Abstract
Two new species of ectotrophic root-infecting fungi pathogenic to warm-season turfgrasses are described. Magnaporthe garrettii P. T. W. Wong & M. L. Dickinson sp. nov. causes a serious patch disease on couch (Cynodon dactylon) bowling greens in South Australia, and Magnaporthe griffinii P. T. W. Wong & A.M. Stirling sp. nov. is associated with a disease complex (“summer decline”) of hybrid couch (C. dactylon × C. transvaalensis) golf greens in New South Wales and Queensland. Both are homothallic, producing perithecia readily on potato dextrose agar. They differ from other Magnaporthe spp. in having uniseriate rather than biseriate or multiseriate ascospores, and the absence of a conidial anamorph. Analysis of nuclear rRNA ITS sequences has shown that M. griffinii is a new taxon with low homology to M. grisea, M. poae, M. rhizophila, M. salvinii and Gaeumannomyces graminis. This could not be carried out with M. garrettii because there were no living cultures available and the genomic DNA extracted from dead mycelia and perithecia was totally degraded. However, the two new species can be readily distinguished by morphological differences in their perithecia and ascospores. Examination of earlier herbarium specimens has shown that M. garrettii was associated with a patch disease of buffalo grass (Stenotaphrum secundatum) in New South Wales and M. griffinii was associated with diseases of South African couch grass (C. tranvaalensis) in South Australia and of kikuyu grass (Pennisetum clandestinum) in New South Wales.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In recent years, patch diseases of couch grass or bermudagrass [Cynodon dactylon (L.) Pers.] and couch hybrids [Cynodon dactylon (L.) Pers. × C. transvaalensis Burtt-Davy] have become more common on golf and bowling greens in Australia. In 2002, a serious patch disease was observed on several couch (cv. Greenlees Park) bowling greens in Adelaide, South Australia (Fig. 1). It appeared in spring (September and October) and increased in size and severity through the warmer months. Field symptoms included patches of unthrifty grass, stunting of stolons, leaf chlorosis and eventual necrosis of stolons. In subsequent years, the numerous small patches (<5 cm) enlarged irregularly and coalesced into large patches that exceeded 0.5 m in diameter.
In Queensland and New South Wales, a disease called “summer decline” by golf superintendents has become increasingly common and severe on hybrid couch, especially cv. Tifgreen, and is becoming more difficult to control (Stirling 2001). The symptoms are irregular patches of unthrifty turf which are most severe in the summer months, especially when mowing heights are low and/or there is a period of heat and moisture stress (Fig. 2). The roots and rhizomes of plants in affected areas become dark brown and rotten so that eventually very few roots are left. So far, commonly used fungicides have not been effective against either of these diseases. This paper reports on the aetiology of these two new diseases and describes the pathogens.
Methods
Isolation of fungi
Root pieces (0.5–1.0 cm) with disease symptoms were surface-sterilised in equal parts of 10% aqueous sodium hypochlorite (3.5% available chlorine) and 95% ethanol for 2 min, rinsed twice in sterile water and plated onto ¼ strength potato dextrose agar + 100 mg/L novobiocin (¼ PDA + N) in 90 mm diam. Petri plates. The agar plates were incubated in the dark at 25°C for 5–10 d before the most commonly occurring fungi were subcultured onto ¼ PDA and PDA. Pure cultures of the fungi were then tested for pathogenicity to couch grass cv. Greenlees Park or the couch hybrid cv. Tifgreen. Specimens and dried agar cultures of the pathogens were lodged in the Plant Pathology Herbarium, Department of Industry and Investment, Orange, NSW (Herb. DAR).
Pathogenicity tests
Pathogenicity tests were carried out in a temperature-controlled glasshouse (12 h at 25°C, 12 h at 30°C). Ten cm diameter plastic pots were filled to 2 cm from the top with steam-sterilised washed river sand and inoculated with the fungus by chopping up a fully-colonised ¼ PDA plate into 1 cm squares and placing them on the soil surface. Three healthy stolons (each 3 cm long and bearing 2 nodes) of couch grass cv. Greenlees Park or the couch hybrid cv. Tifgreen were placed in contact with the agar pieces and covered with a 1 cm layer of sand. Control pots had chopped-up agar colonised by an isolate of non-pathogenic Fusarium oxysporum Schlecht. emend. Snyder & Hansen instead of the test fungi. There were 4 replicates per treatment and treatments were randomised in blocks on a glasshouse bench. Pots were watered daily and fertilised fortnightly with 50 ml per pot of a soluble complete fertiliser, Thrive® (Yates Australia, Padstow, NSW, Australia). Above-ground disease symptoms were recorded monthly for 3 months. At the end of that period, the root systems of all the plants were washed free of sand and observed under a stereo microscope (x 40) for disease symptoms. If diseased roots were present, a few roots were plated out as above to re-isolate the causal fungus.
Cultural characteristics of the pathogens
The pathogens, designated Magnaporthe sp. 1 and Magnaporthe sp. 2, were grown on PDA and ¼ PDA. The agar plates were incubated for up to 3 months at 25°C in the dark and the cultures were observed periodically under a stereo microscope and a compound microscope for colony and mycelial characteristics as well as sporulation. Fungal mycelia and perithecia were mounted in a drop of water or lactophenol acid fuchsin on slides and measurements of hyphal diameter, the dimensions of perithecial characters, asci and 25 random ascospores of each fungus were made using a standard eyepiece micrometer. Photomicrographs were captured with a digital camera attachment (Tucsen Imaging Technology Co. Ltd., Fuzhou, China). After 3 months, when mature perithecia had formed but there were still no conidia, the plates were exposed to alternating dark and diffuse natural light on a laboratory bench (20–25°C) for another month in an attempt to induce conidial production.
To measure the growth rates of the fungi, 5 mm fungal plugs from the advancing margins of the fungal colonies, grown on PDA, were placed at the centres of 90 mm diam. PDA plates (4 replicate plates per fungus) and incubated in the dark at 28°C. The diameters of the fungal colonies were measured every 5–7 days because of the slow growth rate of the fungi and the radial growth rates (mm/day) of the fungi were calculated.
DNA extraction, amplification and sequence analysis
As there were no living cultures of Magnaporthe sp. 1 available, fungal mycelia and perithecia from a dried agar culture of the holotype (DAR 76937) were collected for DNA extraction. For Magnaporthe sp. 2, two isolates (TS99 and TY2) were grown for 4 weeks at 25°C on autoclaved, moist cellophane sheets (50 mm × 50 mm) that had been placed on freshly poured PDA plates. Approximately 20 mg of mycelia were scraped off the cellophane, placed in 2.0 ml Eppendorf tubes and left to dry in silica gel for 2 days. Two 4 mm ball bearings and 50 mg of glass beads were added to the dried samples and ground into a fine powder in a Mixer Mill (Retsch MM300, MEP Instruments Pty Ltd, North Ryde, NSW, Australia) at 25 oscillations/second for 4 min. CTAB buffer (2% CTAB, 1.4 M NaCl, 0.2 M EDTA, 0.1 M Tris–HCl pH 8.0, 0.2% β-mercaptoethanol) was pre-warmed at 60°C and 500 μl aliquots were added to the sample, mixed thoroughly and incubated at 60°C for 30 min. The DNA was then extracted using equal volumes of chloroform, mixed well and then centrifuged at full speed in a microcentrifuge (Eppendorf 5415D, Eppendorf, North Ryde, NSW, Australia) for 5 min. The aqueous portion was decanted into clean tubes and an equal amount of isopropanol was added to precipitate the DNA. The DNA pellets were washed with 70% ethanol and resuspended in 50 μl TE (pH8.0). RNase A was added to give a 50 μg/ml concentration. The DNA concentration was determined by electrophoresis and compared with a known concentration of λ DNA. PCR primers used in this study were ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) (White et al. 1990) which amplifies partial 18S rRNA gene, complete internal transcribed spacer 1 (ITS1), complete 5.8S rRNA gene, complete ITS2, and partial 28S rRNA gene. PCR amplification was carried out in a 20 μl reaction containing 5 ng DNA, 1x GoTaq buffer (Promega, Madison, WI, USA), 0.2 mM dNTPs, 1.5 mM MgCl2, 0.25 μM primers and 0.5 U GoTaq Hot Start Polymerase (Promega, Madison, WI, USA). PCR was performed at 94°C for 3 min and then 35 cycles of 94°C 20 s, 55°C 30 s and 72°C 45 s; final extension at 72°C for 2 min. A 2 μl sample of PCR product was checked on an agarose gel and the remaining product was purified by Wizard SV Gel and PCR Clean-Up system (Promega, Madison, WI, USA). Purified PCR product was sequenced in both directions using the PCR primers at the Australian Genome Research Facility (http://www.agrf.org.au/) according to AGRF’s instructions. The sequence was assembled in BioEdit v 7.0.9 (http://www.mbio.ncsu.edu/bioedit/bioedit.html) and searched for sequence similarity in the GenBank of National Center for Biotechnology Information (NCBI, Bethesda, MD, USA) using BLAST (Altschul et al. 1990).
The new ITS sequence was compared with known ITS sequences retrieved from NCBI (http://www.ncbi.nlm.nih.gov), including sequences of M. rhizophila strain ATCC 96043 (Accession No. DQ528790), M. poae isolate SCR10 (Accession No. DQ528782), M. grisea isolate MG15 Vijayanagaram (Accession No. HQ020359), M. salvinii strain ATCC 32149 (Accession No. FJ746639), and Gaeumannomyces graminis var. tritici strain 9735 (Accession No. AF508789). A multiple sequence alignment was constructed using CLUSTAL W (Thompson et al. 1994) in BioEdit v7.0.9 and a Maximum-Likelihood Tree was constructed using the MEGA5 phylogenetics package (Tamura and Nei 1993) with bootstrap values of 1,000 replicates to evaluate the strength of the internal branches.
Results and discussion
Disease symptoms and isolation of fungi
Washed root systems of the diseased grass from both types of disease patches showed numerous blackened roots, some of which had stelar lesions, similar to those caused by Gaeumannomyces graminis (Sacc.) Arx & D. L. Olivier var. avenae (E.M. Turner) Dennis on bentgrass (Agrostis spp.) and hybrid couch [C. dactylon (L.) Pers. × C. transvaalensis Burtt-Davy] roots (Smiley et al. 2005; Wong et al. 2000) and by G. wongoonoo Wong on buffalo grass (Stenotaphrum secundatum Kunze) roots (Wong 2002). There were also dark runner hyphae on the surface of the roots, similar to those of ectotrophic root-infecting fungi (Clarke and Gould 1993).
Several fungi were commonly isolated from the diseased turf from Adelaide: G. graminis var. graminis, a Phoma sp., a Drechslera sp. and a slow-growing Magnaporthe sp. (designated Magnaporthe sp. 1). From the “summer decline” disease samples from Queensland, Gaeumannomyces incrustans Landschoot & Jackson, Drechslera spp., a Phoma sp. and another slow-growing Magnaporthe sp. (designated Magnaporthe sp. 2) were frequently isolated. Pure cultures of the above fungi were maintained on ¼ PDA slopes at 4°C.
Pathogenicity tests
Two months after inoculating Greenlees Park couch with the putative pathogens isolated from the Adelaide turf, only the grass in the pots inoculated with Magnaporthe sp. 1 showed symptoms of moderate chlorosis and stunting of stolons. The grass in the control pots as well as in those inoculated with G. graminis var. graminis, the Phoma sp., and the Drechslera sp. was healthy and green. After 3 months, the experiment was terminated and the root systems were washed free of sand. When observed under a stereo microscope (x 40), only the root systems of plants inoculated with Magnaporthe sp. 1 showed dark-coloured roots with dark brown stelar lesions, similar to those of diseased field specimens. The surfaces and cortices of the roots were extensively colonised by black ectotrophic mycelium. Occasionally, unlobed appressoria were observed on infected stolons. The root systems of the plants infected with Magnaporthe sp. 1 were reduced in size compared to the healthy controls (Fig. 3) and there was stunting and chlorosis of the shoots. Re-isolation of fungi from the diseased roots on ¼ PDA + N yielded the original fungus, thereby completing Koch’s postulates.
For the pathogenicity tests of the fungi isolated from turf affected by “summer decline”, only Magnaporthe sp. 2 showed characteristic disease lesions with associated dark runner hyphae on roots and unlobed appressoria (Fig. 4) on infected stolons similar to those in field specimens. However, it appeared to be less pathogenic than Magnaporthe sp. 1 as there was no visual reduction in the size of the root systems and the shoots appeared healthy. It is possible that more severe symptoms may have developed had the hybrid couch plants in the glasshouse been subjected to the same type of stresses that are associated with summer decline in the field, such as moisture and heat stress. However, Magnaporthe sp. 2 was consistently re-isolated from the diseased roots on ¼ PDA + N, thereby confirming its pathogenic status.
Cultural characteristics of the pathogens
Both fungi grew very slowly on PDA. Magnaporthe sp. 1 (DAR 76937) grew at the rate of 1.2 mm/day at 28°C on PDA while Magnaporthe sp. 2 (DAR 80512) grew at the rate of 0.8 mm/day at 28°C on PDA. The relatively thin mycelium (1–3 μm) of both fungi was brownish in colour, with lighter brown aerial growth but giving the mature fungal colony an overall dark brown to black colour. The reverse side of the Petri plate was also black. Hyphae at the margins of the colonies were mostly straight and without the “curling back” characteristic of Phialophora spp. or G. graminis (Walker 1972; 1980). No conidia were produced in the cultures even after 3 months of incubation at 25°C or 28°C in the dark or when subsequently exposed to alternating dark and diffuse natural light on a laboratory bench (20–25°C) for another month.
Both fungi produced abundant perithecia (Figs. 5 and 6) on PDA after 4–6 weeks’ incubation at 25°C and 28°C. In 6–8 week old cultures, numerous ejected ascospores were often found on the lids of Petri plates, especially when the plates were placed upside down. Perithecia also developed on the stolons and leaf sheaths of diseased couch kept in a humid chamber for 4–6 weeks at 20–25°C on a laboratory bench, which was exposed to diffused natural light. When the mature perithecia of the fungi were placed in a drop of water on a microscope slide and lightly squashed with a cover slip, large numbers of asci (Figs. 7 and 8) and ascospores (Figs. 9a, b and 10) were observed. Ascospores of both fungi germinated readily on PDA with germ tubes growing out only from the end cells. Cultures from single ascospores of both fungi produced perithecia readily on PDA, showing that the fungi were homothallic. On both field material and on PDA, the perithecia of Magnaporthe sp. 1 (Fig. 5) had characteristically longer necks than those of Magnaporthe sp. 2 (Fig. 6 & Table 2).
ITS sequence and phylogenetic analysis
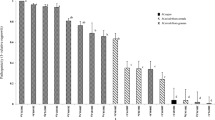
A 550 bp DNA fragment was obtained from ITS PCR of Magnaporthe sp. 2 (isolates TS99 and TY2); however, no product was obtained from Magnaporthe sp. 1 (isolate DAR 76937) because the genomic DNA of the dried culture had been totally degraded. The sequences of TS99 (GenBank Accession No. JQ390311) and TY2 (GenBank Accession No. JQ390312) were identical (GenBank accession no. xxxx); therefore, only the TS99 sequence was used in sequence comparison and tree construction. The percentages of sequence identity among M. rhizophila, M. poae, M. grisea, M. salvinii, G.graminis and TS99 are shown in Table 1. It was found that TS99 has low homology compared with the other sequences. BLAST search (using BLASTN) in GenBank (http://www.ncbi.nlm.nih.gov/) against nucleotide collections showed no identical sequence to TS99 indicating that it is a new species. The closest matches (>90%) are: an uncultured Sordariales clone (Accession No. GQ924037.1), 96%; a fungal endophyte isolate (Accession No. FN392299.1), 96% and an uncultured fungus clone (Accession No. GU053815.1), 94%.
A molecular phylogenetic tree was constructed using the Maximum-Likelihood method in MEGA5 with bootstrap values of 1,000 (Fig. 11). It showed that the Magnaporthe genus is quite divergent. Only M. rhizophila and M. poae are grouped with 100% bootstrap support. M. salvinii is separated from this group with only 77% bootstrap support. M. grisea and M. griffinii (TS99) are outgrouped. As the genus Magnaporthe is somewhat heterogeneous with species having Phialophora, Nakataea and Pyricularia anamorphs, the relationship of M. griffinii and M. garrettii to the other species cannot be resolved at this stage as the latter species have no known anamorphs. However, the fact that M. grisea with a Pyricularia anamorph is also outgrouped and is less closely related to the other Magnaporthe species than G. graminis (Fig. 11) suggests that further work is required to clarify the genus concepts of Magnaporthe and Gaeumannomyces. Morphologically, the ascospores of the two new species are characteristic of Magnaporthe (elongated ascospores with median cells pigmented) rather than Gaeumannomyces (hyaline, filiform ascospores) [Cannon 1994].
Molecular phylogenetic analysis by Maximum-Likelihood method using MEGA5 with bootstrap of 1,000 replicates. Bootstrap values are shown for the internal branches (only values >50% are shown). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. All positions containing gaps and missing data were eliminated. There were a total of 456 positions in the final dataset
Although DNA data were not obtained for M. garrettii to show its relationship in the phylogenetic tree (Fig. 11), its morphological characteristics such as uniseriate ascospores, twice as long asci, much slower growth rate (1.2 mm/day vs ~4 mm/day at 28°C) and lack of a Phialophora anamorph (Table 2) easily separate it from M. rhizophila, the only other homothallic Magnaporthe species. However, it is interesting that the examination of all turfgrass specimens bearing perithecia accessioned as M. rhizophila in Herb. DAR, Orange, NSW, has shown that DAR 51653 on Stenotaphrum secundatum is, in fact, M. garrettii and was the cause of a patch disease of buffalo grass in Bourke, New South Wales. Two other specimens of M. rhizophila were identical to M. griffinii and were the cause of a disease of kikuyu grass in Kanwal, New South Wales (DAR 58461) and a disease of South African couch in Renmark, South Australia (DAR 50508). In all these specimens, when intact asci were present, the ascospores were uniseriate whereas those of M. rhizophila are biseriate. It is, therefore, likely that M. rhizophila and M. poae, have not yet been found in Australia.
Taxonomy
Comparison with other Magnaporthe species found on Poaceae
Although the anamorphs of the two new taxa are unknown, the teleomorphs belong to the genus Magnaporthe according to the concept of Cannon (1994). The four Magnaporthe spp. previously recorded on Poaceae are compared with the two recent isolates in Table 2. Of the described species (Krause and Webster 1972; Yaegashi and Udagawa 1978; Scott and Deacon 1983; Landschoot and Jackson 1989), the pathogens most resemble M. rhizophila Scott & Deacon in ascospore size and in being homothallic (Table 2). However, they grow more slowly (~1 mm/day vs ~4 mm/day at 28°C) and do not have the Phialophora anamorph (Scott and Deacon 1983; Gams 2000) or the biseriate ascospores of M. rhizophila (Scott and Deacon 1983). The asci of the pathogens, bearing uniseriate ascospores (Figs. 7 and 8), are also longer and narrower than the asci in the other Magnaporthe spp. Evidence from earlier herbarium specimens indicates that these two Magnaporthe species have been confused with M. rhizophila in Australia, probably because of their similarity in ascospore size (Table 2). The two pathogens are considered to be distinct from the known species of Magnaporthe (Cannon 1994) and are described below as new.
Magnaporthe garrettii P. T. W. Wong & M.L. Dickinson sp. nov.
MycoBank: (MB 564204)
Etymology: to honour the late Professor S.D. Garrett, Cambridge University, for his significant work on the ecology of root-infecting fungi of Poaceae.
Perithecia in agaro superficiaria et immersa, nigra, praecipue singularia vel 1–5 aggregata, corpore globoso 180–450 μm diam. et colo decrescenti usque ad 260 μm longo, 90–160 μm lato ad basim. Paries perithecii pseudoparenchymatus, cellulis externis fuscatis. Asci unitunicati, numerosi, cylindrici, praecipue recti, octospori, 165–205 × 7–8 μm, stipite breve, apice rotundato, cum annulo refractivo non amyloideo. Ascosporae uniseriatae, cylindricae vel fusiformes, rectae vel leniter curvatae cum extremis rotundatis et septis tribus transversis, 19–25 × 5–7 μm, cellulis duobus centralibus atrobrunneis et cellulis duobus extremis hyalinis. Paraphyses delicatae, hyalinae, filamentosae, septatae, et in peritheciis maturis languescentes. Hyphae in agaro brunneae, rectae vel flexuosae, 1–3 μm latae, in hospite 3–5 μm latae et cum appressoriis lateralibus non-lobatis circa 7 × 10 μm.
Holotypus hic designatus: in cultura agaro sicca ex radicibus Cynodontis dactylonis cv. Greenlees Park, Colonel Light Gardens Bowling Club, Adelaide, South Australia, Australia, 30 October 2004, M. L. Dickinson, DAR 76937.
Perithecia superficial and immersed, globose, black, mostly single, sometimes aggregated in small groups, 180–450 μm diameter with a tapering neck up to 260 μm long, 90–160 μm wide at the base. Perithecial wall pseudoparenchymatous, external cells much darker. Asci unitunicate, numerous, cylindrical, mostly straight, 8-spored, short-stalked, 165–205 × 7–8 μm, with a tapering but rounded apex and a light refractive, non-amyloid apical ring. Ascospores uniseriate, cylindrical to fusiform, straight or slightly curved with rounded ends, 3-septate, with dark brown middle cells and hyaline end cells shorter than the coloured cells, 19–25 × 5–7 μm. Paraphyses delicate, hyaline, filamentous, septate and shrivelling in mature perithecia. Hyphae brown, straight or flexuous, 1–3 μm wide on potato dextrose agar and 3–5 μm on host. Appressoria simple, unlobed, about 7 × 10 μm.
Other specimens examined: On Stenotaphrum secundatum (Walter) Kuntze, Bourke, New South Wales, Australia, 14 May, 1985, R. Stone, DAR 51653 (as M. rhizophila D.B. Scott & Deacon).
The holotype (DAR 76937) has been deposited in the Plant Pathology Herbarium of the Department of Industry and Investment (Herb. DAR), Orange, New South Wales, Australia. It consists of dried PDA cultures with numerous perithecia of the fungus. The fungus was isolated from diseased roots of C. dactylon cv. Greenlees Park from Colonel Light Gardens Bowling Club in Adelaide, South Australia.
Magnaporthe griffinii P. Wong & A.M. Stirling sp. nov.
MycoBank: (MB 564205)
Etymology: to honour Emeritus Professor D.M. Griffin, Australia National University, for his major contribution to the understanding of the role of soil physical factors in the ecology of soil fungi.
Perithecia in agaro superficiaria et immersa, nigra, praecipue singularia vel in gregibus parvis disposita, corpore globose 160–340 μm diam. et colo decrescenti usque ad 120 μm longo, 50–70 μm lato ad basim. Paries perithecii pseudoparenchymatus, cellulis externis fuscatis. Asci unitunicati, numerosi, cylindricae, praecupue recti, octospori, stipite breve, 145–190 × 8–10 μm, apice decrescenti et rotundato cum annulo aefrectivo non amyloideo. Ascosporae uniseriatae vel leviter superpositae, fusiformes, rectae vel leviter curvatae, cum extremis obtusis-acutis et septis tribus transversis, 24–35 × 6–9 μm, cellulis duobus centralibus atrobrunneis et cellulis duobus extremis hyalinis. Paraphyses delicatae, hyalinae, filamentosae, septatae, et in peritheciis maturis languescentes. Hyphae in agaro brunneae, rectae vel flexuosae, 1–3 μm latae, in hospite 3–5 μm latae et cum appressoriis lateralibus non-lobatis circa 8 × 15 μm.
Holotypus hic designatus: in cultura agaro sicca ex radicibus Cynodontis dactylonis × C. transvaalensis cv. Tifgreen, Hyatt Coolum Golf Club, Coolum, Queensland, Australia, 13 March, 2008, M. Whatman, DAR 80512.
Perithecia superficial and immersed, globose, black, mostly single, sometimes aggregated in small groups, 160–340 μm diameter with a short tapering neck less than 120 μm long, 50–70 μm wide at the base. Perithecial wall pseudoparenchymatous, external cells much darker. Asci unitunicate, numerous, cylindrical, mostly straight, 8-spored, short-stalked, 145–190 × 8–10 μm, with a tapering but rounded apex and a light refractive, non-amyloid apical ring. Ascospores uniseriate, slightly overlapping, fusiform, straight or slightly curved with bluntly pointed ends, 3-septate, with dark brown middle cells and hyaline end cells of similar length to coloured cells, 24–35 × 6–9 μm. Paraphyses delicate, hyaline, filamentous, septate and shrivelling in mature perithecia. Hyphae brown, straight or flexuous, 1–3 μm wide on potato dextrose agar and 3–5 μm on the host. Appressoria simple, unlobed, about 8 × 15 μm.
Other specimens examined: On Cynodon transvaalensis Burtt-Davy, Renmark, South Australia, Australia, November 1984, G. E. Walker, DAR 50508 (as M. rhizophila). On Pennisetum clandestinum Hochst. ex Chiov., Kanwal, New South Wales, Australia, 21 November 1986, R. Swan, DAR 58461 (as M. rhizophila). On C. dactylon (L.) Pers. × C. transvaalensis Burtt-Davy, Adelaide, South Australia, Australia, December, 2010, P. Toy, TY2; Brisbane, Queensland, Australia, January, 2000, A.M. Stirling, TS99.
The holotype (DAR 80512) has been deposited in the Plant Pathology Herbarium of the Department of Industry and Investment (Herb. DAR), Orange, New South Wales, Australia. It consists of dried PDA cultures with numerous perithecia of the fungus. The fungus was isolated from diseased roots of hybrid couch cv. Tifgreen (C. dactylon × C. transvaalensis) from Hyatt Coolum Golf Club, Coolum, Queensland, Australia.
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Cannon PF (1994) The newly recognised family Magnaporthaceae and its interrelationships. Syst Ascomycetum 13:25–42
Clarke BB, Gould AB (1993) Turfgrass patch diseases caused by ectotrophic root-infecting fungi. American Phytopathological Society Press, St Paul
Gams W (2000) Phialophora and some similar morphologically little-differentiated anamorphs of divergent ascomycetes. Stud Mycol 45:187–199
Krause RA, Webster RK (1972) The morphology, taxonomy and sexuality of the rice stem rot fungus, Magnaporthe salvinii (Leptosphaeria salvinii). Mycologia 64:103–114
Landschoot PJ, Jackson N (1989) Magnaporthe poae sp. nov., a hyphopodiate fungus with a Phialophora anamorph from grass roots in the United States. Mycol Res 93:59–62
Scott DB, Deacon JW (1983) Magnaporthe rhizophila sp. nov., a dark mycelial fungus with a Phialophora conidial state, from cereal roots from South Africa. Trans Br Mycol Soc 81:71–81
Smiley RW, Dernoeden PH, Clarke BB (2005) Compendium of turfgrass diseases, 3rd edn. American Phytopathology Society Press, St Paul
Stirling AM (2001) Ectotrophic root-infecting fungi on golf turf in Queensland. Horticulture Australia Report (TU00005). pp 26
Tamura K, Nei M (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10:512–526
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Walker J (1972) Type studies on Gaeumannomyces graminis and related fungi. Trans Br Mycol Soc 58:427–457
Walker J (1980) Gaeumannomyces, Linocarpon, Ophiobolus and several other genera of scolecospored ascomycetes and Phialophora conidial states, with a note on hyphopodia. Mycotaxon 11:1–129
White TJ, Bruns TD, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols, a guide to methods and applications. Academic, New York, pp 315–322
Wong PTW (2002) Gaeumannomyces wongoonoo sp. nov., the cause of a patch disease of buffalo grass (St Augustinegrass). Mycol Res 106:857–862
Wong PTW, Tan MK, Beehag GW (2000) Confirmation of take-all patch disease in Tifdwarf couch grass (bermudagrass) by morphological and DNA methods. Australas Plant Pathol 29:19–23
Yaegashi H, Udagawa S (1978) The taxonomic identity of the perfect stage of Pyricularia grisea and its allies. Can J Bot 56:180–183
Acknowledgements
We thank Dr M. Priest for allowing one of us (PTWW) to examine herbarium accessions of Magnaporthe spp. in Herbarium DAR and for accessioning the holotypes, Mr L. Turton and Ms B. Wildner for photographic assistance, Mr A. Leggett for the disease sample from Hyatt Coolum Golf Club and Drs P. M. Martin and J. N. Harris for helpful comments on the manuscript. We are especially grateful to Mr. J. Walker for the Latin descriptions and commenting on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wong, P.T.W., Dong, C., Stirling, A.M. et al. Two new Magnaporthe species pathogenic to warm-season turfgrassses in Australia. Australasian Plant Pathol. 41, 321–329 (2012). https://doi.org/10.1007/s13313-012-0118-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13313-012-0118-6