Abstract
In recent years, transgenic pyramiding or stacking technology has gradually developed with the rapid development of genetically modified (GM) crops. This technology has unique advantages compared with the transgenic technology of a single gene. Transgenic pyramiding technology can simultaneously modify several traits of a crop, particularly the metabolic pathways and yield traits that are usually controlled by multiple genes. A batch of second generation GM crops with stacked traits has been commercialized, which mainly include resistances to pest and herbicide, and exhibit great application potential of transgenic pyramiding breeding. The present stacked GM crops are developed mainly through the integrated use of vector-based pyramiding and molecular marker assisted selection (MAS) based crossing/backcrossing pyramiding. Advances of vector based pyramiding technologies have been achieved on large capacity vectors, multiple-gene assembling, plastid transformation, polyprotein expression system and combinatorial genetic transformation. The combination of transgenic pyramiding and MAS-based pyramiding will help to effectively pyramid more targeted genes together. The advance of single nucleotide polymorphism (SNP) discovery and detecting technology has greatly promoted the MAS-based pyramiding in crop breeding. The rapid development of commercialized stacked GM crops, plant metabolic engineering and crop improvement for disease resistance have proved the successful applications of transgenic pyramiding. The present review discusses the advances of transgenic pyramiding technology, the application of transgenic pyramiding in crop improvement and the prospects and challenges of transgenic pyramiding breeding.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
Transgenic technology has great potential for genetic improvement of crops, in which foreign genes that crops do not possess can be transferred into a crop’s genome . Foreign genes can come from animals, microorganisms, and other species. Gene expression in crops can also be regulated by transgenic technology. Thus, genetically modified (GM) crops can gain new agronomic traits such as resistances to abiotic and biotic stress . In addition, metabolic pathways can be modified by applying transgenic technology. Considering the unique characteristics of transgenic technology contrary to conventional breeding, the research and production of transgenic crops develop rapidly. The global hectarage of GM crops has been increasing every year since the first GM crop was introduced in 1996. According to the International Service for the Acquisition of Agri-biotech Applications (ISAAA) report (James 2013), GM crops reached 175.2 million ha in 2013 (100 times greater than in 1996), distributed across 27 countries and included more than 10 kinds of crops. Although GM crops are developing rapidly, transgenic technology has not yet achieved its full potential. Given the constraints of transgenic technology, many GM crops currently have simple transgenic traits controlled by one or two genes, and thus belong to the first-generation transgenic crops. Research on GM crops with stacked traits is still at an early stage, but has exhibited broad application prospects in crop improvement .
Stacked trait GM crops are superior to the single trait GM crops in various aspects. First, stacked trait GM crops can pyramid multiple genes controlling different traits into a single transgenic crop variety and simultaneously improve several traits in the crop. For example, GM maize Agrisure® Viptera™ 3111 (produced by Syngenta) contains stacked insect-resistance genes (cry1Ab, vip3Aa20, and mcry3A) and herbicide-resistance genes (mepsps and pat) (GM Approval Database 2004). This GM crop is endowed with resistance to Lepidoptera insects , Coleoptera insects , glufosinate and glyphosate. Second, the pyramiding of several genes encoding Bt (Bacillus thuringiensis) toxin proteins with two or more modes of action in a single variety is an effective strategy for insect-resistance management and can delay the occurrence of insects with Bt-toxin resistance (Bates et al. 2005; Ghimire et al. 2011; Zhao et al. 2003). Based on the requests of the Environmental Protection Agency in the USA, 20 % (in the Corn Belt) or 50 % (in the Cotton Belt) of a farmer’s corn acreage should be planted with non-Bt corns as refuge acreage. If corn farmers plant the GM corns with several Bt genes, the refuge acreage can be reduced to 10 %, allowing farmers to grow either 10 or 40 % more Bt corns in the same area. Third, the transgenic pyramiding is suitable for the modification of biological traits controlled by multiple genes, such as metabolic pathways, protein complexes and quantitative traits . The modification of a single gene has no significant effect on these traits.
The focus of this chapter is on the technology for transgenic pyramiding and its applications in crop improvement s. It aims to provide an easily-understandable overview of the latest developments of techniques involved in large-capacity vectors , multiple-gene assembling, plastid transformation , polyprotein expression systems , combinatorial genetic transformation and MAS-based gene pyramiding . Also, applications in commercialized GM crops , plant metabolic engineering , and crop improvements for resistances to biotic and abiotic stresses.
2 Technology for Transgenic Pyramiding Breeding
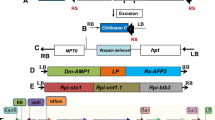
Three methods have been established to pyramid multiple transgenes; namely, multi-gene transformation, retransformation and sexual hybridization of two or more transgenic events (Fig. 13.1). The multi-gene transformation method can be further classified into two types: single-vector transformation and multi-vector co-transformation . For the single-vector transformation method, multiple genes are inserted within a single T-DNA, or multiple T-DNAs are included on a binary vector, the plant transformation of a single-vector is simple, but the assembly of multiple genes into a conventional vector is difficult owing to vector instability, the lack of unique restriction sites and the limited vector capacity. By this method, transgenes in the transgenic plants are tightly linked and co-integrated in the same loci in the plant genome , so they will not segregate in offspring. In multi-vector co-transformation methods, multiple genes are cloned on a different vector, respectively. Multiple strains carrying a different vector are used for simultaneous infection of plant tissues, or multiple genes in different vectors are simultaneously transferred to plant cells. The transgenes from the co-transformation are usually co-integrated at the same position of the transgenic plant chromosomes, which may consequently be inherited together in the progeny (Halpin 2005).
Retransformation is a valid gene pyramiding method, with which multiple transgenes can be sequentially introduced into a plant. For retransformation , every transformation requires a new and different selectable marker from the original event, which is crucial in this process. The drawback of few available selective markers restricts the development of retransformation . The retransformant transgenes are randomly integrated and may segregate in the progeny. Sexual hybridization , which is flexible for the combination of transgenes but time-consuming, is a conventional breeding technology that pyramids transgenes from different events. Considering that the transgenes from different events are not linked and will segregate in the progeny, a large breeding population is required to obtain the plant with stacked genes.
MAS is an efficient technology for gene pyramiding in conventional breeding (Dokku et al. 2013; Hu et al. 2012; Tan et al. 2010). Generally, MAS-based pyramiding can be performed using two strategies: backcrossing and crossing (Fig. 13.2). The backcrossing is used for pyramiding genes into the genetic background of the recurrent parent to improve a few traits of the recurrent parent while the main traits of the recurrent parent remain unchanged. The crossing method is used for pyramiding genes into new genetic backgrounds through the genetic recombination of various parents to develop a new variety.
Schemes of backcrossing /crossing pyramiding based on MAS. P1(1), P2(2), P3(3) and P4(4) represent four different parents carrying different genes (genes are indicated by the numbers in brackets). P1(12), P1(123) and P1(1234) indicate the improved parent P1(1) carrying different pyramided genes. L(1234) indicates the new developed line carrying four pyramided genes
2.1 Transgenic Pyramiding Methods in Commercialized GM Crops
The pyramiding of transgenes in commercialized GM crops is mainly realized through gene stacking based on vector and sexual hybridization . The Genuity® SmartStax™ maize developed by Monsanto was derived from the stacking of four events (MON89034 × TC1507 × MON88017 × 59122) by conventional crossbreeding. Each event originated from the transformation of a vector with multiple genes. The event MON89034 was obtained from the transformation of vector PV-ZMIR254 containing Lepidoptera insect-resistance genes (cry2Ab2 and cry1A.105 from Bacillus thuringiensis). The event MON88017 was achieved from the transformation of vector PV-ZMIR39, which contains glyphosate-resistance gene (cp4 epsps) and Coleoptera insect -resistance gene (cry3Bb1). The event TC1507 was derived from the particle bombardment transformation of a linear fragment PHI8999A that comprises two expression cassettes of Lepidoptera insect-resistance gene (cry1Fa) and glufosinate-resistance gene (pat). The event 59122 contains Coleoptera insect-resistance, particularly corn rootworm-resistance genes (cry34Ab1 and cry35Ab1) and glufosinate-resistance gene (pat). The Agrisure® Viptera™ 3111 maize developed by Syngenta, was also derived from four events (Bt11, GA21, MIR162, and MIR604) by conventional crossbreeding. The event Bt11 was obtained from the transformation of a vector with stacked genes (cry1Ab and pat), and has resistances to Lepidoptera insects and glufosinate herbicide . GA21 was acquired from the transformation of vector pDPG434 carrying glyphosate-resistance gene ( mepses ). The events, MIR162 and MIR604 were obtained from the transformation of vector pNOV1300 containing Lepidoptera insect-resistance gene (vip3Aa20) and pZM26 containing Coleoptera insect-resistance gene (mcry3A).
According to the examples provided, the transgenic pyramiding in commercialized GM crops with stacked traits is realized mainly through conventional crossbreeding of single events deriving from the vector transformation. Each event must be tested and approved before becoming commercially available, which is a complex, time-consuming, and arduous process. Considering that the single events used for crossbreeding have obtained approvals, the stacked products from the conventional crossbreeding do not involve new transformations, and no interaction effect among the stacked traits happens, the registration as a new GM product and new approvals are unnecessary for the stacked products from crossbreeding. Thus, this crossbreeding-based pyramiding can accelerate the commercialization of GM crops and reduce the research cost for GM crops. This accelerated test and approval procedure has become the standard in some countries (Taverniers et al. 2008).
In crop production, more stacked transgenes are not necessarily advantageous. More genes and traits in breeding mean a more complicated and extended breeding process. In addition, each stacked transgene is protected by intellectual property rights, thus more stacked transgenes mean greater cost for farmers who use them. Moreover, the required crop traits are not identical in different crop planting areas. With the crossbreeding method, the desired transgenic traits can be combined to develop the transgenic products suitable to various needs of markets and crop productions. Each of four transgenic events (Bt11, GA21, MIR162 and MIR604) contain different transgenes, and the different combinations of these events developed into various GM varieties. Agrisure™ GT/CB/LL, obtained from the hybridization of events Bt11 and GA21 and contains stacked genes (cry1Ab, pat, and mepsps), providing resistances to Lepidoptera insect , glyphosate and glufosinate. Agrisure® Viptera™ 2100, from the combination of events Bt11 and MIR162 and with stacked genes (cry1Ab, pat, and vip3Aa20), endowing resistances to Lepidoptera and Coleoptera insects and glufosinate. Agrisure® Viptera™ 3110 was developed by stacking GA21 on the basis of Agrisure® Viptera™ 2100, adding a glyphosate-resistance gene (mepsps) . Agrisure® Viptera™ 3100 was bred by stacking MIR604 on the basis of Agrisure® Viptera™ 2100, adding a Lepidoptera insect-resistance gene (mcry3A). Agrisure® Viptera™ 3111 was the result of four events (Bt11, GA21, MIR162 and MIR604) with five stacked genes (cry1Ab, pat, vip3Aa20, mcry3A and mepsps).
Pyramiding by conventional crossbreeding also has disadvantages. Transgenes in different events are not linked and have a tendency to segregate in subsequent breeding progenies. Moreover, transgene loci increase with increasing the crossing parents, resulting in the complex gene segregation. Therefore, a large and sufficient breeding population and more breeding generations are needed for the identification of plants with the stacked genes in offspring. In breeding processes, except for the target transgenic traits, other agronomic traits should also be selected. Thus, if the number of pyramided loci exceeds three, it is more difficult to obtain the plant that is homozygous for all transgene loci.
2.2 Development of Transgenic Pyramiding Technology
2.2.1 Large-Capacity Vectors
The length capacity of vectors for the inserted DNA fragments limits the assembling of multiple genes in a single vector . The maximum length of the inserted fragment of a conventional binary vector is about 50 kb. However, in actual practice, the assembly and transformation of a vector with an inserted DNA fragment of more than 30 kb is difficult. The binary bacterial artificial chromosome (BIBAC) and transformation-competent artificial chromosome (TAC) are two new kinds of vectors that can be used for genomic library construction and plant transformation. In contrast to conventional binary vector, BIBAC and TAC can contain an inserted DNA fragment of up to 200 kb, and are suitable for the transformation of large DNA fragments (Chang et al. 2003; Hamilton et al. 1999; Kong et al. 2006; Lin et al. 2003; Qu et al. 2003; Zhai et al. 2013). At present, these vectors are mainly used in the cloning and functional identification of large genome fragments.
Minichromosome is a newly developed gene expression platform for large DNA fragments. Minichromosome is a micro-artificial chromosome containing the structure and function unit of chromosomes, and exists as an episome in the cell period and cannot match host chromosomes, but can carry genes and transfer genetic information between generations (Goyal et al. 2009). The mammalian minichromosomes have been used as vector systems for gene therapy. However, the minichromosome construction was not successful in plants until the reports of telomere-mediated chromosomal truncation in maize in 2006 (Yu et al. 2006). Maize minichromosomes were obtained from A chromosomes and B chromosomes through telomere-mediated chromosome truncation technology (Yu et al. 2006, 2007). A 2.6 kb Arabidopsis telomere sequence was transformed into maize to cause chromosomal breakage at the site of integration, efficiently resulting in the truncated chromosomes. However, the truncated chromosomes are deficient, and the truncation of A chromosomes causes an obstacle for the growth and development of the plants that carry the deletion, because they lack essential genes. So the recovery of large truncated chromosomes is always difficult through genetic transformation, and moreover the truncated chromosomes could not be transmitted to the next generation for the gametophytic lethality at the haploid level. However, these problems can be overcome through polyploidization of transformants or by using polyploid plants as the target material for genetic transformation. The gametes of tetraploid plants will be diploid and the deficiency of truncated chromosomes would be compensated for by additional copies of the homolog. These problems do not exist in the truncated chromosomes from B chromosomes, because B chromosomes are basically inert, without any known active genes. Yu et al. (2007) demonstrated the transgene manipulation using minichromosomes . A transgenic plant carrying a 35S-lox66-Cre expression transgenic cassette was crossed with a plant carrying the minichromosome , which contains the promoterless lox71-DsRed gene, and the recombination of lox66 site with lox71 site was successfully catalyzed by the Cre recombinase, resulting in the transfer of DsRed and Cre genes and the expression of DsRed gene. As the next-generation genetic vector, plant minichromosomes are in an initial development stage, and are only reported in several plants, including maize, rice and Arabidopsis (Murata et al. 2013; Xu et al. 2012a; Yu et al. 2006, 2007). Larger capacity for the insert DNA fragments of minichromosome can be expected. Although the exact maximum length is unknown, the length may reach the Mb level.
2.2.2 Multiple-Gene Assembling
Gene assembly is a limiting factor for multiple gene transformation. Conventional gene assembly is dependent on the specific restriction endonuclease sites. However, these sites usually cannot satisfy the gene assembly with the increase in the number of genes. So, a number of restriction endonuclease independent multiple-gene assembling technologies have been developed (Fig. 13.3). In theory, the most convenient way for multiple-gene assembly is seamless assembly or de novo synthesis of long chain DNA. At present, DNA fragments of 1–3 kb can be easily synthesized by conventional DNA synthesis technology, and then assembled into the long chain DNA by seamless assembly. The existing long chain DNA assembling technology includes sequence-independent and ligation-independent cloning (SLIC) , circular polymerase extension (CPEC) , Gibson isothermal assembly , homologous recombination method (Gibson et al. 2008, 2009; Li and Elledge 2007, 2012; Quan and Tian 2009) and iterative assembly systems , such as GoldenBraid (Sarrion-Perdigones et al. 2011).
The CPEC method extends overlapping regions between the insert and vector fragments to form a complete circle plasmid using a polymerase extension mechanism, and can not only be used for the cloning of a single gene, but also be used for the cloning of libraries and metabolic pathways. Quan and Tian (2009) successfully assembled four genes of the metabolic pathway for synthesizing a biodegradable plastic material, poly (3HB-co-4HB) in Escherichia coli into the vector using CPEC. The CPEC reaction is completed in a single tube, and only needs 2–5 cycles, depending on the complexity of inserted fragments. The advantage of CPEC is convenience, efficiency and cost-effectiveness. The SLIC method assembles DNA fragments into vector through in vitro homologous recombination and single-strand annealing. The ssDNA overhangs in the insert and vector fragments are generated using exonuclease, and then these fragments can be assembled together by recombination in vitro. The SLIC method also allows an efficient and reproducible assembly of recombinant DNA with as many as 5–10 fragments simultaneously (Li and Elledge 2007, 2012). Gibson isothermal assembly method assembles multiple overlapping DNA fragments by using exonuclease III and antibody-bound Taq DNA polymerase, which allows for one-step thermocycled in vitro recombination. All these technologies can assemble DNA fragments of up to 20 kb, and especially, Gibson isothermal assembly in Saccharomyces cerevisiae can even assemble DNA fragments of 0.5–1 Mb (Gibson et al. 2008, 2009; Li and Elledge 2012; Quan and Tian 2009).
At present, the popular recombinase-based vector systems for multi-gene assembly are Gateway-based systems, such as MultiSite Gateway (Sasaki et al. 2004) and MultiRound Gateway (Buntru et al. 2013). The Gateway system is based on the function of Escherichia coli bacteriophage lambda integrase and two sets of att sites. Two sets of recombination reactions are catalyzed by the attB × attP (BP) and attL × attR (LR) Clonase. BP Clonase is responsible for the recombination of attB sites with attP sites, and LR Clonase catalyzes the recombination of attL sites with attR sites. The attB sites are added to the two terminals of any interest DNA segment through PCR reaction with primers containing attB sites, and then the DNA segment is cloned into the donor vector with attP sites through BP reaction, resulting in an entry clone containing the insert DNA segment flanked with two attL sites. The insert in entry clone can be mobilized into any destination vector with attR site through the recombination of attL site with attR site. To facilitate multi-gene assembly, the Gateway variants, MultiSite Gateway and MultiRound Gateway were developed. Sasaki et al. (2004) assembled four DNA fragments into a single vector through MultiSite Gateway . By using six attB sites, four DNA segments were cloned into four donor vectors with corresponding specific attP sites through the BP reaction, creating four different entry clones, respectively; the four entry clones simultaneously reacted with the destination vector in the presence of LR clonase, resulting in the construct with the three inserts in a defined and oriented order (Sasaki et al. 2004). Compared with the MultiSite Gateway system, that only needs a single LR recombination reaction for multiple entry clones, the MultiRound Gateway system sequentially delivered multiple entry clones into a destination vector by multiple rounds of LR recombination reactions (Chen et al. 2006). These systems are becoming popular in multi-gene assembly because of the advantage of sequence independence, but they suffer from several drawbacks in that the assembly process is intricate, time-consuming and expensive. Chen et al. (2010) developed a multi-gene assembly system named MISSA (for multiple-round in vivo site-specific assembly), combining the Cre/LoxP recombinase system, phage λ site-specific recombinase system, and conjugal transfer of genes among bacteria together. MISSA consists of donor vectors and acceptor vectors, and the corresponding donor bacteria and acceptor bacteria. The gene transfer from a donor vector to an acceptor vector can be accomplished through in vivo conjugational transfer between the corresponding acceptor bacteria and donor bacteria. Therefore, target genes only need to be assembled into the donor vectors, and then multiple genes in the donor vectors can be transferred to the acceptor vectors individually through several rounds of conjugational transfer between the donor bacteria and the acceptor bacteria. Furthermore, the basic vectors of several acceptor vectors of this system are developed from BIBAC and TAC ; thus, MISSA is not suitable only for the assembly of multiple-gene, but also for the assembly of large DNA fragment.
GoldenBraid is an iterative assembly system for multi-gene based on the use of a second type, IIS restriction enzymes. The second type IIS restriction enzyme recognizes asymmetric DNA sequences (the recognition site) and cleaves DNA at a defined position (the cleavage site) several base pairs away from the recognition site, no specific sequence requiring in the cleavage site, leaving a short overhang in the digested segments. The segments with complementing overhangs can be assembled together in the defined order, so the cleavage sites can be designed as the boundaries of DNA segments. In the GoldenBraid system, the parts (e.g. promoters, coding sequences, terminators, etc.) are created by PCR amplification, adding appropriate extensions to the primers to ensuring appropriate overhangs, and cloned into the entry clones flanked by the Bsa I sites. The digested part segments by Bsa I are assembled together with the digested destination plasmid containing a LacZ cassette flanked by the Bsa I sites in divergent orientation by incubating in a tube, creating a device (e.g. transcriptional units), where the Bsa I sites have disappeared. In order to assemble multiple devices together, another second type IIS restriction enzyme (e.g. Bsm BI) is added to the destination plasmids, so that Bsa I-assembled devices (the first order assembly) could similarly be assembled in second order destination plasmids. The GoldenBraid system consists of a set of four destination plasmids (pDGBs), All pDGBs contain a LacZ selection cassette flanked by four second Type IIS restriction sites (Bsa I, Bsm BI), but positioned in the inverted positions and orientations. The relative position of second type IIS restriction sites inside pDGBs inserts a double loop (braid) topology in the cloning design, so that the assemblies from first level become entry plasmids for second level assemblies and vice versa. Using this system, Sarrion-Perdigones et al. (2011) successfully assembled five devices in a T-DNA of binary vectors.
2.2.3 Plastid Transformation
By contrast with nuclear genes, plastid genes are organized as operons co-expressed as a single transcriptional unit. This gene organization can coordinate the expression of several genes in an operon. Thus, plastid transformation is particularly suitable for the study of metabolic pathways that are synergistically regulated by multiple genes. Bohmert-Tatarev et al. (2011) designed a plastid transformation system of tobacco (Nicotiana tabacum) for the production of renewable and biodegradable plastic polyhydroxybutyrate (PHB). Three genes in the PHB pathway were assembled in the psbA operon expression cassette to be transformed into the tobacco plastid genome . The BPH contents in T0 and T1 plants reached up to 18.8 and 17.3 % (dry weight) in leaf tissue samples, respectively. Homogentisate phytyltransferase, tocopherol cyclase, and γ-tocopherol methyltransferase are three key enzymes of tocochromanol pathway that provides tocopherols and tocotrienols, which are collectively referred as vitamin E. The genes that encode these three proteins were assembled in an operon construct to be transformed into the tomato chloroplast genome. The content of tocochromanol in transgenic plants is 10 times higher than that of the wild control (Lu et al. 2013).
Plastid transformation technology has been proved efficient in model plants, such as tobacco (Svab and Maliga 1993), tomato (Ruf et al. 2001) and potato (Sidorov et al. 1999); Kumar et al. (2004) also reported the high-efficient plastid transformation of cotton. However, plastid transformation is still not well developed in major crops like rice , maize , wheat and rape. Major challenges for the application of plastid transformation in crops include lower transformation efficiency, the difficulties to generate homoplastomic plants and to express transgenes in non-green plastids. In rice, two transformants were obtained on 100–120 bombarded plates, and no transplastomic plant of homoplasmy was obtained (Lee et al. 2006); The chloroplast transformants of oilseed rape were yielded at a frequency of 4 in 1000 bombarded cotyledon petioles (Hou et al. 2003). The transformation efficiency of wheat was estimated as 2 transplastomic plants per 42 bombarded plates of scutella and 1 transgenic line per 15 bombarded plates of immature inflorescences (Cui et al. 2011). However, up to 14 transplastomic lines of tobacco were obtained per bombarded leaf (Daniell et al. 2001). The vector inefficiency is one of the major obstacles of plastid transformation technology. The DNA fragments in vectors are transferred into the plastid genome through homologous recombination of the targeting sequence in vectors with the homologous sequence in plastids. So, the specificity of targeting sequence in vectors may be the important element influencing the transformation efficiency (Skarjinskaia et al. 2003). In addition, the explant, bombardment parameter and marker gene are also important for transformation efficiency and homoplasmy. The efforts for improving the plastid transformation of major crops should be concentrated on these respects.
2.2.4 Polyprotein Expression System
The polyprotein expression system based on linker peptide is another method of coordinating multi-gene expression . In this system, multiple genes linked by the linker peptide sequences are regulated by a promoter as a single open reading frame. After translation, the polyprotein is cleaved into its constituent protein units through the self-splicing function of linker peptide. The linker peptide 2A is a short peptide of 20 amino acids from foot-and-mouth disease virus, and mediates co-translational cleavage at its own carboxy terminus by an apparently enzyme-independent type of reaction. The efficiency of peptide 2A polyprotein expression system has been proven in human, mammalian, fungus and yeast (de Felipe et al. 2003; Fang et al. 2005; Kim et al. 2011; Ryan and Drew 1994; Suzuki et al. 2000), and also has been tested in plants. Ma and Mitra (2002) incorporated CAT and GUS gene into a single open reading frame with a copy of the FMDV 2A protein gene, and transformed tobacco; the freed CAT and GUS proteins were detected in transgenic plants with the cleavage efficiency ranging from 80–100 %. el Amrani et al. (2004) demonstrated that each protein from a 2A-polyprotein in plant cells only has its own independent targeting signals, and can correctly target various subcellular locations via either co-translational or posttranslational mechanisms. LP4 is another linker peptide, which comes from Raphanus sativus seeds, and has a recognition site and is cleaved by a protease. The advantage of LP4 as a polyprotein linker exists in that the polyprotein is completely cleaved with few redundant amino acids left at two terminals of proteins. LP4/2A is a hybrid peptide that contains the first 9 amino acids of LP4 and 20 amino acids from 2A, and can efficiently produce the individual proteins that can accurately target to the respective cellular compartments in Arabidopsis (François et al. 2002, 2004). Sun et al. (2012) demonstrated that the expression level of the two genes linked by LP4/2A was higher than those linked by 2A in tobacco. Several polyprotein transformations in major crops were reported in recent years. Two antimicrobial protein encoding genes, Dm-AMP1 and Rs-AFP2, linked by LP4 peptide sequence, were introduced into rice , and the proteins Dm-AMP1 and Rs-AFP2 were detected in the leaf extracts of transgenic plants; the disease resistance against Magnaporthe oryzae and Rhizoctonia solani of transgenic plants were improved by 90 and 79 %, respectively, as compared to the untransformed plants (Jha and Chattoo 2009). Ha et al. (2010) successfully obtained the transgenic rice accumulating 1.3 μg/g total carotenoids in seed endosperms through the transformation of two carotenoid biosynthetic genes, Psy and Crtl, linked by 2A peptide.
2.2.5 Combinatorial Genetic Transformation
Metabolic pathways usually involve multiple genes. Therefore, the function of every gene and gene combination in the entire metabolic pathway should be thoroughly understood. This knowledge is beneficial in searching for the limiting gene and gene combination when modifying metabolic pathways by metabolic engineering technology. White maize variety M37W lacks carotenoids in the endosperm because of the absence of phytoene synthase (PSY1). To accumulate carotenoids in maize, the carotenoid-synthesis pathways should be reconstructed in the maize endosperm. Five genes for carotenoid synthesis, namely, Zmpsy1 (Zea mays phytoene synthase 1), PacrtI (Pantoea ananatis phytoene desaturase), Gllycb (Gentiana lutea lycopene γ-cyclase), Glbch (Gentiana lutea γ-carotene hydroxylase), and ParacrtW (Paracoccus γ-carotene ketolase), were assembled to five vectors, respectively, and co-transformed into maize variety M37W using bombardment technology. A series of transgenic lines containing one to five genes were obtained. The analysis of carotenoid content in the seeds of these transgenic lines indicated that all transgenic lines containing Zmpsy1 gene (except the transgenic line Ph-2 without Zmpsy1 gene) could synthesize carotenoid, but varying in the carotenoid content. The line Ph-3, which contains Zmpsy1 and PacrtI, had the highest carotenoid and β-carotene content, suggesting that the combination of Zmpsy1 and PacrtI are the most effective for the modification of the carotenoid synthesis pathway in maize (Zhu et al. 2008). This transformation method is known as the combinatorial genetic transformation designed to generate the population of transgenic plants containing random transgene combinations through one transformation experiment; and is appropriate for systematic analysis of metabolic pathway and other genetic networks that require the coordinated expression of multiple genes (Farre et al. 2012).
2.2.6 New Development of MAS Technology
The successful use of MAS technology depends on three factors: functionally clarified genes, tightly linked molecular marker s and cost-effective identification technology of molecular markers . Single nucleotide polymorphism (SNP) markers are rapidly becoming the choice for genetic and breeding applications because of abundant polymorphism and automated detecting. However, the high cost and low efficiency of SNP discovery and detecting had been the main hurdles, especially for species with no reference sequence, until the next-generation sequencing (NGS) technology emerged. These NGS methods include reduced-representation sequencing using reduced-representation libraries (RRLs) or complexity reduction of polymorphic sequences (CRoPS), restriction-site-associated DNA sequencing (RAD-seq) and low coverage genotyping (Davey et al. 2011). In recent years, a large number of SNP markers have been developed in many crops using NGS technology, moreover some crops were resequenced (Davey et al. 2011; Seeb et al. 2011; Xu et al. 2012b). SNP markers have been widely used in genetic mapping , association analysis, genotyping, MAS-based breeding, and so on (Chen et al. 2011; Ferguson et al. 2012; Tiwari et al. 2014; Yang et al. 2012). Many intragenic SNPs are the source of genotypic mutation , and reflect the allelic variations . These SNP markers are designated as genic molecular markers (GMMs) or functional markers, and greatly facilitate the pyramiding of alleles as compared with SSR markers. In the base sequence of rice grain size gene GS3, the C to A mutation in the second exon is associated with enhanced grain length. A cleaved amplified polymorphic sequence (CAPS) based on C-A polymorphism was developed to identify the GS3 alleles controlling various grain sizes in rice (Fan et al. 2009).
3 A Case of Pyramiding of Five Genes for Multiple Resistances in Rice
Rice production is affected by various environmental stresses, including diseases (rice blast), insects (rice leaf folder, stem borer and brown plant hopper) and weeds. So, the objective is to pyramid resistance genes against disease, insect and herbicide into a single variety, developing a variety carrying multiple resistances.
The rice restorer line R022 is an elite rice restorer line that contains brown planthopper resistance genes Bph14 and Bph15. A backcrossing breeding scheme was designed to improve the resistance of R022 against rice blast, leaf folder, stem borer and herbicide. In this scheme, R022 was used as the recurrent parent. The transgenic rice T1C-19 was used as the donor of cry1C (encoding Bacillus thuringiensis toxin protein) and glufosinate resistance gene bar (encoding phosphinthricin acetyltransferase). T1C-19 was developed through the transformation of a construct containing cry1C and bar in the binary vector plasmid pC-1C* (Tang et al. 2006). The rice restorer line R2047 was used as the donor of rice blast resistance gene Pi9. Five genes, cry1C, bar, Pi9, Bph14 and Bph15 were stepwise pyramided into R022 through MAS-based crossing.
The F1 generation was obtained by crossing between female parent R022 and male parent T1C-19, followed by three consecutive backcrosses using R022 as recurrent parent and three selfings to obtain BC3F4 lines. In each generation of backcrossing , the herbicide glufosinate was used for the identification of bar gene, the co-dominant molecular marker s were used for the identification of genes, cry1C, Pi9, Bph14 and Bph15. Finally, the line W425 in BC3F4 was identified according to their agronomic traits and genetic backgrounds, and was named KR022. KR022 was confirmed to contain four homozygous genes, cry1C, bar, Bph14 and Bph15 through glufosinate identification and PCR assay, and showed the strong resistance against rice leaf folder and stem borer, brown planthopper and glufosinate. No damage caused by leaf folder, stem borer and glufosinate was observed in KR022 and its hybrid rice in two years of field study, while the control R022 suffered serious damage by leaf folder and stem borer, and thoroughly withered in the glufosinate treatment. KR022 inherits the brown planthopper resistance from R022 (Wan et al. 2014).
Similarly, rice blast resistance genes Pi9 from R2047 was pyramided into the R022 background through backcrossing . In the BC3F1, the plants containing homozygous Bph14 and Bph15, heterozygous Pi9 were identified to pollinate KR022 plants to produce F1, and then the F1 plants were backcrossed by KR022 and followed by three successive selfings. Finally, the line carrying five genes, Pi9, cry1C, bar, Bph14 and Bph15, was selected in BC3F3. The improved line showed the enhanced blast resistance (resistance score of 3) as compared with KR022 (resistance score of 9).
4 Application of Transgenic Pyramiding Breeding
4.1 Commercialized Transgenic Pyramiding Breeding
In recent years, stacked traits have rapidly increased in global GM crops . A record of 43.7 million ha of stacked traits was grown globally in 2012, which is equivalent to 26 % of the global 170 million ha of GM crops in 2012 and 7.5 times higher than the 5.8 million ha of stacked traits in 2003. Stacked traits mainly exist in GM maize and cotton. In 2012, 40 million ha of GM maize were stacked, contributing 91.5 % to 43.7 million ha of stacked traits, and stacked GM cottons were 3.7 million ha (Table 13.1). Table 13.2 lists some of the worlds commercialized GM maize and cotton with stacked traits.
At present, the stacked traits of GM crops are mainly insect resistance and herbicide resistance. Insect resistance primarily comes from the Bacillus thuringiensis genes encoding toxic proteins, such as cry1Ab, cry1Ac, cry2Ae, vip3A (a), cry1F, cry3A, mcry3A, cry34Ab1 and cry35Ab1. Herbicide resistance comes from the glyphosate-resistance gene (epsps) encoding 5-enolpyruvylshikimate-3-phosphate synthase, the glufosinate-resistance genes (pat and bar) encoding phosphinothricin N-acetyltransferase.
Several stacked GM varieties have been introduced. Agrisure® Viptera™ 3111 maize , bred by Syngenta, possessed genes, cry1Ab, vip3Aa20, mcry3A, pat and epsps. cry1Ab and vip3Aa20 offer the resistance to Lepidoptera insect aboveground; mcry3A offers the resistance to Coleoptera insect , particularly corn rootworm pest underground. The pyramiding of cry1Ab, vip3Aa20 and mcry3A broadens the resistance spectrum of Agrisure® Viptera™ 3111. According to the data published by Syngenta (http://www.syngenta.com/), Agrisure® Viptera™ 3111 can resist 14 kinds of aboveground and underground insects.
Two modes of resistance to herbicides are offered by pat and epsps gene, and the pyramiding of these two genes provides a more flexible herbicide use of glufosinate and glyphosate. Agrisure® Viptera™ 3111 has gained the approvals for food in eight countries or districts, for feed in five countries, and for cultivation in two countries. Genuity® VT Triple Pro™ maize containing epsps, cry1A.105, cry2Bb2 and cry3Bb1, developed by Monsanto, is endowed with resistances to glyphosate herbicide and Lepidoptera and Coleoptera insect . Genuity® VT Triple Pro™ has been approved for food use in ten countries or districts, for feed use in eight countries or districts, and for cultivation in five countries. Genuity® SmartStax™ maize was developed by Monsanto and Tow AgroSciences and possesses eight effect genes, including cry1A105, cry2Ab1, cry3Bb1, cry1Fa2, cry34Ab1, cry35Ab1, epsps and pat. These genes confer Genuity® SmartStax™ the resistances against Lepidoptera and Coleoptera insects and the herbicide glyphosate and glufosinate. Genuity® SmartStax™ has been approved for food or feed cultivation in eight countries or districts. Widestrike™ Roundup Ready Flex™ Cotton, jointly developed by Monsanto and Dow AgroSciences, possesses four effect genes, including epsps, cry1F, cry1Ac and bar, which endow Widestrike™ Roundup Ready Flex™ with the resistances against Lepidoptera insect and the herbicide glyphosate and glufosinate. Widestrike™ Roundup Ready Flex™ has obtained approvals for food or feed cultivation in four countries.
4.2 Application of Transgenic Pyramiding Breeding in Metabolic Engineering
Although plant metabolic engineering is still at an early stage and has not been widely applied in commercialized GM crops , more successful cases of plant metabolic engineering research are reported. Table 13.3 lists a few engineered plants from the transgenic pyramiding breeding in recent years, in which some have potential for commercial application.
4.2.1 Polyunsaturated Fatty Acid Synthesis
Very long chain polyunsaturated fatty acids (VLCPUFAs) are nutritional substances with important function in human health (SanGiovanni and Chew 2005). Eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA) are two of the most important VLCPUFAs for the human body. These two fatty acids occur primarily in ocean algae, and accumulate in the fat of deep-sea fishes. The commercial VLCPUFA are mainly refined from deep-sea fishes, but the deep-sea fish supply has been limited for overfishing, and thus cannot satisfy the market demand. Oilseeds are easily obtained with high yields and generally contain unsaturated fatty acids, such as oleic acid (OA), linoleic acid (LA), and α-linolenic acid (ALA). Oilseeds can be used as substrates for VLCPUFAs synthesis, but lack the capacity of elongation/unsaturation. Thus, to synthesize VLCPUFAs, biologists have tried to reconstruct its synthesis pathway in oilseeds through metabolic engineering.
In 2004, VLCPUFA synthesis pathway was reconstructed in Arabidopsis thaliana through the transformation of genes encoding a Δ9-elongase from Isochrysis galbana, a Δ8- desaturase from Euglena gracilis, and a Δ5- desaturase from Mortierella alpinato. EPA successfully accumulated for the first time in the engineering of Arabidopsis thaliana (Qi et al. 2004). Subsequently, Abbadi et al. (2004) transformed tobacco and linseed with genes encoding a Δ6-fatty acid desaturase from Physcomitrella patens, a Δ5-fatty acid desaturase from Phaeodactylum tricornutum, and a Δ6- prolonged desaturase from Physcomitrella patens, and detected approximately 0.5 % EPA in the transgenic seed oils. DHA was synthesized for the first time in seed oils in 2005. Arabidopsis thaliana was transformed with a Δ6/Δ5 desaturase from zebrafish. A transgenic plant accumulating 1.6 % AA and 3.2 % EPA was selected to be retransformed with genes encoding a Δ4-desaturase and a Δ5- desaturase from Pavlova salina, and 0.2–0.5 % DHA was detected in transgenic seed oils (Robert et al. 2005). These studies demonstrated the feasibility of DHA and EPA synthesis in engineering plants, but their yields should be improved. Wu et al. (2005) reported the transgenic production of significant amounts of EPA and AA in Brassica juncea by stepwise metabolic engineering strategy. Through a series of transformations with increasing the number of transgenes encoding desaturase and prolonged desaturase from different organisms, the transgenic plants were demonstrated to produce VLCPUFAs with up to 25 % AA and 15 % EPA of total seed fatty acids. Moreover, DHA synthesis pathway was reconstituted in plant seeds through adding genes in Δ4 pathway and successfully synthesized DHA despite of 0.2 % of total fatty acids only. To overcome the poor DHA synthesis, Petrie et al. (2012) designed a transgenic synthesis pathway from OA to DHA, which involved seven genes encoding desaturases and prolonged enzymes, including the Δ12-desaturase from Lachancea kluyveri, the Δ15-/ω3-desaturase from Pichia pastoris, the Δ6-desaturase from Micromonas pusilla, the Δ5-elongases from Pyramimonas cordata, and the Δ5- and Δ4-desaturases from Pavlova salina. These seven genes were transformed into Arabidopsis thaliana, and DHA averaged 13.3 % in seed oils of transgenic plants. This content level, even exceeds the DHA content in common commercial fish oil, and thus satisfies the demand for the commercialized VLCPUFA product.
4.2.2 Carotene Synthesis
Vitamin A is an indispensable nutrient, and has an important physiological function in maintaining the normal growth and development of the human body. Vitamin A deficiency can induce many diseases, such as nyctalopia, and still remains a problem in the poor developing countries (WHO 2009). Vitamin A can be obtained from animal foods as retinol, and obtained from plant and animal foods as carotene that can be transformed into pro-vitamin A.
Rice is the primary world grain crop. However, rice cannot supply vitamin A to the human body because of the lack of synthesis of pro-vitamin A in the edible parts of rice. Thus, biologists attempted to reconstruct the carotene synthesis pathway in rice by overexpressing key genes in the carotene synthesis pathway in rice endosperms. Ye et al. (2000) used three genes encoding the key enzymes in β-carotene synthesis, namely phytoene desaturase (crtI) from Erwinia uredovora, phytoene synthase (psy) from daffodil, and lycopene β-cyclase (lcy) from Narcissus pseudonarcissus. These genes were introduced into rice under the control of an endosperm-specific glutenin promoter Gtl by double-vector co-transformation . The transgenic rice has yellow endosperms for the accumulation of β-carotene, and is called Golden Rice. However, the β-carotene content is only 1.6 μg/g in this Golden Rice. To increase the β-carotenoid content in transgenic rice endosperms, genes encoding phytoene synthases, the limiting step for β-carotene accumulation, were tested from different species. A maize phytoene synthase encoding gene (psy) that substantially increases carotenoid accumulation in a model plant system was identified to transform rice combined with the Erwinia uredovora carotene desaturase encoding gene (crtI) to develop Golden Rice 2. As a result, the total carotenoids in the endosperm of Golden Rice 2 increased up to 23-fold (maximum 37 μg/g) compared with the original Golden Rice, and β-carotene has a preferential accumulation (Paine et al. 2005). According to the American RDA, 100 g of Golden rice 2 consumed per day can satisfy 55–70 % of the vitamin requirements of an adult male (Tang et al. 2009).
The metabolic engineering of carotenoids has also been successful in potato and maize . A mini-pathway of β-carotene synthesis was reconstructed in potato through expressing three genes encoding phytoene synthase (CrtB), phytoene desaturase (CrtI), and lycopene beta-cyclase (CrtY) from Erwinia under the control of tuber-specific promoter. Carotenoid largely accumulated in the transgenic potato tuber. The carotenoid content in the tuber increased 20-fold, and reached 114 μg/g dry weight, and the β-carotene content increased 3600-fold and reached 47 μg/g dry weight (Diretto et al. 2007). The overexpression of crtB and crtI from Erwinia under the control of the super γ-zein promoter in maize also resulted in an increase of total carotenoids of up to 34-fold with a preferential accumulation of β-carotene in the transgenic plants (Aluru et al. 2008). Naqvi et al. (2009) developed an elite transgenic corn containing four transgenes for the metabolism of β-carotene, ascorbate and folate, including two genes in the pathways of the β-carotene synthesis (psy1 from maize controlled by the wheat LMW glutenin promoter and crtI from Pantoea ananatis under the control of barley D-hordein promoter), one gene involved in the synthesis of ascorbate (dhar encoding rice dehydroascorbate reductase under the control of barley D-hordein promoter), and one gene involved in the metabolism of folate (folE encoding Escherichia coli. GTP cyclohydrolase under the control of barley D-hordein promoter). The contents of β-carotene, ascorbate, and folate in the seeds of transgenic maize increased up to 169-fold (59.32 μg/g DW), 6-fold (106.94 μg/g DW) and 2-fold (1.94 μg/g DW) compared with the wild control, respectively.
4.2.3 Starch Synthesis
Starch composes the main edible part of food crops, and is the main human energy source. Starch in plants exists in two forms: amylose and amylopectin, those contents and proportion involve both crop yield and palatability. Starch synthesis is a complex pathway controlled by many genes encoding ADP-glucose pyrophosphorylase (AGP), granule-bound starch synthase (GBSS), soluble starch synthase (SS), starch branching enzyme (SBE), starch debranching enzyme (DBE), etc. The expression of two starch branching enzymes, SBE IIa and SBE IIb were suppressed in barley through RNAi-mediated silencing technology . The transgenic lines, where both SBE IIa and SBE IIb expression were reduced by >80 %, was observed with a high amylose phenotype (>70 %) (Regina et al. 2010). All genes encoding starch branching enzymes (SBE I, SBE IIa, SBE IIb) were further simultaneously suppressed in barley, resulting in the production of amylose-only starch granules in the endosperm of transgenic lines. In these lines, a very high content of resistance starch (RS) (65 %) was observed, which is 2.2-fold higher than control (29 %) (Carciofi et al. 2012). In maize , the expression of six genes involving starch synthesis was modified, these genes include the overexpressed Bt2, Sh2, Sh1 and GbssIIa (to enhance the activity of sucrose synthase, AGPase and granule-bound starch synthase) and the suppressed SbeI and SbeIIb. The transgenic plants expressing all six genes showed a 2.8–7.7 % increase in the endosperm starch content and a 37.8–43.7 % increase in the proportion of amylose. Moreover, the 100-grain weight and ear weight of transgenic plants had up to 20.1–34.7 % and 13.9–19.0 % increases, respectively (Jiang et al. 2013).
4.3 Gene Pyramiding for Resistances against Disease, Drought and Salt
Disease resistance improvement of crops may be the most successfully used field of gene pyramiding . A number of works have been done on this aspect. Since the 1990s, the International Rice Research Institute (IRRI) has developed a series of rice near-isogenic lines (NILs) (named IRBB) carrying stacked various rice bacterial blight (BB) resistance genes (Huang et al. 1997). These NILs have been widely used in rice breeding programs. Recently, IRBB60 (Xa4/xa5/xa13/Xa21) was used as the donor of BB resistance genes to improve the BB resistance of an elite rice cultivar in India (Dokku et al. 2013). Jin 23B is a cytoplasmic male sterile (CMS) rice maintainer line. Its sterile line, Jin 23A, has been widely used in hybrid rice production in China. However, Jin 23B is highly susceptible to rice blast. In a backcross breeding scheme, three blast resistance genes, Pi1, Pi2, and D12, were introgressed into Jin 23B through MAS, and the improved line Jin 23B (Pi1/Pi2/D12) was obtained from BC4F3 families. The blast resistance of Jin 23B (Pi1/Pi2/D12) and its hybrid rice , Jinyou 402 (Pi1/Pi2/D12), significantly increased compared with their counterparts, Jin 23B and its hybrid rice , Jinyou 402. The resistance scores of leaf blast at tillering stage for Jin 23B (Pi1/Pi2/D12) and Jinyou 402 (Pi1/Pi2/D12) were 0.33 and 0.72, respectively, which are significantly lower than the scores for Jin 23B and Jinyou 402 (4.01 and 4.02, respectively). Moreover, the hybrid rice from Jin 23B (Pi1/Pi2/D12) under the disease condition had the same yield as under the normal condition (Jiang et al. 2012). In wheat , eight QTLs/genes were pyramided into a popular elite wheat cultivar PBW343 through MAS, endowing the improved PBW343 four grain quality traits and the resistance against three rusts (Tyagi et al. 2014).
Abiotic stress , including drought and salinity, has become more serious for crops in recent years. Stress resistance has been repeatedly achieved in crops through the transformation of a single gene. For example, Monsanto had developed a drought-resistance GM maize variety carrying a cold shock protein B gene cspB, Genuity® DroughtGard™ (GM Approval Database of ISAAA). However, the gene pyramiding is still necessary for further enhancement of resistance to abiotic stress . The transgenic maize carrying stacked betA (encoding choline dehydrogenase from Escherichia coli) and TsVP (encoding V-H+-PPase from Thellungiella halophila) had been produced by cross-pollination. It contained higher relative water content (RWC), greater solute accumulation and lower cell damage under drought stress treatment, and grew more vigorously with less growth retardation, shorter anthesis-silking intervals and higher yields than their parental lines, which had either betA or TsVP, and the wild-type (Wei et al. 2011). Nguyen et al. (2013) developed the transgenic maize carrying the stacked mtlD (bacterial mannitol-1-phosphate dehydrogenase encoding gene) and HVA1 (Hordeum vulgare) through co-transformation of two vectors. The transgenic plants (mtlD + HVA1) showed higher leaf RWC and greater plant survival as compared with the transgenic plants of a single transgene and with their control plants under drought stress , and also showed higher fresh and dry shoot matter and dry root matter as compared with the transgenic plants of a single transgene and with their control plants under salt stress.
5 Conclusions and Prospects
With the coming of the post-genome era, crop breeding has gradually developed from conventional breeding based on phenotype selection to molecular design breeding based on gene selection. As a directed improvement technology, transgenic pyramiding breeding will provide a powerful technological support for crop molecular design breeding. However, transgenic pyramiding breeding also comes up against many technology challenges for meeting the molecular design breeding requirements. In contrast to conventional breeding, transgenic crops should not only be evaluated in agronomic characters such as yield and resistance, but also in their biosafety. Biosafety assessment is a time-consuming and laborious process, and also increases the product cost and prolongs the breeding process. More genes involved in transgenic pyramiding will result in a more complex biosafety assessment. Therefore, to simplify the process of biosafety assessment regarding breeding strategy and technology within the allowable range of biosafety management is important.
Current transgenic crops involve few effect genes only, and all these genes constitutively overexpress in transgenic crops. However, with the development of transgenic pyramiding breeding, the number of transgenes will increase, and their expression patterns, including expression level, tissue-specificity, developmental stage-specificity, etc., will become more complicated. Therefore, it is necessary to ensure every transgene truly expressing according to the breeding objective. Although the current multiple-gene transformation technology can pyramid eight to ten genes or transformed the fragments of 20 kb or more in length, the technology should be further developed to become a conventional breeding technology.
Crop variety is a special commodity, which has commodity properties, such as market demand and cost. Moreover, biological product properties, such as ecological adaptability, stress resistance, yield traits , quality traits , and biosafety , can also be found in the variety. As the second-generation transgenic crops, the commercialization of transgenic crops with stacked pest and herbicide resistance has started, and will be in a fast developing period. A few metabolic engineering crops of carotenoids and VLCPUFAs have achieved the requirement for commercialization. Moreover, by elucidating the metabolic pathways in plants, more metabolic engineering technologies will mature. In recent years, with the rapid development of plant genomics research, a number of genes controlling important agronomy characters have been identified and cloned, and their functions have been illuminated. These achievements can provide the new directions for the transgenic pyramiding breeding of crops.
References
Abbadi A, Domergue F, Bauer J et al (2004) Biosynthesis of very-long-chain polyunsaturated fatty acids in transgenic oilseeds: constraints on their accumulation. Plant Cell 16(10):2734–2748
Aluru M, Xu Y, Guo R et al (2008) Generation of transgenic maize with enhanced provitamin A content. J Exp Bot 59(13):3551–3562
Azadi P, Otang NV, Chin DP et al (2010) Metabolic engineering of Lilium × formolongi using multiple genes of the carotenoid biosynthesis pathway. Plant Biotechnol Rep 4(4):269–280
Bates SL, Zhao JZ, Roush RT et al (2005) Insect resistance management in GM crops:past, present and future. Nat Biotechnol 23(1):57–62
Bohmert-Tatarev K, McAvoy S, Daughtry S et al (2011) High levels of bioplastic are produced in fertile transplastomic tobacco plants engineered with a synthetic operon for the production of polyhydroxybutyrate. Plant Physiol 155(4):1690–1708
Buntru M, Gärtner S, Staib L et al (2013) Delivery of multiple transgenes to plant cells by an improved version of MultiRound Gateway technology. Transgenic Res 22(1):153–167
Carciofi M, Blennow A, Jensen SL et al (2012) Concerted suppression of all starch branching enzyme genes in barley produces amylose-only starch granules. BMC Plant Biol 12(1):223
Chang YL, Henriquez X, Preuss D et al (2003) A plant-transformation-competent BIBAC library from the Arabidopsis thaliana Landsberg ecotype for functional and comparative genomics. Theor Appl Genet 106(2):269–276
Chen QJ, Zhou HM, Chen J et al (2006) A Gateway-based platform for multigene plant transformation. Plant Mol Biol 62(6):927–936
Chen QJ, Xie M, Ma XX et al (2010) MISSA is a highly efficient in vivo DNA assembly method for plant multiple-gene transformation. Plant Physiol 153(1):41–51
Chen H, He H, Zou Y et al (2011) Development and application of a set of breeder-friendly SNP markers for genetic analyses and molecular breeding of rice (Oryza sativa L.). Theor Appl Genet 123(6):869–879
Cheng B, Wu G, Vrinten P et al (2010) Towards the production of high levels of eicosapentaenoic acid in transgenic plants: the effects of different host species, genes and promoters. Transgenic Res 19(2):221–229
Cui C, Song F, Tan Y et al (2011) Stable chloroplast transformation of immature scutella and inflorescences in wheat (Triticum aestivum L.). Acta Biochim Biophys Sin 43(4):284–291
Daniell H, Lee SB, Pahchal T et al (2001) Expression of the native cholera toxin B subunit gene and assembly as functional oligomers in transgenic tobacco chloroplasts. J Mol Biol 311(5):1001–1009
Davey JW, Hohenlohe PA, Etter PD et al (2011) Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat Rev Genet 12(7):499–510
de Felipe P, Hughes LE, Ryan MD et al (2003) Co-translational, intraribosomal cleavage of polypeptides by the foot-and-mouth disease virus 2A peptide. J Biol Chem 278(13):11441–11448
Diretto G, Al-Babili S, Tavazza R et al (2007) Metabolic engineering of potato carotenoid content through tubers-specific overexpression of a bacterial mini-pathway. PLoS One 2(4), e350
Dokku P, Das KM, Rao GJN (2013) Pyramiding of four resistance genes of bacterial blight in Tapaswini, an elite rice cultivar, through marker-assisted selection. Euphytica 192(1):87–96
el Amrani A, Barakate A, Askari BM et al (2004) Coordinate expression and independent subcellular targeting of multiple proteins from a single transgene. Plant Physiol 135(1):16–24
Fan C, Yu S, Wang C et al (2009) A causal C–A mutation in the second exon of GS3 highly associated with rice grain length and validated as a functional marker. Theor Appl Genet 118(3):465–472
Fang J, Qian JJ, Yi S et al (2005) Stable antibody expression at therapeutic levels using the 2A peptide. Nat Biotechnol 23(5):584–590
Farre G, Naqvi S, Sanahuja G et al (2012) Combinatorial genetic transformation of cereals and the creation of metabolic libraries for the carotenoid pathway. Methods Mol Biol 847:419–435
Ferguson ME, Hearne SJ, Close TJ et al (2012) Identification, validation and high-throughput genotyping of transcribed gene SNPs in cassava. Theor Appl Genet 124(4):685–695
Flores T, Karpova O, Su X et al (2008) Silencing of the GmFAD3 gene by siRNA leads to low a-linolenic acids (18:3) offad3-mutant phenotype in soybean Glycine max (Merr.). Transgenic Res 17:839–850
François IE, De Bolle MF, Dwyer G et al (2002) Transgenic expression in Arabidopsis of a polyprotein construct leading to production of two different antimicrobial proteins. Plant Physiol 128(4):1346–1358
François IE, Van Hemelrijck W, Aerts AM et al (2004) Processing in Arabidopsis thaliana of a heterologous polyprotein resulting in differential targeting of the individual plant defensins. Plant Sci 166(1):113–121
Fujisawa M, Takita E, Harada H et al (2009) Pathway engineering of Brassica napus seeds using multiple key enzyme genes involved in ketocarotenoid formation. J Exp Bot 60(4):1319–1332
Ghimire MN, Huang F, Leonard R et al (2011) Susceptibility of Cry1Ab-susceptible and -resistant sugarcane borer to transgenic corn plants containing single or pyramided Bacillus thuringiensis genes. Crop Prot 30(1):74–81
Gibson DG, Benders GA, Axelrod KC (2008) One-step assembly in yeast of 25 overlapping DNA fragments to form a complete synthetic Mycoplasma genitalium genome. Proc Nat Acad Sci 105(51):20404–20409
Gibson DG, Young L, Chuang RY (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6(5):343–345
GM Approval Database (2004) ISAAA. http://www.isaaa.org/gmapprovaldatabase/. Accessed 12 Feb 2004
Goyal A, Bhowmik PK, Basu SK (2009) Minichromosomes: the second generation genetic engineering tool. Plant Omics J 2(1):1–8
Ha SH, Liang YS, Jung H et al (2010) Application of two bicistronic systems involving 2A and IRES sequences to the biosynthesis of carotenoids in rice endosperm. Plant Biotechnol J 8(8):928–938
Halpin C (2005) Gene stacking in transgenic plants–the challenge for 21st century plant biotechnology. Plant Biotechnol 3(2):141–155
Hamilton CM, Frary A, Xu Y (1999) Construction of tomato genomic DNA libraries in a binary-BAC (BIBAC) vector. Plant J 18(2):223–229
Hou BK, Zhou YH, Wan LH et al (2003) Chloroplast transformation in oilseed rape. Transgenic Res 12(1):111–114
Hu J, Li X, Wu C et al (2012) Pyramiding and evaluation of the brown planthopper resistance genes Bph14 and Bph15 in hybrid rice. Mol Breed 29(1):61–69
Huang JC, Zhong YJ, Liu J et al (2013) Metabolic engineering of tomato for high-yield production of astaxanthin. Metab Eng 17(100):59–67
Huang N, Angeles ER, Domingo J et al (1997) Pyramiding of bacterial blight resistance genes in rice: marker-assisted selection using RFLP and PCR. Theor Appl Genet 95(3):313–320
James C (2013) Global status of commercialized Biotech/GM Crops. ISAAA, Ithaca
Jha S, Chattoo BB (2009) Transgene stacking and coordinated expression of plant defensins confer fungal resistance in rice. Rice 2(4):143–154
Jiang H, Feng Y, Bao L et al (2012) Improving blast resistance of Jin 23B and its hybrid rice by marker-assisted gene pyramiding. Mol Breed 30(4):1679–1688
Jiang L, Yu X, Qi X et al (2013) Multigene engineering of starch biosynthesis in maize endosperm increases the total starch content and the proportion of amylose. Transgenic Res 22(6):1133–1142
Kim JH, Lee SR, Li LH et al (2011) High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PLoS One 6(4), e18556
Kong FN, Jiang SM, Shi LX (2006) Construction and characterization of a transformation-competent artificial chromosome (TAC) library of Zizania latifolia (Griseb.). Plant Mol Biol Report 24(2):219–227
Kumar S, Dhingra A, Daniell H (2004) Stable transformation of the cotton plastid genome and maternal inheritance of transgenes. Plant Mol Biol 56(2):203–216
Lange BM, Mahmoud SS, Wildung MR et al (2011) Improving peppermint essential oil yield and composition by metabolic engineering. Proc Nat Acad Sci 108(41):16944–16949
Lee SM, Kang K, Chung H et al (2006) Plastid transformation in the monocotyledonous cereal crop, rice (Oryza sativa) and transmission of transgenes to their progeny. Mol Cells 21(3):401–410
Li MZ, Elledge SJ (2007) Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat Meth 4(3):251–256
Li MZ, Elledge SJ (2012) SLIC: a method for sequence-and ligation-independent cloning. Methods Mol Biol 852:51–59
Lin L, Liu YG, Xu X (2003) Efficient linking and transfer of multiple genes by a multigene assembly and transformation vector system. Proc Nat Acad Sci 100:5962–5967
Lu Y, Rijzaani H, Karcher D et al (2013) Efficient metabolic pathway engineering in transgenic tobacco and tomato plastids with synthetic multigene operons. Proc Nat Acad Sci 110(8):E623–E632
Ma C, Mitra A (2002) Expressing multiple genes in a single open reading frame with the 2A region of foot-and-mouth disease virus as a linker. Mol Breed 9(3):191–199
Murata M, Shibata F, Hironaka A et al (2013) Generation of an artificial ring chromosome in Arabidopsis by Cre/LoxP-mediated recombination. Plant J 74(3):363–371
Naqvi S, Farré G, Zhu C et al (2011) Simultaneous expression of Arabidopsis ρ-hydroxyphenylpyruvate dioxygenase and MPBQ methyltransferase in transgenic corn kernels triples the tocopherol content. Transgenic Res 20(1):177–181
Naqvi S, Zhu C, Farre G et al (2009) Transgenic multivitamin corn through biofortification of endosperm with three vitamins representing three distinct metabolic pathways. Proc Nat Acad Sci 106(19):7762–7767
Nguyen TX, Nguyen T, Alameldin H et al (2013) Transgene pyramiding of the HVA1 and mtlD in T3 maize (Zea mays L.) plants confers drought and salt tolerance, along with an increase in crop biomass. Int J Agron, doi:10.1155/2013/598163
Paine JA, Shipton CA, Chaggar S et al (2005) Improving the nutritional value of Golden Rice through increased pro-vitamin A content. Nat Biotechnol 23(4):482–487
Petrie JR, Shrestha P, Zhou XR et al (2012) Metabolic engineering plant seeds with fish oil-like levels of DHA. PLoS One 7(11), e49165
Qi B, Fraser T, Mugford S et al (2004) Production of very long chain polyunsaturated omega-3 and omega-6 fatty acids in plants. Nat Biotechnol 22(6):739–745
Qu S, Coaker G, Francis D (2003) Development of a new transformation-competent artificial chromosome (TAC) vector and construction of tomato and rice TAC libraries. Mol Breed 12(4):297–308
Quan J, Tian J (2009) Circular polymerase extension cloning of complex gene libraries and pathways. PLoS One 4(7), e6441
Regina A, Kosar-Hashemi B, Ling S et al (2010) Control of starch branching in barley defined through differential RNAi suppression of starch branching enzyme IIa and IIb. J Exp Bot 61(5):1469–1482
Robert SS, Singh SP, Zhou XR et al (2005) Metabolic engineering of Arabidopsis to produce nutritionally important DHA in seed oil. Func Plant Biol 32(6):473–479
Ruf S, Hermann M, Berger I et al (2001) Stable genetic transformation of tomato plastids and expression of a foreign protein in fruit. Nat Biotechnol 19(9):870–875
Ruiz-Lopez N, Haslam RP, Napier JA et al (2014) Successful high-level accumulation of fish oil omega-3 long-chain polyunsaturated fatty acids in a transgenic oilseed crop. Plant J 77(2):198–208
Ruiz-Lopez N, Haslam RP, Usher SL et al (2013) Reconstitution of EPA and DHA biosynthesis in Arabidopsis: iterative metabolic engineering for the synthesis of n − 3 LC-PUFAs in transgenic plants. Metab Eng 17(100):30–41
Ryan MD, Drew J (1994) Foot-and-mouth disease virus 2A oligopeptide mediated cleavage of an artificial polyprotein. EMBO J 13(4):928–933
SanGiovanni JP, Chew EY (2005) The role of omega-3 long-chain polyunsaturated fatty acids in health and disease of the retina. Prog Retin Eye Res 24(1):87–138
Sarrion-Perdigones A, Falconi EE, Zandalinas SI et al (2011) GoldenBraid: an iterative cloning system for standardized assembly of reusable genetic modules. PLoS One 6(7), e21622
Sasaki Y, Sone T, Yoshida S et al (2004) Evidence for high specificity and efficiency of multiple recombination signals in mixed DNA cloning by the Multisite Gateway system. J Biotechnol 107(3):233–243
Seeb JE, Carvalho G, Hauser L et al (2011) Single‐nucleotide polymorphism (SNP) discovery and applications of SNP genotyping in nonmodel organisms. Mol Ecol Res 11(s1):1–8
Sidorov VA, Kasten D, Pang SZ et al (1999) Stable chloroplast transformation in potato: use of green fluorescent protein as a plastid marker. Plant J 19(2):209–216
Skarjinskaia M, Svab Z, Maliga P (2003) Plastid transformation in Lesquerella fendleri, an oilseed Brassicacea. Transgenic Res 12(1):115–122
Sun H, Lang Z, Zhu L et al (2012) Acquiring transgenic tobacco plants with insect resistance and glyphosate tolerance by fusion gene transformation. Plant Cell Rep 31(10):1877–1887
Suzuki N, Geletka LM, Nuss DL (2000) Essential and dispensable virus-encoded replication elements revealed by efforts to develop hypoviruses as gene expression vectors. J Virol 74(16):7568–7577
Svab Z, Maliga P (1993) High-frequency plastid transformation in tobacco by selection for a chimeric aadA gene. Proc Nat Acad Sci 90(3):913–917
Tan MA, Hutten RC, Visser RG et al (2010) The effect of pyramiding Phytophthora infestans resistance genes R Pi-mcd1 and R Pi-ber in potato. Theor Appl Genet 121(1):117–125
Tan H, Yang X, Zhang F et al (2011) Enhanced seed oil production in canola by conditional expression of Brassica napus LEAFY COTYLEDON1 and LEC1-LIKE in developing seeds. Plant Physiol 156(3):1577–1588
Tang W, Chen H, Xu C et al (2006) Development of insect-resistant transgenic indica rice with a synthetic cry1C* gene. Mol Breed 18(1):1–10
Tang G, Qin J, Dolnikowski GG et al (2009) Golden Rice is an effective source of vitamin A. Am J Clin Nutr 89(6):1776–1783
Taverniers I, Papazova N, Bertheau Y et al (2008) Gene stacking in transgenic plants: towards compliance between definitions, terminology, and detection within the EU regulatory framework. Envir Biosaf Res 7(4):197–218
Tiwari VK, Wang S, Sehgal S et al (2014) SNP discovery for mapping alien introgressions in wheat. BMC Genomics. doi:10.1186/1471-2164-15-273
Tyagi S, Mir RR, Kaur H et al (2014) Marker-assisted pyramiding of eight QTLs/genes for seven different traits in common wheat (Triticum aestivum L.). Mol Breed. doi:10.1007/s11032-014-0027-1
Wan B, Zha Z, Li J et al (2014) Development of elite rice restorer lines in the genetic background of R022 possessing tolerance to brown planthopper, stem borer, leaf folder and herbicide through marker-assisted breeding. Euphytica 195(1):129–142
Wei AY, He CM, Li B et al (2011) The pyramid of transgenes TsVP and BetA effectively enhances the drought tolerance of maize plants. Plant Biotechnol J 9(2):216–229
WHO (2009) Global prevalence of vitamin A deficiency in populations at risk 1995–2005. WHO global database on vitamin A deficiency. Geneva, World Health Organization, 2009. Available from: http://www.who.int/vmnis/vitamina/en/
Wu G, Truksa M, Datla N et al (2005) Stepwise engineering to produce high yields of very long-chain polyunsaturated fatty acids in plants. Nat Biotechnol 23(8):1013–1017
Xu C, Cheng Z, Yu W (2012a) Construction of rice mini-chromosomes by telomere-mediated chromosomal truncation. Plant J 70(6):1070–1079
Xu X, Liu X, Ge S et al (2012b) Resequencing 50 accessions of cultivated and wild rice yields markers for identifying agronomically important genes. Nat Biotechnol 30(1):105–111
Yang H, Tao Y, Zheng Z et al (2012) Application of next-generation sequencing for rapid marker development in molecular plant breeding: a case study on anthracnose disease resistance in Lupinus angustifolius L. BMC Genomics. doi:10.1186/1471-2164-13-318
Ye X, Al-Babili S, Klöti A et al (2000) Engineering the provitamin A (β-carotene) biosynthetic pathway into (carotenoid-free) rice endosperm. Science 287(5451):303–305
Yu W, Lamb JC, Han F et al (2006) Telomere-mediated chromosomal truncation in maize. Proc Nat Acad Sci 103(46):17331–17336
Yu W, Han F, Gao Z (2007) Construction and behavior of engineered minichromosomes in maize. Proc Nat Acad Sci 104(21):8924–8929
Zhai J, Wang Y, Sun C (2013) A plant-transformation-competent BIBAC library of ginseng (Panax ginseng C. A. Meyer) for functional genomics research and characterization of genes involved in ginsenoside biosynthesis. Mol Breed 31(3):685–692
Zhang J, Martin JM, Beecher B et al (2010) The ectopic expression of the wheat Puroindoline genes increase germ size and seed oil content in transgenic corn. Plant Mol Biol 74(4–5):353–365
Zhao JZ, Cao J, Li Y et al (2003) Transgenic plants expressing two Bacillus thuringiensis toxins delay insect resistance evolution. Nat Biotechnol 21(12):1493–1497
Zhu C, Naqvi S, Breitenbach J et al (2008) Combinatorial genetic transformation generates a library of metabolic phenotypes for the carotenoid pathway in maize. Proc Nat Acad Sci 105(47):18232–18237
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Wan, B. (2015). Transgenic Pyramiding for Crop Improvement. In: Al-Khayri, J., Jain, S., Johnson, D. (eds) Advances in Plant Breeding Strategies: Breeding, Biotechnology and Molecular Tools. Springer, Cham. https://doi.org/10.1007/978-3-319-22521-0_13
Download citation
DOI: https://doi.org/10.1007/978-3-319-22521-0_13
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-22520-3
Online ISBN: 978-3-319-22521-0
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)