecture and pathogenicity in the rice blast fungus. Fungal Genet Biol 24:228–239
Lee FN, Jackson MA, Walker NR (2000) Characteristics of Pyricularia grisea “microsclerotia” produced in shaked culture. In: Norman RJ, Beyrouty CA (eds) BR wells rice research studies 1999. University of Arkansas, Fayetteville, AR, pp 475–479
Lee K, Singh P, Chung WC, Ash J, Kim TS, Hang L, Park S (2006) Light regulation of asexual development in the rice blast fungus, Magnaporthe oryzae. Fungal Genet Biol 43:694–706
Lee SK, Song MY, Seo YS et al (2009) Rice Pi5-mediated resistance to Magnaporthe oryzae requires the presence of two coiled-coil-nucleotide-binding-leucine-rich repeat genes. Genetics 181:1627–1638
Leung H, Borromeo E, Bernardo M, Notteghem JL (1988) Genetic analysis of virulence in the rice blast fungus Magnaporthe grisea. Phytopathology 78:1227–1233
Li L, Xue CY, Bruno K, Nishimura M, Xu JR (2004) Two PAK kinase genes, CHM1 and MST20, have distinct functions in Magnaporthe grisea. Mol Plant Microbe Interact 17:547–556
Li L, Ding SL, Sharon A, Orbach M, Xu JR (2007) Mir1 is highly upregulated and localized to nuclei during infectious hyphal growth in the rice blast fungus. Mol Plant Microbe Interact 20:448–458
Li W, Wang BH, Wu J et al (2009) The Magnaporthe oryzae avirulence gene AvrPiz-t encodes a predicted secreted protein that triggers the immunity in rice mediated by the blast resistance gene Piz-t. Mol Plant Microbe Interact 22:411–420
Li GT, Zhou XY, Kong LG, Wang YL, Zhang H, Zhu H, Mitchell TK, Dean RA, Xu JR (2011) MoSfl1 is important for virulence and heat tolerance in Magnaporthe oryzae. PLoS One 6:e19951
Lin F, Chen S, Que ZQ, Wang L, Liu XQ, Pan QH (2007) The blast resistance gene Pi37 encodes a nucleotide binding site-leucine-rich repeat protein and is a member of a resistance gene cluster on rice chromosome 1. Genetics 177:1871–1880
Linder MB, Szilvay GR, Nakari-Setala T, Penttila ME (2005) Hydrophobins: the protein-amphiphiles of filamentous fungi. FEMS Microbiol Rev 29:877–896
Litvintseva AP, Henson JM (2002) Cloning, characterization, and transcription of three laccase genes from Gaeumannomyces graminis var. tritici, the take-all fungus. Appl Environ Microbiol 68:1305–1311
Liu XQ, Lin F, Wang L, Pan QH (2007) The in silico map-based cloning of Pi36, a rice coiled-coil-nucleotide-binding site-leucine-rich repeat gene that confers race-specific resistance to the blast fungus. Genetics 176:2541–2549
Liu JL, Wang XJ, Mitchell T, Hu YJ, Liu XL, Dai LY, Wang GL (2010a) Recent progress and understanding of the molecular mechanisms of the rice-Magnaporthe oryzae interaction. Mol Plant Pathol 11:419–427
Liu WD, Xie SY, Zhao XH, Chen X, Zheng WH, Lu GD, Xu JR, Wang ZH (2010b) A homeobox gene is essential for conidiogenesis of the rice blast fungus Magnaporthe oryzae. Mol Plant Microbe Interact 23:366–375
Liu WD, Zhou XY, Li GT, Li L, Kong LG, Wang CF, Zhang HF, Xu JR (2011) Multiple plant surface signals are sensed by different mechanisms in the rice blast fungus for appressorium formation. PLoS Pathog 7
Marcel S, Sawers R, Oakeley E, Angliker H, Paszkowski U (2010) Tissue-adapted invasion strategies of the rice blast fungus Magnaporthe oryzae. Plant Cell 22:3177–3187
Mehrabi R, Ding S, Xu JR (2008) MADS-box transcription factor Mig1 is required for infectious growth in Magnaporthe grisea. Eukaryot Cell 7:791–799
Mendgen K, Hahn M (2002) Plant infection and the establishment of fungal biotrophy. Trends Plant Sci 7:352–356
Mengiste T (2012) Plant immunity to necrotrophs. Annu Rev Phytopathol 50(50):267–294
Mentlak TA, Kombrink A, Shinya T et al (2012) Effector-mediated suppression of chitin-triggered immunity by Magnaporthe oryzae is necessary for rice blast disease. Plant Cell 24:322–335
Mosquera G, Giraldo MC, Khang CH, Coughlan S, Valent B (2009) Interaction transcriptome analysis identifies Magnaporthe oryzae BAS1-4 as biotrophy-associated secreted proteins in rice blast disease. Plant Cell 21:1273–1290
Mostowy S, Cossart P (2012) Septins: the fourth component of the cytoskeleton. Nat Rev Mol Cell Biol 13:183–194
Nguyen QB, Kadotani N, Kasahara S, Tosa Y, Mayama S, Nakayashiki H (2008) Systematic functional analysis of calcium-signalling proteins in the genome of the rice-blast fungus, Magnaporthe oryzae, using a high-throughput RNA-silencing system. Mol Microbiol 68:1348–1365
Noguchi MT, Yasuda N, Fujita Y (2006) Evidence of genetic exchange by parasexual recombination and genetic analysis of pathogenicity and mating type of parasexual recombinants in rice blast fungus, Magnaporthe oryzae. Phytopathology 96:746–750
Oh Y, Donofrio N, Pan HQ, Coughlan S, Brown DE, Meng SW, Mitchell T, Dean RA (2008) Transcriptome analysis reveals new insight into appressorium formation and function in the rice blast fungus Magnaporthe oryzae. Genome Biol 9(5):R85
Okuyama Y, Kanzaki H, Abe A et al (2011) A multifaceted genomics approach allows the isolation of the rice Pia-blast resistance gene consisting of two adjacent NBS-LRR protein genes. Plant J 66:467–479
Olmedo M, Ruger-Herreros C, Corrochano LM (2010a) Regulation by blue light of the fluffy gene encoding a major regulator of conidiation in Neurospora crassa. Genetics 184:651–658
Olmedo M, Ruger-Herreros C, Luque EM, Corrochano LM (2010b) A complex photoreceptor system mediates the regulation by light of the conidiation genes con-10 and con-6 in Neurospora crassa. Fungal Genet Biol 47:352–363
Osbourn AE, Clarke BR, Lunness P, Scott PR (1994) An oat species lacking avenacin is susceptible to infection by Gaeumannomyces graminis var. tritici. Physiol Mol Plant Pathol 45:457
Ou SH (1985) Rice diseases, 2nd edn. Commonwealth Mycological Institute, Kew, Surrey
Park HS, Yu JH (2012) Genetic control of asexual sporulation in filamentous fungi. Curr Opin Microbiol 15:669–677
Park G, Xue C, Zheng L, Lam S, Xu JR (2002) MST12 regulates infectious growth but not appressorium formation in the rice blast fungus Magnaporthe grisea. Mol Plant Microbe Interact 15:183–192
Park G, Xue C, Zhao X, Kim Y, Orbach M, Xu J-R (2006) Multiple upstream signals converge on the adaptor protein Mst50 in Magnaporthe grisea. Plant Cell 18:2822–2835
Patkar RN, Xue YK, Shui GH, Wenk MR, Naqvi NI (2012) Abc3-mediated efflux of an endogenous digoxin-like steroidal glycoside by Magnaporthe oryzae is necessary for host invasion during blast disease. PLoS Pathog 8:e1002888
Qu SH, Liu GF, Zhou B, Bellizzi M, Zeng LR, Dai LY, Han B, Wang GL (2006) The broad-spectrum blast resistance gene Pi9 encodes a nucleotide-binding site-leucine-rich repeat protein and is a member of a multigene family in rice. Genetics 172:1901–1914
Ribot C, Hirsch J, Batzergue S, Tharreau D, Notteghem JL, Lebrun MH, Morel JB (2008) Susceptibility of rice to the blast fungus, Magnaporthe grisea. J Plant Physiol 165:114–124
Rodrigues FA, Benhamou N, Datnoff LE, Jones JB, Belanger RR (2003) Ultrastructural and cytochemical aspects of silicon-mediated rice blast resistance. Phytopathology 93:535–546
Rossman AY, Howard RJ, Valent B (1990) Pyricularia grisea, the correct name for the rice blast disease fungus. Mycologia 82:509–512
Ruger-Herreros C, Rodriguez-Romero J, Fernandez-Barranco R, Olmedo M, Fischer R, Corrochano LM, Canovas D (2011) Regulation of conidiation by light in Aspergillus nidulans. Genetics 188:809–U897
Saccardo PA (1880) Fungorum extra-europaeorum Pugillus. Michelia 2:136–149
Saitoh H, Fujisawa S, Ito A, Mitsuoka C, Berberich T, Tosa Y, Asakura M, Takano Y, Terauchi R (2009) SPM1 encoding a vacuole-localized protease is required for infection-related autophagy of the rice blast fungus Magnaporthe oryzae. FEMS Microbiol Lett 300:115–121
Saitoh H, Fujisawa S, Mitsuoka C et al (2012) Large-scale gene disruption in Magnaporthe oryzae identifies MC69, a secreted protein required for infection by monocot and dicot fungal pathogens. PLoS Pathog 8(5):e1002711
Saleh D, Xu P, Shen Y et al (2012) Sex at the origin: an Asian population of the rice blast fungus Magnaporthe oryzae reproduces sexually. Mol Ecol 21:1330–1344
Saunders DG, Dagdas YF, Talbot NJ (2010a) Spatial uncoupling of mitosis and cytokinesis during appressorium-mediated plant infection by the rice blast fungus Magnaporthe oryzae. Plant Cell 22:2417–2428
Saunders DGO, Aves SJ, Talbot NJ (2010b) Cell cycle-mediated regulation of plant infection by the rice blast fungus. Plant Cell 22:497–507
Sesma A, Osbourn AE (2004) The rice leaf blast pathogen undergoes developmental processes typical of root-infecting fungi. Nature 431:582–586
Shang JJ, Tao Y, Chen XW et al (2009) Identification of a new rice blast resistance gene, Pid3, by genomewide comparison of paired nucleotide-binding site-leucine-rich repeat genes and their pseudogene alleles between the two sequenced rice genomes. Genetics 182:1303–1311
Sharma TR, Rai AK, Gupta SK, Singh NK (2010) Broad-spectrum blast resistance gene Pi-k(h) cloned from rice line Tetep designated as Pi54. J Plant Biochem Biotechnol 19:87–89
Shattil SJ, Kim C, Ginsberg MH (2010) The final steps of integrin activation: the end game. Nat Rev Mol Cell Biol 11:288–300
Shi Z, Christian D, Leung H (1998) Interactions between spore morphogenetic mutations affect cell types, sporulation, and pathogenesis in Magnaporthe grisea. Mol Plant Microbe Interact 11:199–207
Silué D, Tharreau D, Talbot NJ, Clergeot PH, Notteghem JL, Lebrun MH (1998) Identification and characterization of apf1 in a non-pathogenic mutant of the rice blast fungus Magnaporthe grisea which is unable to differentiate appressoria. Physiol Mol Plant Pathol 53:239–251
Skamnioti P, Gurr SJ (2007) Magnaporthe grisea cutinase2 mediates appressorium differentiation and host penetration and is required for full virulence. Plant Cell 19:2674–2689
Skamnioti P, Gurr SJ (2009) Against the grain: safeguarding rice from rice blast disease. Trends Biotechnol 27:141–150
Soanes DM, Kershaw MJ, Cooley RN, Talbot NJ (2002) Regulation of the MPG1 hydrophobin gene in the rice blast fungus Magnaporthe grisea. Mol Plant Microbe Interact 15:1253–1267
Soanes DM, Alam I, Cornell M et al (2008) Comparative genome analysis of filamentous fungi reveals gene family expansions associated with fungal pathogenesis. PLoS One 3(6):e2300
Soanes DM, Chakrabarti A, Paszkiewicz KH, Dawe AL, Talbot NJ (2012) Genome-wide transcriptional profiling of appressorium development by the rice blast fungus Magnaporthe oryzae. PLoS Pathog 8:e1002514
Springer ML, Yanofsky C (1992) Expression of con genes along the three sporulation pathways of Neurospora crassa. Genes Dev 6:1052–1057
Sreedhar L, Kobayashi DY, Bunting TE, Hillman BI, Belanger FC (1999) Fungal proteinase expression in the interaction of the plant pathogen Magnaporthe oryzae with its host. Gene 235:121–129
Stergiopoulos I, de Wit PJGM (2009) Fungal effector proteins. Annu Rev Phytopathol 47:233–263
Sun CB, Suresh A, Deng YZ, Naqvi NI (2006) A multidrug resistance transporter in magnaporthe is required for host penetration and for survival during oxidative stress. Plant Cell 18:3686–3705
Sweigard JA, Chumley FG, Valent B (1992) Disruption of a Magnaporthe grisea cutinase gene. Mol Gen Genet 232:183–190
Sweigard JA, Chumley FG, Carroll AM, Farrall L, Valent B (1995) Identification, cloning, and characterization of PWL2, a gene for host species specificity in the rice blast fungus. Plant Cell 7:1221–1233
Talbot NJ (2003) On the trail of a cereal killer: exploring the biology of Magnaporthe grisea. Annu Rev Microbiol 57:177–202
Talbot NJ, Kershaw MJ (2009) The emerging role of autophagy in plant pathogen attack and host defence. Curr Opin Plant Biol 12:444–450
Talbot NJ, Ebbole DJ, Hamer JE (1993) Identification and characterization of MPG1, a gene involved in pathogenicity from the rice blast fungus Magnaporthe grisea. Plant Cell 5:1575–1590
Talbot NJ, Kershaw MJ, Wakley GE, De Vries OMH, Wessels JGH, Hamer JE (1996) MPG1 encodes a fungal hydrophobin involved in surface interactions during infection-related development of Magnaportha grisea. Plant Cell 8:985–999
Talbot NJ, McCafferty HRK, Ma M, Moore K, Hamer JE (1997) Nitrogen starvation of the rice blast fungus Magnaporthe grisea may act as an environmental cue for disease symptom expression. Physiol Mol Plant Pathol 50:179–198
Tamasloukht M, Sejalon-Delmas N, Kluever A, Jauneau A, Roux C, Becard G, Franken P (2003) Root factors induce mitochondrial-related gene expression and fungal respiration during the developmental switch from asymbiosis to presymbiosis in the arbuscular mycorrhizal fungus Gigaspora rosea. Plant Physiol 131:1468–1478
Tanzer MM, Arst HN, Skalchunes AR, Coffin M, Darveaux BA, Heiniger RW, Shuster JR (2003) Global nutritional profiling for mutant and chemical mode-of-action analysis in filamentous fungi. Funct Integr Genomics 3:160–170
Thines E, Weber RWS, Talbot NJ (2000) MAP kinase and protein kinase A-dependent mobilization of triacylglycerol and glycogen during appressorium turgor generation by Magnaporthe grisea. Plant Cell 12:1703–1718
Thinlay X, Finckh MR, Bordeos AC, Zeigler RS (2000) Effects and possible causes of an unprecedented rice blast epidemic on the traditional farming system of Bhutan. Agric Ecosyst Environ 78:237–248
Thongkantha S, Jeewon R, Vijaykrishna D, Lumyong S, McKenzie EHC, Hyde KD (2009) Molecular phylogeny of Magnaporthaceae (Sordariomycetes) with a new species Ophioceras chiangdaoense from Dracaena loureiroi in Thailand. Fungal Divers 34:157–173
Tredway LP (2006) Genetic relationships among Magnaporthe oryzae isolates from turfgrass hosts and relative susceptibility of “Penncross” and “Penn A-4” creeping bentgrass. Plant Dis 90:1531–1538
Tsurushima T, Don LD, Kawashima K, Murakami J, Nakayashiki H, Tosa Y, Mayama S (2005) Pyrichalasin H production and pathogenicity of Digitaria-specific isolates of Pyricularia grisea. Mol Plant Pathol 6:605–613
Tsurushima T, Minami Y, Miyagawa H, Nakayashiki H, Tosa Y, Mayama S (2010) Induction of chlorosis, ROs generation and cell death by a toxin isolated from Pyricularia oryzae. Biosci Biotechnol Biochem 74:2220–2225
Tucker SL, Talbot NJ (2001) Surface attachment and pre-penetration stage development by plant pathogenic fungi. Annu Rev Phytopathol 39:385–418
Tucker SL, Besi MI, Galhano R, Franceschetti M, Goetz S, Lenhert S, Osbourn A, Sesma A (2010) Common genetic pathways regulate organ-specific infection-related development in the rice blast fungus. Plant Cell 22:953–972
Ulrich K, Augustin C, Werner A (2000) Identification and characterization of a new group of root-colonizing fungi within the Gaeumannomyces-Phialophora complex. New Phytol 145:127–136
Urban M, Bhargava T, Hamer JE (1999) An ATP-driven efflux pump is a novel pathogenicity factor in rice blast disease. EMBO J 18:512–521
Valent B, Chumley FG (1991) Molecular genetic analysis of the rice blast fungus, Magnaporthe grisea. Annu Rev Phytopathol 29:443–467
Valent B, Farrall L, Chumley F (1991) Magnaporthe grisea genes for pathogenicity and virulence identified through a series of backcrosses. Genetics 127:87–101
Vasilyeva LN (1998) Nizshie Rasteniya, Griby i Mokhoobraznye Dalnego Vostoka Rossii. Pirenomitsety i Lokuloaskomitsety, Sankt Petersburg
Veneault-Fourrey C, Barooah M, Egan M, Wakley G, Talbot NJ (2006) Autophagic fungal cell death is necessary for infection by the rice blast fungus. Science 312:580–583
Walker J (1972) Type studies on Gaeumannomyces graminis and related fungi. Trans Br Mycol Soc 58:427–457
Walker J (1980) Gaeumannomyces, Linocarpon, Ophiobolus and several other genera of scolecospored ascomycetes and Phialophora conidial states, with a note on hyphopodia. Mycotaxon 11:1–129
Wang Z-X, Yano M, Yamanouchi U, Iwamoto M, Monna L, Hayasaka H, Katayose Y, Sasaki T (1999) The Pib gene for rice blast resistance belongs to the nucleotide binding and leucine-rich repeat class of plant disease resistance genes. Plant J 19:55–64
Wang ZY, Thornton CR, Kershaw MJ, Debao L, Talbot NJ (2003) The glyoxylate cycle is required for temporal regulation of virulence by the plant pathogenic fungus Magnaporthe grisea. Mol Microbiol 47:1601–1612
Wilson RA, Jenkinson JM, Gibson RP, Littlechild JA, Wang ZY, Talbot NJ (2007) Tps1 regulates the pentose phosphate pathway, nitrogen metabolism and fungal virulence. EMBO J 26:3673–3685
Wilson RA, Gibson RP, Quispe CF, Littlechild JA, Talbot NJ (2010) An NADPH-dependent genetic switch regulates plant infection by the rice blast fungus. Proc Natl Acad Sci USA 107:21902–21907
Wong PTW (2002) Gaeumannomyces wongoonoo sp nov., the cause of a patch disease of buffalo grass (St Augustine grass). Mycol Res 106:857–862
Wong PTW, Dong C, Stirling AM, Dickinson ML (2012) Two new Magnaporthe species pathogenic to warm-season turfgrassses in Australia. Australas Plant Pathol 41:321–329
Xiao JZ, Ohshima A, Kamakura T, Ishiyama T (1994) Extracellular glycoprotein(s) associated with cellular differentiation in Magnaporthe grisea. Mol Plant Microbe Interact 7:639
Xu JR, Hamer JE (1996) MAP kinase and cAMP signalling regulate infection structure formation and pathogenic growth in the rice blast fungus Magnaporthe grisea. Genes Dev 10:2696–2706
Xu JR, Urban M, Sweigard JA, Hamer JE (1997) The CPKA Gene of Magnaporthe grisea is essential for appressorial penetration. Mol Plant Microbe Interact 10:187–194
Xu JR, Staiger CJ, Hamer JE (1998) Inactivation of the mitogen-activated protein kinase Mps1 from the rice blast fungus prevents penetration of host cells but allows activation of plant defense responses. Proc Natl Acad Sci USA 95:12713–12718
Xue C, Park G, Choi W, Zheng L, Dean RA, Xu J-R (2002) Two novel fungal virulence genes specifically expressed in appressoria of the rice blast fungus. Plant Cell 14:2107–2119
Xue MF, Yang J, Li ZG et al (2012) Comparative analysis of the genomes of two field isolates of the rice blast fungus Magnaporthe oryzae. PLoS Genet 8(8):e1002869
Yang J, Zhao XY, Sun J, Kang ZS, Ding SL, Xu JR, Peng YL (2010) A novel protein com1 is required for normal conidium morphology and full virulence in Magnaporthe oryzae. Mol Plant Microbe Interact 23:112–123
Yao JM, Wang YC, Zhu YG (1992) A new variety of the pathogen of maize take-all. Acta Mycologica Sinica 11:99–104
Yoshida K, Saitoh H, Fujisawa S et al (2009) Association genetics reveals three novel avirulence genes from the rice blast fungal pathogen Magnaporthe oryzae. Plant Cell 21:1573–1591
You MP, Lanoiselet V, Wang CP, Shivas RG, Li YP, Barbetti MJ (2012) First report of rice blast (Magnaporthe oryzae) on rice (Oryza sativa) in Western Australia. Plant Dis 96:1228–1228
Yu J, Hu S, Wang J et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296:79–92
Yuan B, Zhai C, Wang WJ, Zeng XS, Xu XK, Hu HQ, Lin F, Wang L, Pan QH (2011) The Pik-p resistance to Magnaporthe oryzae in rice is mediated by a pair of closely linked CC-NBS-LRR genes. Theor Appl Genet 122:1017–1028
Zeigler RS (1998) Recombination in Magnaporthe grisea. Annu Rev Phytopathol 36:249–276
Zhai C, Lin F, Dong ZQ, He XY, Yuan B, Zeng XS, Wang L, Pan QH (2011) The isolation and characterization of Pik, a rice blast resistance gene which emerged after rice domestication. New Phytol 189:321–334
Zhang HF, Liu KY, Zhang X et al (2011a) Two phosphodiesterase genes, PDEL and PDEH, regulate development and pathogenicity by modulating intracellular cyclic AMP levels in Magnaporthe oryzae. PLoS One 6(2):e17241
Zhang HF, Xue CY, Kong LG, Li GT, Xu JR (2011b) A Pmk1-interacting gene is involved in appressorium differentiation and plant infection in Magnaporthe oryzae. Eukaryot Cell 10:1062–1070
Zhang N, Zhao S, Shen QR (2011c) A six-gene phylogeny reveals the evolution of mode of infection in the rice blast fungus and allied species. Mycologia 103:1267–1276
Zhao X, Kim Y, Park G, Xu J-R (2005) A mitogen-activated protein kinase cascade regulating infection-related morphogenesis in Magnaporthe grisea. Plant Cell 17:1317–1329
Zhao XH, Mehrabi R, Xu JR (2007) Mitogen-activated protein kinase pathways and fungal pathogenesis. Eukaryot Cell 6:1701–1714
Zhao S, Clarke BB, Shen QR, Zhang LS, Zhang N (2012) Development and application of a TaqMan real-time PCR assay for rapid detection of Magnaporthe oryzae. Mycologia 104:1250–1259
Zhou B, Qu SH, Liu GF, Dolan M, Sakai H, Lu GD, Bellizzi M, Wang GL (2006) The eight amino-acid differences within three leucine-rich repeats between Pi2 and Piz-t resistance proteins determine the resistance specificity to Magnaporthe grisea. Mol Plant Microbe Interact 19:1216–1228
Zhou ZZ, Li GH, Lin CH, He CZ (2009) Conidiophore stalk-less1 encodes a putative zinc-finger protein involved in the early stage of conidiation and mycelial infection in Magnaporthe oryzae. Mol Plant Microbe Interact 22:402–410
Zhou X, Liu W, Wang C, Xu Q, Wang Y, Ding S, Xu JR (2011) A MADS-box transcription factor MoMcm1 is required for male fertility, microconidium production and virulence in Magnaporthe oryzae. Mol Microbiol 80:33–53
Zhou XY, Zhang HF, Li GT, Shaw B, Xu JR (2012) The cyclase-associated protein cap1 is important for proper regulation of infection-related morphogenesis in Magnaporthe oryzae. PLoS Pathog 8
Zipfel C (2008) Pattern-recognition receptors in plant innate immunity. Curr Opin Immunol 20:10–16
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2013 Springer-Verlag Berlin Heidelberg
About this chapter
Cite this chapter
Illana, A., Rodriguez-Romero, J., Sesma, A. (2013). Major Plant Pathogens of the Magnaporthaceae Family. In: Horwitz, B., Mukherjee, P., Mukherjee, M., Kubicek, C. (eds) Genomics of Soil- and Plant-Associated Fungi. Soil Biology, vol 36. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-39339-6_4
Download citation
DOI: https://doi.org/10.1007/978-3-642-39339-6_4
Published:
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-39338-9
Online ISBN: 978-3-642-39339-6
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)