Abstract
STAT (signal transducers and activators of transcription) are latent cytoplasmic transcription factors that function as downstream effectors of cytokine and growth factor receptor signaling. The canonical JAK/STAT signaling pathway involves the activation of Janus kinases (JAK) or growth factors receptor kinases, phosphorylation of STAT proteins, their dimerization and translocation into the nucleus where STATs act as transcription factors with pleiotropic downstream effects. STAT signaling is tightly controlled with restricted kinetics due to action of its negative regulators. While STAT1 is believed to play an important role in growth arrest and apoptosis, and to act as a tumor suppressor, STAT3 and 5 are involved in promoting cell cycle progression, cellular transformation, and preventing apoptosis. Aberrant activation of STATs, in particular STAT3 and STAT5, have been found in a large number of human tumors, including gliomas and may contribute to oncogenesis. In this chapter, we have (1) summarized the mechanisms of STAT activation in normal and malignant signaling; (2) discussed evidence for the critical role of constitutively activated STAT3 and STAT5 in glioma pathobiology; (3) disclosed molecular and pharmacological strategies to interfere with STAT signaling for potential therapeutic intervention in gliomas.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Cytokine and growth factor receptor signaling
- Protein tyrosine kinases
- STAT proteins
- Gliomas
- Transcription regulation
10.1 Introduction
STATs (signal transducers and activators of transcription) are a family of latent transcription factors that are activated in response to intracellular stimuli. Seven STAT proteins have been identified (STATs 1, 2, 3, 4, 5a, 5b, 6), and the corresponding gene products share a high degree of similarity. Each STAT protein has a DNA binding domain, a src-homology 2 (SH2) domain necessary for homo- or heterodimerization and a conserved tyrosine residue 705, phosphorylated by tyrosine kinases (Kisseleva et al. 2002 ).
Over 40 different cytokines or growth factors can activate STAT signaling pathway. Upon binding of cytokines to cognate receptors on the surface of cells, receptors dimerize and thereby activate receptor-associated tyrosine kinases such as Janus kinases (JAKs) that phosphorylate the receptor cytoplasmic portion. STATs are also activated by growth factor receptors with the intrinsic tyrosine kinase activity exemplified by epidermal growth factor receptor (EGFR) or platelet-derived growth factor receptor (PDGFR). These kinases phosphorylate specific tyrosine residues in the cytoplasmic tail of the receptor, providing docking sites for the SH2 domain of inactive STAT monomers that are recruited to the activated receptors. Phosphorylated STAT dimers translocate to the nucleus and bind to a consensus DNA element upstream of regulated genes (Bromberg 2001; Bromberg and Darnell 2000 ). Genes that are known to be regulated by STAT are involved in many fundamental biological processes, such as proliferation, apoptosis, angiogenesis and immune response (Yu and Jove 2004 ).
The activation duration of individual STATs is temporary and usually lasts from a few minutes to several hours under normal physiological conditions. However, numerous studies have demonstrated constitutive activation of STATs, in particular STAT1, STAT3, and STAT5, in a large number of diverse human tumor cell lines and tumors, including gliomas. Aberrant activation of STATs is a consequence of constitutively activated cytokine receptor, most common by autocrine or paracrine expression of their respective ligands, mutation or overexpression of tyrosine kinase encoding genes or loss of endogenous inhibitors. Aberrations in these pathways, as those caused by the recently identified JAK2V617F mutation and translocations of the JAK2 gene, are underlying causes of leukemias and other myeloproliferative disorders (Jatiani et al. 2010 ).
Significant progress in defining normal physiological functions of individual STAT proteins was derived from Stat knockout mice and/or by tissue-specific deletions. Stat3-deficient mice die during early embryogenesis in contrast to other Stat knockout mice. Conditional Stat3 gene targeting using Cre-loxP system (Takeda et al. 1998 ) allowed for analysis of tissue specific Stat3-deficient mice demonstrating its crucial role in a variety of biological processes such as wound healing, T-cell development, mammary gland development, cell growth, apoptosis or cell motility. All those studies demonstrated that STAT signaling is involved in a broad spectrum of fundamental processes such as embryonic development, organogenesis, innate and adaptive immunity, cell differentiation, growth, and apoptosis (Akira 2000 ). Constitutive activation of the STAT family members such as STAT3 and STAT5, and/or loss of STAT1 signaling, are found in a large group of diverse tumors.
10.2 A Brief Summary of Mechanisms of STAT Activation in Normal and Malignant Signaling
10.2.1 Mechanisms of STAT Activation
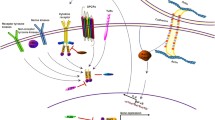
Upon binding of cytokines to cognate surface receptors, receptors dimerize and thereby activate receptor-associated tyrosine kinases, such as JAKs that phosphorylate the receptor cytoplasmic portion. Receptors with intrinsic tyrosine kinase activity, such as PDGFR or EGFR, autophosphorylate the receptor cytoplasmic tail (Reddy et al. 2000 ). Tyrosine-phosphorylated receptors provide docking sites for the recruitment of cytoplasmic monomeric STAT proteins via their SH2 domains. In some cases, non-receptor tyrosine kinases of the Src family can participate in STAT activation. Oncogenic derivatives of cytoplasmic tyrosine kinases such as v-Src or Bcr-Abl can phosphorylate STATs independently of receptor engagement (Danial and Rothman 2000 ). The critical tyrosine (Y) residue in all STATs is required for SH2-phosphotyrosine interaction. Tyrosine phosphorylation regulates the dimerization of STATs which is prerequisite for the establishment of a classical JAK/STAT signaling pathway (Fig. 10.1). Other routes of STAT activation occurring through G-protein signaling and JAK activation have been reported following angiotensin binding to its 7-transmembrane receptor (Marrero et al. 1995; Vila-Coro et al. 1999 ) or RANTES (regulated on activation, normal T cell expressed and secreted) and MIP (macrophage inflammatory protein)-1α binding to shared chemokine receptors (Wong and Fish 1998 ).
A scheme of STAT signaling pathway. Stimulation of cells with growth factors or cytokines results in dimerization of their cognate receptors and activation of intrinsic receptor tyrosine kinase or receptor associated kinases such as JAKs or Src. These kinases phosphorylate specific tyrosine residues of the cytoplasmic tail of the receptor, providing docking sites for the SH2 domain of inactive, cytoplasmic STAT monomers that are recruited to the activated receptors. Oncogenic kinases such as Src or Abl can also phosphorylate STATs independently of receptor engagement. Phosphorylated STAT dimers translocate to the nucleus where through their DNA-binding domain bind with a consensus DNA element upstream of regulated genes. Genes that are known to be regulated by STAT are involved in many fundamental biological processes, such as proliferation, apoptosis, angiogenesis and immune response
Most STATs (except STAT2 and 6) were found phosphorylated also on serine residues in a stimulus-regulated manner. The C-terminal part of STAT1 and STAT3, contains a serine (S) residue 727 phosphorylation site that enhances transcriptional activity (Decker and Kovarik 2000; Wen et al. 1995 ). Studies on murine fibroblasts revealed that candidate serine kinases for the phosphorylation of Stat3 include the various mitogen-activated protein kinase family members (Decker and Kovarik 2000; Lo et al. 2003; Turkson et al. 1999 ). Inhibition of p38 and JNK (c-jun N-terminal kinase) activities suppresses constitutive Stat3 serine phosphorylation and Stat3-mediated gene regulation in Src-transformed fibroblasts (Turkson et al. 1999 ). Recent studies demonstrated that PKCε (protein kinase C), a phosphatidylserine (PS)-dependent serine/threonine kinase, interacts with STAT3, integrates with MAPK (mitogen-activated protein kinase) cascade to phosphorylate Ser727, and increases both DNA-binding and transcriptional activity of STAT3 in skin, prostate cancers, T98G and MO59K glioma cells (Aziz et al. 2010 ).
An active STAT dimer is formed through reciprocal interactions between the SH2 domain of one monomer and the phosphorylated tyrosine of other (Heim et al. 1995 ). Phosphorylated STATs dimerize via reciprocal phosphotyrosine-SH2 domain interactions as homodimers, as seen with all STATs (except STAT2), or as heterodimers, as seen with STAT1/2, STAT1/3, and STAT5a/5b. Within minutes, the dimers translocate to the nucleus, interact with other transcriptional modulators bound to specific promoter sequences and induce gene expression. Dimer-dimer interactions can occur via the NH2-terminal portion of STATs to form tetrameric STAT molecules. Tetramerization of STATs contributes to stabilized DNA-binding activity on weak promoters (John et al. 1999 ). The DNA-binding domain in the center of the molecule determines DNA sequence specificity of individual STATs.
10.2.2 Negative Regulators of STAT Signaling
The activation of individual STAT proteins in normal physiological conditions is tightly controlled and usually lasts from a few minutes to several hours. STAT signaling is assumed to be terminated by dephosphorylation through nuclear tyrosine phosphatases (Lehmann et al. 2003 ) and/or through proteolytic degradation (Haspel and Darnell 1999 ). Because STAT1, STAT3, and STAT5 are often activated by the same ligand and/or intracellular tyrosine kinase, it has been suggested that cytoplasmic and nuclear proteins interact with common and unique elements to modulate STAT-specific responses.
A number of modulators of STAT signaling pathways have been described. The suppressors of cytokine signaling (SOCS) protein family can suppress STAT signaling by binding to and inhibiting JAKs (Croker et al. 2008 ). Eight proteins, cytokine-inducible SH2-domain-containing protein (Fuh et al. 2009 ) and suppressors of cytokine signaling 1–7 (SOCS-1-7), have been identified in the SOCS family so far. Expression of SOCS-1-3 and CIS is induced by cytokine or growth factor stimulation, and some of these proteins are transcriptionally regulated by STATs themselves, which directly antagonizes STAT activation as part of a classic feedback loop. In particular, SOCS1 is strongly involved in the IFNγ signaling and can associate with all known JAKs directly inhibiting their catalytic activity. SOCS3 in contrast do not directly interact with JAK kinases but needs to be recruited to phosphotyrosine residues of activated receptors, in particular gp130, leptin, growth hormone and erythropoietin receptors (Nicholson et al. 2000 ). In addition, all SOCS members are thought to act as E3 ubiquitin ligases and to mediate proteasomal degradation of associated proteins (Yoshimura 2005 ). SOCS3 is frequently silenced by hypermethylation in human cancers (He et al. 2003 ).
The PIAS (protein inhibitors of activated STAT) represent another group of proteins blocking STAT signaling. Unlike SOCS proteins, which are expressed upon cytokine stimulation, PIAS1 is constitutively expressed in a number of cell lines. PIAS bind only to activated STAT dimers and inhibit their DNA binding or their transactivating capacity by multiple molecular mechanisms, including the recruitment of histone deacetylases, the promotion of the sumoylation of STATs, the induction of the dissociation of dimers, or the sequestration of transcription factors to subnuclear structures (Schmidt and Muller 2003; Shuai and Liu 2005 ). Overexpression of PIAS1 and PIAS3, specific nuclear inhibitors of STAT1 and STAT3, respectively, suppressed gene transcription mediated by these STATs (Chung et al. 1997; Liu et al. 1998 ). The constitutive expression of these molecules implies that their physiological function differs from that of SOCS proteins, which are induced in a classical negative feedback loop upon cytokine stimulation (Greenhalgh and Hilton 2001 ).
10.2.3 Transcriptional Targets of STATs
Differences in physiological properties of distinct STATs are largely attributed to their preferential activation by specific ligands, their binding to and activating specific genes. All of the STAT proteins bind a palindromic consensus sequence TTC(N)2–4GAA. The optimal binding sites for STAT1 and STAT3 have the same consensus TTCC(C/G)GGAA (Horvath et al. 1995 ), and the experimentally verified binding sites in genes share a consensus TTC(N)3GAA (Ehret et al. 2001 ). All STAT homodimers (with the exception of STAT2) differentially bind more than ten related IFNγ-activated sequence (Letimier et al.) elements that are characterized by the consensus sequence, TTNCNNNAA. A complex comprised of STAT1, STAT2, and IFN regulatory factor (IRF) nine binds to the IFN-α/β–stimulated response element (ISRE) (AGTTN3TTTC) (O’Shea et al. 2002 ).
For a few genes, including Socs3 regulation by both Stats via the same binding site was demonstrated (Ehret et al. 2001 ). However, the expression of a larger number of genes, including Myc (Ramana et al. 2000; Zhang et al. 2003 ), Bcl2l1 (Catlett-Falcone et al. 1999; Fujio et al. 1997 ), Icam1 and Ccl2 (Fujio et al. 1997; Naik et al. 1997; Valente et al. 1998 ) was differentially affected by treatments that activate specific Stat. Binding sites specific for a particular Stat or a subset of them have been experimentally demonstrated. For example, the IFNγ-activated sequences(Letimier et al. 2007 ) of mouse Ly6E gene (Khan et al. 1993 ) was shown to bind preferentially to Stat1 homodimer, but not Stat3 homodimer or Stat1-Stat3 heterodimer (Horvath et al. 1995 ). The SIE (sis-inducible element) in the human FOS (Wagner et al. 1990 ) was shown to bind STAT1 and STAT3 (Horvath et al. 1995 ), but not STAT4 or STAT5 (Leong et al. 2003 ).
Studies on STAT null mice and transcriptional profiling has helped to clarify genes whose expression is controlled by STATs, and provided evidence that these transcription factors are clearly important for regulating a wide array of genes (Chen et al. 2003; Hoey et al. 2003; Lund et al. 2004 ). Immune, growth and apoptosis regulatory activities associated with STAT1-dependent gene transcription are mostly associated with the transcriptional regulation of IRF-1, MHC, FcγRI, Fas and FasL, TRAIL, cyclin-dependent kinase inhibitors, p21waf1 and caspase genes (Battle and Frank 2002; Ivashkiv and Hu 2004 ). Many studies have shown that the anti-apoptotic gene encoding Bcl-xL protein is a downstream target of Stat3 (Bromberg et al. 1999 ) and Stat5 (Gesbert and Griffin 2000; Socolovsky et al. 1999 ). The cell cycle control gene c-Myc has been shown to be induced in response to Stat3 signaling in v-Src-transformed NIH3T3 fibroblasts as well as through Stat5 activation (Bowman et al. 2001; Lord et al. 2000 ).
Recent results challenge a simple view that distinct STATs regulate discrete and non-overlapping sets of genes. In non-stimulated rat C6 glioma cells Stat1 and Stat3 proteins are expressed and phosphorylated to some extent (Fig. 10.2). Both proteins are strongly phosphorylated after a treatment with either IFNγ or IL-6. The IFN-α/β–stimulated response element (ISRE)-driven transcription was inhibited by the ISRE oligodeoxynucleotide decoys (ODNs) (Fig. 10.2). Furthermore, our studies employing ODNs and siRNA interfering with either Stat1 or Stat3 activity/expression suggest that Stat1 supports the basal expression of Stat3 target genes Bcl2l1 and possibly c-Myc in proliferating C6 cells (Adach-Kilon et al. 2011 ). Also, studies on STAT3−/− cells showed that the immediate early genes Fos and Egr1 become STAT1 transcriptional targets in the absence of STAT3 (Schiavone et al. 2011 ). It suggests that the abrogation of STAT3 expression directs the STAT1 to transcribe new target genes, known to drive mitogen responses and tumor transformation.
STAT signaling in rat C6 glioma cells. (a). The level of total and phosphorylated Stat1 and Stat3 in rat C6 glioma cells untreated or exposed to interferon (IFN)-γ or IL-6 30 min after stimulation. Immunoblots were re-probed with an antibody recognizing β-actin to ensure equal loading. (b). A scheme of the experiment with ODN decoys. (c). Representative pictures show cells 4 h after transfection with the FITC-labelled ISRE decoys (13.4 nM) visualized by fluorescence microscopy. (d). The luciferase activity was measured in untreated cells or 24 h after co-transfection pISRE-luc with the indicated decoy at various concentrations. The bars indicate mean values of luciferase activity in a representative experiment (in triplicate). Arrows mark the ODN concentration (13.4 nM) selected for experiments. ISRE, IFN-α/β-stimulated response element
Until recently, a technology did not permit the discrimination of genes that are regulated directly compared to indirect regulation. Recent analyses employing DNA chromatin immunoprecipitation followed by microarray technique (Chip-chip) brought new insight into the accurate determination of Stat1-DNA binding sites. Those studies revealed that Stat1 binds to many sites on chromosome 22 and uncovered many new candidate target genes, not associated previously with IFN-responsive genes which are induced only in certain cell types and under specific conditions (Hartman et al. 2005; Wormald et al. 2006 ). Chromatin immunoprecipitation studies demonstrated that in the case of STAT4, known targets include Ifng, Il18r1, Il12rb2, Il2ra and Furin in Th1 lymphocytes (Letimier et al. 2007; O’Sullivan et al. 2004; Pesu et al. 2006; Thieu et al. 2008 ), whereas STAT6-bound genes include Il4, Gata3, and Ccl17 (Kubo et al. 1997; Wirnsberger et al. 2006 ).
Interactions with basic transcription co-regulators may modulate the functional specificity of individual STAT. Induction of a subset of IL-6-responsive genes was found to be dependent on Brg1 (Brahma-related gene 1). BRG1 protein is the ATPase subunits of the SWI/SNF complexes, chromatin remodeling proteins involved in altering local chromatin structure and facilitation of recruitment of essential transcription factors. BRG1 was required for STAT3 recruitment to the IFN regulatory factor (IRF) 1 promoter, downstream histone modifications, and IL-6-induced chromatin remodeling. Therefore, it has been suggested that BRG1 plays a role in mediating STAT accessibility at multiple cytokine-responsive promoters and may influence access of different STAT proteins to the same target (Ni and Bremner 2007 ). Interestingly, while both hBrm (human Brahma) and BRG1 interact with STAT1 in vitro, under normal conditions only hBrm is recruited by STAT1 to IFNγ-activated sequences of individual genes; while phosphorylated STAT1 mainly binds to BRG1 under stress conditions. Under basal conditions, hBrm exists in a mSin3/HDAC co-repressor complex associated with a compact chromatin structure. Upon heat-shock, the phosphorylated STAT1 binds and recruits BRG1 to the GAS, leading to induction of gene expression. This hBrm/BRG1 switch occurs at the GAS in specific cell types upon exposure to IFNγ (Zhang et al. 2010 ).
Furthermore, a wide variety of factors interacting with STATs exemplified by NF-κB, SMADs, c-Jun (Baker et al. 2008 ), Sp1 (Look et al. 1995 ), BRCA1 (Ouchi et al. 2000 ), etc. may antagonistically or synergistically influence the functional specificity of individual STAT.
10.3 Dysfunction of STAT Signaling in Gliomas
10.3.1 Constitutive Activation of STAT3 in Gliomas
Clinical and experimental studies show various expressions of STATs in gliomas. Immunohistological studies showed STAT1 expression in a majority of glioblastomas, but in tumor tissue the signal was mostly localized in the cytoplasm suggesting the predominant presence of an inactive form of STAT1. Within the infiltration area strong STAT1 expression was found in reactive astrocytes and in microglial components (Haybaeck et al. 2007 ). STAT3 was shown to be constitutively active, as assessed by tyrosine phosphorylation status in malignant gliomas tumors and cell lines when compared with normal human astrocytes, white matter, and normal tissue adjacent to tumor (Mizoguchi et al. 2006; Rahaman et al. 2002; Schaefer et al. 2002 ). Experimental mouse gliomas express constitutively activated STAT3 (Weissenberger et al. 2004 ). Many studies reported constitutive STAT3 activation, corresponding to high expression of STAT3 target genes such as Bcl2l1, Bcl-2 or Mcl-1 in glioblastoma cell lines (Iwamaru et al. 2007; Rahaman et al. 2002 ).
There is a controversy concerning potential correlation between activation STAT3 and histological glioma grade. While a nuclear staining for pSTAT3 reflecting its activation was detected by immunohistochemistry in 259 glioma samples of different grades, the positive rate was <9 % in high grade gliomas (Wang et al. 2004 ). On the other hand, immunohistochemical analysis of 55 glioma samples of different grades showed that STAT3 was constitutively activated in 28 % of low (I-II) and 60 % in high (III-IV) grade gliomas (Lo et al. 2008 ). Activation of STAT3 was essentially identical in 82 malignant astrocytic gliomas (55 glioblastomas and 27 anaplastic astrocytomas) (Mizoguchi et al. 2006 ). Measurements of the amounts of pSTAT3 in 15 human gliomas, five diffuse astrocytomas (WHO grade II), five anaplastic astrocytomas (WHO grade III), five glioblastoma multiforme (GBMs) (WHO grade IV) and in normal human brain by Western blot analysis showed the level of pSTAT3 increased with malignancy being highest in GBM (Weissenberger et al. 2004 ). Interestingly, in low-grade human gliomas pSTAT3 was located in the nucleus, while in GBM activated STAT3 was localized to the nucleus and to plasma membrane (Weissenberger et al. 2004 ).
Recent investigation of prognostic relevance of activated STAT3 in a larger collection of patients with glioblastoma provided evidence that activation of STAT3 is linked with clinically more aggressive behavior. Glioblastoma patients with high or very high numbers of pSTAT3-positive tumor cells had significantly shorter overall survival than those with no or low numbers (Birner et al. 2010 ). The proportion of grade III glioma cases with high or very high numbers of pSTAT3-positive tumor cells was similar to that in grade IV glioblastoma suggesting lack of association of STAT3 activation with tumor grade III glioma progression (Birner et al. 2010 ).
Immunohistochemical staining revealed markedly increased expression of STAT5b in GBM (57.1 %) compared with that in normal cortex (22.2 %) and diffuse astrocytoma (27.3 %) (Liang et al. 2009 ). Phosphorylated STAT5 was detected in primary gliomas, predominantly in the nucleus (Cao et al. 2010 ).
Studies using multiparameter flow cytometric cell sorting of ex vivo tumor specimens demonstrate the up-regulation of STAT3 and STAT5 in microglia sorted from GBM tumor specimens but not from meningiomas (Kostianovsky et al. 2008 ). Up-regulation of STATs in immune cells infiltrating gliomas may be involved in a complex network of inhibitory pathways, responsible for the GBM-mediated suppression of monocyte/microglial function (Hussain et al. 2007 ).
10.3.2 STAT Activation in Gliomas Results from Dysfunction in Control Mechanisms
While mutations of cytokine receptors and JAKs are rare, gain-of-function mutations of growth factor receptor kinases are quite common in human cancers including gliomas. The gene encoding EGFR and its constitutively activated variant, EGFRvIII, are often amplified and overexpressed in human adult gliomas. EGFRvIII is a product of rearrangement with an in-frame deletion of 801 bp of the coding sequence of the extracellular domain, resulting in a deletion of residues 6–273 and a glycine insertion as residue 6. EGFRvIII deletion results in a ligand-independent, constitutively active, and cell surface–retained receptor. Both EGFR and EGFRvIII are tumorigenic for gliomas and major targets for glioma therapy (Friedman and Bigner 2005; Nishikawa et al. 1994 ). Blocking of EGFR, but not PDGFR or SRC kinase activity, by pharmacological compounds markedly reduced the constitutive activation of STAT3 in U251 glioma cells suggesting EGFR activation contributes to the constitutive activation of STAT3 in those cells (Rahaman et al. 2002 ). Several studies demonstrated that three STAT3-activating kinases, JAK2, EGFR, and EGFRvIII, contribute to STAT3 activation. Constitutive STAT3 activation coexisted with EGFR expression in 27.2 % of primary high-grade gliomas. Combination of an anti-EGFR agent – Iressa and a JAK2/STAT3 inhibitor – JSI-124, synergistically suppressed STAT3 activation and potently killed glioblastoma cell lines expressing EGFR or EGFRvIII (Lo et al. 2008 ).
In addition to the tyrosine kinase EGFR, other tyrosine kinases known to activate STAT3 may be important in activating signaling pathways relevant to gliomagenesis. These proteins include SRC (Yu et al. 1995 ) and the endothelial receptor vascular endothelial growth factor receptor-2 (VEGFR2) (Korpelainen et al. 1999 ). Unlike STAT activation by PDGFR and EGFR, which activate both STAT1 and STAT3, VEGFR2 did not activate STAT1.
GBM samples contain significantly higher levels of IL6 protein compared to those of control brains (Weissenberger et al. 2004 ). In U251glioblastoma cells STAT3 activation was in part caused by autocrine IL-6 , as neutralizing IL-6 antibodies reduced STAT3 activation by 70 % (Rahaman et al. 2002 ). A recent study demonstrates expression of the interleukin 6 receptor α (IL6Rα) and glycoprotein 130 (gp130) in glioma stem cells (GSCs). Targeting IL6Rα or IL6 ligand expression in those cells with short hairpin RNAs (shRNAs) attenuated STAT3 activation indicating that STAT3 is a downstream mediator of pro-survival IL6 signals in GSCs (Wang et al. 2009 ).
Altered expression of negative regulators of STAT may contribute to their constitutive activation. SOCS1 and SOCS3 are aberrantly expressed in GBM cell lines and primary tissues. The promoter of SOCS1-2-3 was methylated in 24, 6.5 and 35 % of GBM, respectively, that resulted in reduced SOCSs expression (SOCS1-2-3 mRNA was reduced by 5, 3 and 7-folds, respectively) when compared with unmethylated GBM. Hypermethylation of SOCS3 promoter was significantly associated with an unfavorable clinical outcome (Martini et al. 2008 ). The other study demonstrated that 10 tested GBM cell lines lacked SOCS1 expression, whereas GBM cell lines and primary GBM tumor samples constitutively expressed SOCS3. SOCS1 gene repression was linked to hypermethylation of the SOCS1 genetic locus in GBM cells. Reintroduction of SOCS1 or blocking SOCS3 expression sensitized cells to radiation and decreased the levels of activated ERK in GBM cells (Zhou et al. 2007 ). Interestingly, assessment of the relationship between SOCS3 and EGFR aberrations revealed that SOCS3 promoter hypermethylation was inversely related to both the EGFR gene dosage as well as the EGFR protein expression (Lindemann et al. 2011 ). About 89 % of glioblastoma samples were found to be PIAS3 negative and pSTAT3 positive. The ectopic expression of PIAS3 in a glioblastoma cell line caused the inhibition of the transcriptional activity of STAT3 (Brantley et al. 2008 ).
10.4 Functions of STAT3 in Gliomas
10.4.1 STAT3 as Oncogene
There is an accepted concept that STAT1 and STAT3, despite their similar structures, have antagonistic effects on cellular proliferation and apoptosis, with STAT3 acting like an oncogene and STAT1 playing a role of tumor suppressor (Battle and Frank 2002; Bromberg and Darnell 2000; Stephanou and Latchman 2005 ). A first evidence for a potential oncogenic function of STAT3 came from findings showing its constitutive activation in Src-transformed cell lines (Bromberg et al. 1998; Turkson et al. 1998 ). Further studies showed that STAT3C, a constitutively active mutant of STAT3, can transform cultured fibroblasts which form tumors when injected into mice (Bromberg et al. 1999 ). Therefore, STAT3 is considered to be one of the major mediators of tumorigenesis and numerous studies show how interfering with STAT3 signaling affects growth, survival and tumorigenicity of many tumors, including gliomas.
U87-derived cell lines stably expressing a dominant negative mutant DN-STAT3 in hypoxia inducible manner failed to grow in mice due to impaired cell proliferation, survival and reduced angiogenesis. Mice implanted with DN-STAT3 expressing clones survived significantly longer than control mice (Dasgupta et al. 2009 ). Knockdown of STAT3 expression by RNAi suppressed growth, induced apoptosis and differentiation in glioblastoma stem cells (Li et al. 2010 ). Effects of STAT3 on cell cycle and proliferation were mediated through its ability to regulate the expression of Cyclin D1 and c-Myc. Double-stranded decoy oligodeoxynucleotides which correspond closely to the STAT3 response element within the c-Fos promoter blocked STAT3 signaling and subsequently inhibited cell proliferation by inducing apoptosis and cell-cycle arrest in two glioma cell lines U251 and A172 (Gu et al. 2005; Iwamaru et al. 2007; Lo et al. 2008 ). On the other hand, we found no reduction in viability of C6 glioma cells after a treatment with the STAT3 decoys or STAT3 silencing by siRNA (Adach-Kilon et al. 2011 ).
Suppression of apoptosis by STAT3 is mediated through expression of various survival genes that are regulated by STAT3 such as Bcl2l1, Bcl-2, survivin, Mcl-1. Pharmacological inhibition of STAT3 activation in glioma cells leads to down-regulation of survival-related genes and apoptosis (Iwamaru et al. 2007; Konnikova et al. 2003; Rahaman et al. 2002 ).
STAT3 plays also an important role in regulating tumor cell invasion. STAT3 is known to directly up-regulate the expression of matrix metalloproteinases: MMP-2 and MMP-9, involved in the degradation of extracellular matrix. Inhibition of STAT3 activity impaired migratory and invasive potential of numerous glioma cell lines and decreased MMP-2 and MMP-9 transcription, and their proteolytic activity (Chen et al. 2010; Senft et al. 2010 ).
STAT3 was shown to be a direct transcriptional activator of the VEGF gene which is the most potent angiogenesis inducing signal. In gliomas VEGF is expressed in cells with activated STAT3 (Lo et al. 2008 ) and activated STAT3 dramatically increases the transcription from the VEGF gene promoter (Schaefer et al. 2002 ). As enhanced expression of VEGFR leads to activation of STAT3, it suggests its participation in a VEGF/VEGFR autocrine loop facilitating angiogenesis in malignant gliomas (Schaefer et al. 2002 ).
Infiltration of immune cells into tumors, their switch to the alternative, pro-tumorigenic phenotype and formation of an inflammatory microenvironment support glioma progression (Gabrusiewicz et al. 2011 ). Cancer-associated inflammation is marked by the presence of specific inflammatory mediators, including numerous cytokines and chemokines. Recent evidence suggest a crucial role of STATs, in particular STAT3, in tumor-induced immunosuppression. Interference with STAT3 expression/activity in tumor-infiltrating immune cells reduced tumor progression in animal models (Yu et al. 2007, 2009 ). STAT3 signaling in innate immune cells is required for the immunsuppressive and tumor promoting effects of myeloid-derived suppressor cells and tumor-associated macrophages in several experimental models. STAT3 also modulates expansion of T regulatory cells in tumors and is necessary for the development of TH17 T cells (Yu et al. 2009 ). Inhibition of STAT3 activity by JSI-124 (a STAT3 inhibitor) promoted maturation of tumor infiltrating CD11c + dendritic cells and activation of tumor-conditioned cytotoxic T cells. Brain infiltrating lymphocytes isolated from JSI-124-treated mice exhibited enhanced expression of surface maturation markers such as MHC class II, CD40, CD80, CD86 on tumor infiltrating CD11c + dendritic cells. Moreover, in vivo JSI-124 treatment reduced CD11b+/Gr1+ myeloid suppressor cells and CD4+/CD25+ regulatory T cells in a murine GL261 intracranial glioma model (Fujita et al. 2008 ). These data further support the hypothesis that systemic inhibition of STAT3 signaling can reverse the immunosuppressive environment in gliomas.
Furthermore, ex vivo STAT3 inhibition has been reported to activate T cell, monocytes and microglia isolated from glioma patients. Glioma-infiltrating microglia/macrophages expressed MHCII but lacked expression of the co-stimulatory molecules CD80, CD86 and CD40 critical for T-cell activation and were unable to stimulate T lymphocytes. Inhibition of STAT3 by WP1066 (a JAK inhibitor) resulted in up-regulation of CD80 and CD86 on both normal donor PBMCs and on tumor infiltrating microglia/macrophages isolated from GBM patients. Moreover, the treatment stimulated the production of immune-stimulatory cytokines IL2, IL4, IL12, IL15 and induced proliferation of effector T cells (Hussain et al. 2007 ). Altogether, STAT3 targeting in tumor microenvironment seems to be an effective strategy to overcome glioma-induced immunosuppression and to induce anti-tumor immunity in gliomas.
10.4.2 STAT3 as a Tumor Suppressor in Gliomas
Although most studies have highlighted the oncogenic function of STAT3 in non-brain tumors, recent data suggest that STAT3 may play both tumor suppressive and oncogenic function in glioma pathogenesis, depending on the genetic background of the tumor (de la Iglesia et al. 2008a, b, 2009 ). STAT3 has been shown to promote cell differentiation along the astrocytic lineage (Bonni et al. 1997; Rajan and McKay 1998 ). Stat3 −/− mouse astrocytes displayed increased proliferation and invasiveness as compared to control astrocytes, indicating that STAT3 inhibits astrocyte proliferation and invasiveness. Although loss of Stat3 gene was not sufficient to transform astrocytes, combined with knockdown of the tumor suppressor PTEN led to malignant transformation (de la Iglesia et al. 2008b ). Deficiency of PTEN triggered the cascade of events that inhibits STAT3 signaling in murine astrocytes and human glioblastoma tumors (de la Iglesia et al. 2008b ). Reactivation of STAT3 in PTEN-deficient but not in PTEN-expressing glioblastoma cells inhibited their proliferation, invasiveness and spreading on myelin (de la Iglesia et al. 2008a, b ). In a panel of human brain tumors, PTEN loss correlates tightly with down-regulation of LIFRβ and low levels of phosphorylated STAT3. These findings provide correlative evidence that PTEN loss and inhibition of LIFRβ-STAT3 signaling are linked in gliomas (de la Iglesia et al. 2008b ).
We have performed a global analysis of phospho-STAT3 binding sites in rat C6 glioma cells by chIP-on chip and crossed putative STAT3 target genes with the profiles of gene expression in control and JAK/STAT3 inhibitor treated cells. The distribution of genes containing peaks for phospho-STAT3, which demonstrates STAT3 binding to promoters, correlated (p < 10−10 in the Kolmogorov-Smirnov test) with changes in gene expression induced by inhibition of STAT3 phosphorylation (mostly with expression increases). It suggests that STAT3 binding contributes mostly to negative regulation of target gene expression in proliferating C6 glioma cells (unpublished).
In contrast to STAT3 acting as a tumor suppressor in PTEN deficient cells, STAT3 acts as an oncogene in EGFRvIII-expressing cells. STAT3 was required for the malignant transformation of astrocytes that are both PTEN-deficient and express EGFRvIII, and physically associates with EGFRvIII within the nucleus. It has been suggested that nuclear EGFRvIII acts as a switch to convert STAT3 from the tumor suppressive to pro-oncogenic protein (de la Iglesia et al. 2008a ). Thus, the role of STAT3 as a tumor suppressive and oncogenic protein depends on the genetic background of the tumor.
10.4.3 STAT3 in Glioma Cancer Initiating Cells
A subpopulation of cells with stem-like features, glioma-initiating cells or glioma stem cells were identified in GBM (Singh et al. 2003, 2004; Yuan et al. 2004 ). Those cells are characterized by their ability to undergo self-renewal and to differentiate into neuronal, astroglial and oligodendroglial cells, are highly tumorigenic (Galli et al. 2004; Vescovi et al. 2006 ). STAT3 mediates self-renewal of pluripotent embryonic stem cells (Niwa et al. 1998; Raz et al. 1999 ) and is implicated in neurogenesis and gliogenesis in neural stem cells (Gu et al. 2005 ). Isolated glioma-initiating cells grow as anchorage-independent spheres and express STAT3 phosphorylated on activating tyrosine and serine residues. Inhibition of STAT3 signaling in those cells with either small molecule inhibitors or RNAi inhibited cell growth and spheres formation (Sherry et al. 2009; Villalva et al. 2010 ), and sensitized cells to an anti-tumor drug – Temozolomide (Villalva et al. 2010 ). STAT3 has emerged as the important regulator of immunosuppressive pathway in glioma initiating cells (Hatiboglu et al. 2010 ). The inhibition of STAT3 did not alter the immunological phenotype of those cells, but reduced Treg induction, restored T-cell function and induced T-cell proliferation (Wei et al. 2010 ).
10.5 Molecular and Pharmacological Strategies to Interfere with STAT Signaling for Potential Therapeutic Intervention in Gliomas
A number of studies have focused on development of STAT3 inhibitors due to their expected therapeutic potential in cancer therapy. Therapeutic strategies for blocking STAT3 activity are based either on direct targeting of STAT3 protein or indirect targeting of the upstream components of the STAT3 signaling pathway (Table 10.1). Due to difficulties in designing specific inhibitors of particular kinases, problems with drug specificities and side effects, recent investigator’s efforts focused on developing direct STAT3 inhibitors. These include dominant negative STAT3 expression vectors, oligonucleotides decoys, small interfering RNA, peptides, peptidomimetics and small molecules inhibitors. Mechanism of action of these compounds is usually based on disruption of STAT3 dimerization or STAT3 DNA binding activity (Turkson 2004 ).
Recently several direct STAT3 inhibitors have been successfully applied in glioblastoma. Two non-peptide, cell-permeable, small molecules, termed LLL3 and LLL12 were developed using structure-based drug design (Fuh et al. 2009; Lin et al. 2010 ). Computer modeling with docking simulation showed that these compounds bind directly to the phosphoryl tyrosine 705 binding sites of STAT3 monomer. Both compounds inhibit STAT3 phosphorylation and downstream STAT3 target genes that results in reduced cell viability and induction of apoptosis (Fuh et al. 2009; Lin et al. 2010 ). LLL12 is a more potent inhibitor of cell viability than previously described JAK2 inhibitor WP1066 (Iwamaru et al. 2007 ), with half maximal inhibitory concentration values 0.21 and 0.86 μM for U87-MG and U373-MG, respectively (Lin et al. 2010 ). Furthermore, both inhibitors demonstrated very potent activity in vivo. The U87 glioblastoma tumor-bearing mice treated with LLL3 or LLL12 had smaller intracranial tumors and LLL3 treated mice exhibited prolonged survival relative to vehicle treated mice (28.5 vs 16 days) (Fuh et al. 2009 ).
A dimerization-disrupting phosphopeptide sequence derived from the SH2 domain-binding region of STAT3, PY*LKTK (where Y* represents phosphotyrosine) and its tripeptide derivatives PY*L and AY*L, were developed as inhibitors of STAT3 activation and biological function. Specific peptidomimetics selectively disrupt STAT3 DNA-binding activity in vitro and interfere with STAT3 DNA binding and a reporter gene activity in Src-transformed fibroblasts (Turkson et al. 2001, 2004 ).
Dimerization of STAT3 proteins can be blocked by peptide aptamers, short, usually 12 to 20 amino acids in length, peptides that specifically bind to a target protein. Peptide aptamers specifically interacting with the STAT3 dimerization domain were selected from a peptide library by an adaptation of the yeast two-hybrid procedure (Nagel-Wolfrum et al. 2004 ). Purified recombinant peptide aptamer, tagged with a protein transduction motif of nine arginine and fused with thioredoxin as a scaffold protein, was cell-permeable and selectively induced growth inhibition and apoptosis of glioblastoma cells (Borghouts et al. 2008 ). Currently, peptides aptamers represent one of the most effective approaches to disrupt STAT3 function in vitro with half maximal inhibitory concentration values for glioblastoma cells <1 μM. No prior knowledge of the structure of the target protein is required and their binding ability is not limited to preexisting small molecular weight compound binding pocket (Borghouts et al. 2008 ). However, if the peptide aptamers are considered for in vivo usage, limitations such as stability and permeability of the fusion protein must be optimized.
Another approach developed for direct STAT3 targeting are oligodeoxynucleotides decoys, a short, synthetic DNA carrying the cognate DNA-binding sites of transcription factor (Yu and Jove 2004 ). STAT3 decoy-ODN inhibited the binding of STAT3 to DNA and altered the downstream gene expression in U251 and A172 glioma cell lines (Gu et al. 2008 ). Intratumorally administrated STAT3 decoy-ODN significantly suppressed the growth of glioma by inhibiting proliferation and promoting apoptosis in xenografts (Shen et al. 2009 ). Our studies on different tumor cells, including rat C6 and human T98 glioma cells demonstrated lack of significant STAT3 decoy-ODN effects on the expression of endogenous, STAT dependent genes and cell survival (Adach-Kilon et al. 2011 and unpublished). Although such nucleic acids based strategy works well in cell culture and in animal model, it might be limited in terms of their clinical development as therapeutic agents.
Indirect inhibition of STAT3 can be achieved by disruption of the ligand-receptor interaction at the extracellular surface or by blocking upstream tyrosine kinases that are responsible for its activation. A plethora of small molecule inhibitors of JAK, Src or EGFR has been shown to effectively block STAT3 signaling, inhibit glioma proliferation and induce apoptosis (Heimberger and Priebe 2008 ). These STAT3 inhibitors display marked efficacy in murine glioma models, including intracerebral tumors. The mechanism of this in vivo efficacy of the STAT3 blockade agents is a combination of direct tumor cytotoxicity and immune cytotoxic clearance (Hussain et al. 2007; Iwamaru et al. 2007 ).
One of the first reports showing that STAT3 is a valid target for therapy of glioblastoma came from a study with AG490, the JAK1/2 inhibitor. A treatment with AG490 reduced a constitutive STAT3 activation, inhibited proliferation of cultured U251 GBM cells and induced apoptosis by reducing the steady-state level of Bcl-xL, Bcl-2 and Mcl-1 anti-apoptotic proteins (Rahaman et al. 2002 ). Inhibition of the JAK/STAT3 signaling pathway impedes the migratory and invasive potential of human glioblastoma cells. Treatment with AG490 reduced migratory and invasive potential of five different glioblastoma cell lines that was paralleled by a decrease in transcription of MMP-2 and MMP-9, and their reduced proteolytic activity (Senft et al. 2010 ).
WP1066 was designed by modifying the structure of AG490 and inhibited STAT3 activation by blocking JAK2. It showed selective cytotoxicity toward cultured malignant glioma U87-MG and U373-MG cells in lower doses than AG490 and significantly inhibited the growth of subcutaneous tumors generated from U87-MG in mice (Iwamaru et al. 2007 ). Although the subcutaneous glioma model was used, the concentration of the compound in the brain was 10 times higher than in the plasma after intraperitoneal treatment, indicating a good penetrance of the blood–brain barrier. Furthermore, WP1066 reversed immune tolerance in immune cells isolated from GBM patients (Hussain et al. 2007 ).
Interestingly, the traditional herbal medicine cucrubitacin I (JSI-124) is a selective inhibitor of JAK kinase and STAT3 signaling. JSI-124 induced G2/M arrest and apoptosis of GBM cell lines via down-regulation of Cyclin B1 and Cdc2 expression (Lo et al. 2008; Su et al. 2008 ). Combination of JSI-124 and dasatinib (Src family kinase inhibitor) synergistically decreased cell proliferation and viability, and had a significant effect on cell migration exceeding those observed with either drug alone (Premkumar et al. 2010 ). Another naturally occurring, dietary compound, displaying the inhibitory potential toward JAK signaling is curcumin. Curcumin was shown to suppress malignant glioma growth in vitro with induction of G2/M arrest and attenuation of migratory and invasive behavior. In vivo, curcumin reduced growth of intracranial gliomas and proliferation of tumors cells but failed to increase survival of glioma-bearing mice (Weissenberger et al. 2010 ).
The multikinase inhibitors such as sorafenib, sunitib or vandetanib that are already in clinical trials were shown to mediate the antitumor effects by reducing STAT3 activity. ZD6474 (ZACTIMA, vandetanib) is an inhibitor of various receptor tyrosine kinases, in particular VEGFR2 and EGFR. ZD6474 inhibited growth and survival through attenuation of STAT3 phosphorylation, Akt, and Bcl-xL expression in glioma cells expressing EGFRvIII but not in cells with non-detectable EGFRvIII (Yiin et al. 2010 ). Vandetanib was used in the phase I of clinical trial in children with newly diagnosed diffuse intrinsic pontine glioma (Broniscer et al. 2010 ). Sorafenib (BAY43-9006, Nexavar) is an oral multikinase inhibitor originally developed to Raf and receptor tyrosine kinase signaling. Sorafenib (≤10 μmol/L) inhibited cell proliferation and induced apoptosis of U87 and U251 glioma cell lines, and two primary cultures (PBT015 and PBT022) from human glioblastomas. The effects of sorafenib were associated with inhibition of STAT3 phosphorylation. Overexpression of a constitutively activated STAT3 partially blocked the effects of sorafenib. The level of phosphorylated JAK1 was reduced in U87 and U251 cells, whereas phosphorylated JAK2 was down-regulated in primary cultures (Yang et al. 2010 ). Glioma pathogenesis involves abnormalities in many cellular pathways, thus molecularly targeted therapies with multikinase inhibitors may provide clinical benefits in the treatment of glioblastomas.
Currently, the major approaches to treating glioma are surgical resection, radiotherapy, and adjuvant chemotherapy. The combination of oral cytotoxic chemotherapy with concomitant radiotherapy has been shown to improve survival of patients with glioblastoma (Stupp et al. 2005 ). However, GBM cells often develop resistance to ionizing radiation (IR) and chemotherapeutics, rendering therapy ineffective. Therefore, increasing cell sensitivity to radiation and chemotherapy could significantly increase therapeutic outcome. Attempts to sensitize GBM cells to radiation have focused on the use of broadly acting inhibitors of kinases known to be mutated or amplified in gliomas. In particular, targeting EGFR has been shown to sensitize GBM cells to IR (Stea et al. 2003; Zhou et al. 2007 ). Expression of DN-STAT3 sensitized U87 cells to the cytotoxic effects of IR (Zhou et al. 2007 ). Moreover, suppression of STAT3 with siRNA enhanced radiation-induced growth inhibition in a U251 glioma model. Simultaneous inhibition of STAT3 and ErbB2 combined with radiotherapy led to the most significant reduction of tumor growth (Gao et al. 2010 ). Inhibition of STAT3 with JSI-124 sensitized malignant glioma cells to TMZ (temozolomide), the most commonly used agent in the therapy of GBM and alykalating agents (cisplatin or 1,3 bis(2chlorylethyl)-1-nitrosourea) (Lo et al. 2008 ). When STAT3 expression/activity was blocked by either shRNA or Sttatic (a direct STAT3 inhibitor) in glioma initiating stem cells, the treatment with IC20 concentration of TMZ decreased proliferation rate by 50 % in comparison to untreated cells (Villalva et al. 2010 ).
While an array of STAT3 inhibitors that induce antitumor effects cultured cells and animal models have been identified, most of the STAT3 inhibitors reported to date have not undergone an in vivo efficacy, pharmacology or toxicity testing (Yue and Turkson 2009 ). Overall, there is a need for re-examination of the ongoing strategies to target STAT3 intended not only for refinement, but also to incorporate newest technologies to transform current compounds into clinically useful anticancer therapeutics.
Abbreviations
- Bcl-2:
-
B-cell lymphoma 2
- Bcl-xL :
-
B-cell lymphoma-extra large
- BRG1:
-
Brahma-related gene 1
- EGFR:
-
Epidermal growth factor receptor
- GAS:
-
IFNg-activated sequence
- GBM:
-
Glioblastoma multiforme
- GSC:
-
Glioma stem cells
- IFNg :
-
Interferon g
- IRF:
-
IFN regulatory factor
- ISRE:
-
IFN-a/b–stimulated response element
- JAK:
-
Janus kinase
- Mcl-1:
-
Induced myeloid leukemia cell differentiation protein
- MMP:
-
Metalloproteinase
- ODN:
-
Oligodeoxynucleotide
- PDGFR:
-
Platelet-derived growth factor receptor
- PIAS:
-
Protein inhibitors of activated STAT
- PTEN:
-
Phosphatase and tensin homolog
- SH2:
-
Src homology domain 2
- SOCS:
-
Suppressors of cytokine signaling
- STAT:
-
Signal transducers and activators of transcription
- TMZ:
-
Temozolomide
- VEGF:
-
Vascular endothelial growth factor
- VEGFR:
-
Vascular endothelial growth factor receptor
References
Adach-Kilon A, Swiatek-Machado K, Kaminska B, Dabrowski M (2011) Signal transducer and activator of transcription 1 (Stat1) maintains basal mRNA expression of pro-survival Stat3-target genes in glioma C6 cells. J Cell Biochem 112:3685–3694
Akira S (2000) Roles of STAT3 defined by tissue-specific gene targeting. Oncogene 19:2607–2611
Aziz MH, Hafeez BB, Sand JM, Pierce DB, Aziz SW, Dreckschmidt NE, Verma AK (2010) Protein kinase Cvarepsilon mediates Stat3Ser727 phosphorylation, Stat3-regulated gene expression, and cell invasion in various human cancer cell lines through integration with MAPK cascade (RAF-1, MEK1/2, and ERK1/2). Oncogene 29:3100–3109
Baker BJ, Qin H, Benveniste EN (2008) Molecular basis of oncostatin M-induced SOCS-3 expression in astrocytes. Glia 56:1250–1262
Battle TE, Frank DA (2002) The role of STATs in apoptosis. Curr Mol Med 2:381–392
Birner P, Toumangelova-Uzeir K, Natchev S, Guentchev M (2010) STAT3 tyrosine phosphorylation influences survival in glioblastoma. J Neurooncol 100:339–343
Bonni A, Sun Y, Nadal-Vicens M, Bhatt A, Frank DA, Rozovsky I, Stahl N, Yancopoulos GD, Greenberg ME (1997) Regulation of gliogenesis in the central nervous system by the JAK-STAT signaling pathway. Science 278:477–483
Borghouts C, Kunz C, Delis N, Groner B (2008) Monomeric recombinant peptide aptamers are required for efficient intracellular uptake and target inhibition. Mol Cancer Res 6:267–281
Bowman T, Broome MA, Sinibaldi D, Wharton W, Pledger WJ, Sedivy JM, Irby R, Yeatman T, Courtneidge SA, Jove R (2001) Stat3-mediated Myc expression is required for Src transformation and PDGF-induced mitogenesis. Proc Natl Acad Sci U S A 98:7319–7324
Brantley EC, Nabors LB, Gillespie GY, Choi YH, Palmer CA, Harrison K, Roarty K, Benveniste EN (2008) Loss of protein inhibitors of activated STAT-3 expression in glioblastoma multiforme tumors: implications for STAT-3 activation and gene expression. Clin Cancer Res 14:4694–4704
Bromberg JF (2001) Activation of STAT proteins and growth control. Bioessays 23:161–169
Bromberg J, Darnell JE Jr (2000) The role of STATs in transcriptional control and their impact on cellular function. Oncogene 19:2468–2473
Bromberg JF, Horvath CM, Besser D, Lathem WW, Darnell JE Jr (1998) Stat3 activation is required for cellular transformation by v-src. Mol Cell Biol 18:2553–2558
Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, Darnell JE Jr (1999) Stat3 as an oncogene. Cell 98:295–303
Broniscer A, Baker JN, Tagen M, Onar-Thomas A, Gilbertson RJ, Davidoff AM, Pai Panandiker AS, Leung W, Chin TK, Stewart CF, Kocak M, Rowland C, Merchant TE, Kaste SC, Gajjar A (2010) Phase I study of vandetanib during and after radiotherapy in children with diffuse intrinsic pontine glioma. J Clin Oncol 28:4762–4768
Cao S, Wang C, Zheng Q, Qiao Y, Xu K, Jiang T, Wu A (2010) STAT5 regulates glioma cell invasion by pathways dependent and independent of STAT5 DNA binding. Neurosci Lett 487:228–233
Catlett-Falcone R, Landowski TH, Oshiro MM, Turkson J, Levitzki A, Savino R, Ciliberto G, Moscinski L, Fernandez-Luna JL, Nunez G, Dalton WS, Jove R (1999) Constitutive activation of Stat3 signaling confers resistance to apoptosis in human U266 myeloma cells. Immunity 10:105–115
Chen Z, Lund R, Aittokallio T, Kosonen M, Nevalainen O, Lahesmaa R (2003) Identification of novel IL-4/Stat6-regulated genes in T lymphocytes. J Immunol 171:3627–3635
Chen F, Xu Y, Luo Y, Zheng D, Song Y, Yu K, Li H, Zhang L, Zhong W, Ji Y (2010) Down-regulation of Stat3 decreases invasion activity and induces apoptosis of human glioma cells. J Mol Neurosci 40:353–359
Chung CD, Liao J, Liu B, Rao X, Jay P, Berta P, Shuai K (1997) Specific inhibition of Stat3 signal transduction by PIAS3. Science 278:1803–1805
Croker BA, Kiu H, Nicholson SE (2008) SOCS regulation of the JAK/STAT signalling pathway. Semin Cell Dev Biol 19:414–422
Danial NN, Rothman P (2000) JAK-STAT signaling activated by Abl oncogenes. Oncogene 19:2523–2531
Dasgupta A, Raychaudhuri B, Haqqi T, Prayson R, Van Meir EG, Vogelbaum M, Haque SJ (2009) Stat3 activation is required for the growth of U87 cell-derived tumours in mice. Eur J Cancer 45:677–684
de la Iglesia N, Konopka G, Lim KL, Nutt CL, Bromberg JF, Frank DA, Mischel PS, Louis DN, Bonni A (2008a) Deregulation of a STAT3-interleukin 8 signaling pathway promotes human glioblastoma cell proliferation and invasiveness. J Neurosci 28:5870–5878
de la Iglesia N, Konopka G, Puram SV, Chan JA, Bachoo RM, You MJ, Levy DE, Depinho RA, Bonni A (2008b) Identification of a PTEN-regulated STAT3 brain tumor suppressor pathway. Genes Dev 22:449–462
de la Iglesia N, Puram SV, Bonni A (2009) STAT3 regulation of glioblastoma pathogenesis. Curr Mol Med 9:580–590
Decker T, Kovarik P (2000) Serine phosphorylation of STATs. Oncogene 19:2628–2637
Ehret GB, Reichenbach P, Schindler U, Horvath CM, Fritz S, Nabholz M, Bucher P (2001) DNA binding specificity of different STAT proteins. Comparison of in vitro specificity with natural target sites. J Biol Chem 276:6675–6688
Friedman HS, Bigner DD (2005) Glioblastoma multiforme and the epidermal growth factor receptor. N Engl J Med 353:1997–1999
Fuh B, Sobo M, Cen L, Josiah D, Hutzen B, Cisek K, Bhasin D, Regan N, Lin L, Chan C, Caldas H, DeAngelis S, Li C, Li PK, Lin J (2009) LLL-3 inhibits STAT3 activity, suppresses glioblastoma cell growth and prolongs survival in a mouse glioblastoma model. Br J Cancer 100:106–112
Fujio Y, Kunisada K, Hirota H, Yamauchi-Takihara K, Kishimoto T (1997) Signals through gp130 upregulate bcl-x gene expression via STAT1-binding cis-element in cardiac myocytes. J Clin Invest 99:2898–2905
Fujita M, Zhu X, Sasaki K, Ueda R, Low KL, Pollack IF, Okada H (2008) Inhibition of STAT3 promotes the efficacy of adoptive transfer therapy using type-1 CTLs by modulation of the immunological microenvironment in a murine intracranial glioma. J Immunol 180:2089–2098
Gabrusiewicz K, Ellert-Miklaszewska A, Lipko M, Sielska M, Frankowska M, Kaminska B (2011) Characteristics of the alternative phenotype of microglia/macrophages and its modulation in experimental gliomas. PLoS One 6:e23902
Galli R, Binda E, Orfanelli U, Cipelletti B, Gritti A, De Vitis S, Fiocco R, Foroni C, Dimeco F, Vescovi A (2004) Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res 64:7011–7021
Gao L, Li F, Dong B, Zhang J, Rao Y, Cong Y, Mao B, Chen X (2010) Inhibition of STAT3 and ErbB2 suppresses tumor growth, enhances radiosensitivity, and induces mitochondria-dependent apoptosis in glioma cells. Int J Radiat Oncol Biol Phys 77:1223–1231
Gesbert F, Griffin JD (2000) Bcr/Abl activates transcription of the Bcl-X gene through STAT5. Blood 96:2269–2276
Greenhalgh CJ, Hilton DJ (2001) Negative regulation of cytokine signaling. J Leukoc Biol 70:348–356
Gu F, Hata R, Ma YJ, Tanaka J, Mitsuda N, Kumon Y, Hanakawa Y, Hashimoto K, Nakajima K, Sakanaka M (2005) Suppression of Stat3 promotes neurogenesis in cultured neural stem cells. J Neurosci Res 81:163–171
Gu J, Li G, Sun T, Su Y, Zhang X, Shen J, Tian Z, Zhang J (2008) Blockage of the STAT3 signaling pathway with a decoy oligonucleotide suppresses growth of human malignant glioma cells. J Neurooncol 89:9–17
Hartman SE, Bertone P, Nath AK, Royce TE, Gerstein M, Weissman S, Snyder M (2005) Global changes in STAT target selection and transcription regulation upon interferon treatments. Genes Dev 19:2953–2968
Haspel RL, Darnell JE Jr (1999) A nuclear protein tyrosine phosphatase is required for the inactivation of Stat1. Proc Natl Acad Sci U S A 96:10188–10193
Hatiboglu MA, Wei J, Wu AS, Heimberger AB (2010) Immune therapeutic targeting of glioma cancer stem cells. Target Oncol 5:217–227
Haybaeck J, Obrist P, Schindler CU, Spizzo G, Doppler W (2007) STAT-1 expression in human glioblastoma and peritumoral tissue. Anticancer Res 27:3829–3835
He B, You L, Uematsu K, Zang K, Xu Z, Lee AY, Costello JF, McCormick F, Jablons DM (2003) SOCS-3 is frequently silenced by hypermethylation and suppresses cell growth in human lung cancer. Proc Natl Acad Sci U S A 100:14133–14138
Heim MH, Kerr IM, Stark GR, Darnell JE Jr (1995) Contribution of STAT SH2 groups to specific interferon signaling by the Jak-STAT pathway. Science 267:1347–1349
Heimberger AB, Priebe W (2008) Small molecular inhibitors of p-STAT3: novel agents for treatment of primary and metastatic CNS cancers. Recent Pat CNS Drug Discov 3:179–188
Hoey T, Zhang S, Schmidt N, Yu Q, Ramchandani S, Xu X, Naeger LK, Sun YL, Kaplan MH (2003) Distinct requirements for the naturally occurring splice forms Stat4alpha and Stat4beta in IL-12 responses. EMBO J 22:4237–4248
Horvath CM, Wen Z, Darnell JE Jr (1995) A STAT protein domain that determines DNA sequence recognition suggests a novel DNA-binding domain. Genes Dev 9:984–994
Hussain SF, Kong LY, Jordan J, Conrad C, Madden T, Fokt I, Priebe W, Heimberger AB (2007) A novel small molecule inhibitor of signal transducers and activators of transcription 3 reverses immune tolerance in malignant glioma patients. Cancer Res 67:9630–9636
Ivashkiv LB, Hu X (2004) Signaling by STATs. Arthritis Res Ther 6:159–168
Iwamaru A, Szymanski S, Iwado E, Aoki H, Yokoyama T, Fokt I, Hess K, Conrad C, Madden T, Sawaya R, Kondo S, Priebe W, Kondo Y (2007) A novel inhibitor of the STAT3 pathway induces apoptosis in malignant glioma cells both in vitro and in vivo. Oncogene 26:2435–2444
Jatiani SS, Cosenza SC, Reddy MV, Ha JH, Baker SJ, Samanta AK, Olnes MJ, Pfannes L, Sloand EM, Arlinghaus RB, Reddy EP (2010) A non-ATP-competitive dual inhibitor of JAK2 and BCR-ABL kinases: elucidation of a novel therapeutic spectrum based on substrate competitive inhibition. Genes Cancer 1:331–345
John S, Vinkemeier U, Soldaini E, Darnell JE Jr, Leonard WJ (1999) The significance of tetramerization in promoter recruitment by Stat5. Mol Cell Biol 19:1910–1918
Khan KD, Shuai K, Lindwall G, Maher SE, Darnell JE Jr, Bothwell AL (1993) Induction of the Ly-6A/E gene by interferon alpha/beta and gamma requires a DNA element to which a tyrosine-phosphorylated 91-kDa protein binds. Proc Natl Acad Sci U S A 90:6806–6810
Kisseleva T, Bhattacharya S, Braunstein J, Schindler CW (2002) Signaling through the JAK/STAT pathway, recent advances and future challenges. Gene 285:1–24
Konnikova L, Kotecki M, Kruger MM, Cochran BH (2003) Knockdown of STAT3 expression by RNAi induces apoptosis in astrocytoma cells. BMC Cancer 3:23
Korpelainen EI, Karkkainen M, Gunji Y, Vikkula M, Alitalo K (1999) Endothelial receptor tyrosine kinases activate the STAT signaling pathway: mutant Tie-2 causing venous malformations signals a distinct STAT activation response. Oncogene 18:1–8
Kostianovsky AM, Maier LM, Anderson RC, Bruce JN, Anderson DE (2008) Astrocytic regulation of human monocytic/microglial activation. J Immunol 181:5425–5432
Kubo M, Ransom J, Webb D, Hashimoto Y, Tada T, Nakayama T (1997) T-cell subset-specific expression of the IL-4 gene is regulated by a silencer element and STAT6. EMBO J 16:4007–4020
Lehmann U, Schmitz J, Weissenbach M, Sobota RM, Hortner M, Friederichs K, Behrmann I, Tsiaris W, Sasaki A, Schneider-Mergener J, Yoshimura A, Neel BG, Heinrich PC, Schaper F (2003) SHP2 and SOCS3 contribute to Tyr-759-dependent attenuation of interleukin-6 signaling through gp130. J Biol Chem 278:661–671
Leong PL, Andrews GA, Johnson DE, Dyer KF, Xi S, Mai JC, Robbins PD, Gadiparthi S, Burke NA, Watkins SF, Grandis JR (2003) Targeted inhibition of Stat3 with a decoy oligonucleotide abrogates head and neck cancer cell growth. Proc Natl Acad Sci U S A 100:4138–4143
Letimier FA, Passini N, Gasparian S, Bianchi E, Rogge L (2007) Chromatin remodeling by the SWI/SNF-like BAF complex and STAT4 activation synergistically induce IL-12Rbeta2 expression during human Th1 cell differentiation. EMBO J 26:1292–1302
Li GH, Wei H, Lv SQ, Ji H, Wang DL (2010) Knockdown of STAT3 expression by RNAi suppresses growth and induces apoptosis and differentiation in glioblastoma stem cells. Int J Oncol 37:103–110
Liang QC, Xiong H, Zhao ZW, Jia D, Li WX, Qin HZ, Deng JP, Gao L, Zhang H, Gao GD (2009) Inhibition of transcription factor STAT5b suppresses proliferation, induces G1 cell cycle arrest and reduces tumor cell invasion in human glioblastoma multiforme cells. Cancer Lett 273:164–171
Lin L, Hutzen B, Li PK, Ball S, Zuo M, DeAngelis S, Foust E, Sobo M, Friedman L, Bhasin D, Cen L, Li C, Lin J (2010) A novel small molecule, LLL12, inhibits STAT3 phosphorylation and activities and exhibits potent growth-suppressive activity in human cancer cells. Neoplasia 12:39–50
Lindemann C, Hackmann O, Delic S, Schmidt N, Reifenberger G, Riemenschneider MJ (2011) SOCS3 promoter methylation is mutually exclusive to EGFR amplification in gliomas and promotes glioma cell invasion through STAT3 and FAK activation. Acta Neuropathol 122:241–251
Liu B, Liao J, Rao X, Kushner SA, Chung CD, Chang DD, Shuai K (1998) Inhibition of Stat1-mediated gene activation by PIAS1. Proc Natl Acad Sci U S A 95:10626–10631
Lo RK, Cheung H, Wong YH (2003) Constitutively active Galpha16 stimulates STAT3 via a c-Src/JAK- and ERK-dependent mechanism. J Biol Chem 278:52154–52165
Lo HW, Cao X, Zhu H, Ali-Osman F (2008) Constitutively activated STAT3 frequently coexpresses with epidermal growth factor receptor in high-grade gliomas and targeting STAT3 sensitizes them to Iressa and alkylators. Clin Cancer Res 14:6042–6054
Look DC, Pelletier MR, Tidwell RM, Roswit WT, Holtzman MJ (1995) Stat1 depends on transcriptional synergy with Sp1. J Biol Chem 270:30264–30267
Lord JD, McIntosh BC, Greenberg PD, Nelson BH (2000) The IL-2 receptor promotes lymphocyte proliferation and induction of the c-myc, bcl-2, and bcl-x genes through the trans-activation domain of Stat5. J Immunol 164:2533–2541
Lund RJ, Chen Z, Scheinin J, Lahesmaa R (2004) Early target genes of IL-12 and STAT4 signaling in th cells. J Immunol 172:6775–6782
Marrero MB, Schieffer B, Paxton WG, Heerdt L, Berk BC, Delafontaine P, Bernstein KE (1995) Direct stimulation of Jak/STAT pathway by the angiotensin II AT1 receptor. Nature 375:247–250
Martini M, Pallini R, Luongo G, Cenci T, Lucantoni C, Larocca LM (2008) Prognostic relevance of SOCS3 hypermethylation in patients with glioblastoma multiforme. Int J Cancer 123:2955–2960
Mizoguchi M, Betensky RA, Batchelor TT, Bernay DC, Louis DN, Nutt CL (2006) Activation of STAT3, MAPK, and AKT in malignant astrocytic gliomas: correlation with EGFR status, tumor grade, and survival. J Neuropathol Exp Neurol 65:1181–1188
Nagel-Wolfrum K, Buerger C, Wittig I, Butz K, Hoppe-Seyler F, Groner B (2004) The interaction of specific peptide aptamers with the DNA binding domain and the dimerization domain of the transcription factor Stat3 inhibits transactivation and induces apoptosis in tumor cells. Mol Cancer Res 2:170–182
Naik SM, Shibagaki N, Li LJ, Quinlan KL, Paxton LL, Caughman SW (1997) Interferon gamma-dependent induction of human intercellular adhesion molecule-1 gene expression involves activation of a distinct STAT protein complex. J Biol Chem 272:1283–1290
Ni Z, Bremner R (2007) Brahma-related gene 1-dependent STAT3 recruitment at IL-6-inducible genes. J Immunol 178:345–351
Nicholson SE, De Souza D, Fabri LJ, Corbin J, Willson TA, Zhang JG, Silva A, Asimakis M, Farley A, Nash AD, Metcalf D, Hilton DJ, Nicola NA, Baca M (2000) Suppressor of cytokine signaling-3 preferentially binds to the SHP-2-binding site on the shared cytokine receptor subunit gp130. Proc Natl Acad Sci U S A 97:6493–6498
Nishikawa R, Ji XD, Harmon RC, Lazar CS, Gill GN, Cavenee WK, Huang HJ (1994) A mutant epidermal growth factor receptor common in human glioma confers enhanced tumorigenicity. Proc Natl Acad Sci U S A 91:7727–7731
Niwa H, Burdon T, Chambers I, Smith A (1998) Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev 12:2048–2060
O’Shea JJ, Gadina M, Schreiber RD (2002) Cytokine signaling in 2002: new surprises in the Jak/Stat pathway. Cell 109(Suppl):S121–S131
O’Sullivan A, Chang HC, Yu Q, Kaplan MH (2004) STAT4 is required for interleukin-12-induced chromatin remodeling of the CD25 locus. J Biol Chem 279:7339–7345
Ouchi T, Lee SW, Ouchi M, Aaronson SA, Horvath CM (2000) Collaboration of signal transducer and activator of transcription 1 (STAT1) and BRCA1 in differential regulation of IFN-gamma target genes. Proc Natl Acad Sci U S A 97:5208–5213
Pesu M, Muul L, Kanno Y, O’Shea JJ (2006) Proprotein convertase furin is preferentially expressed in T helper 1 cells and regulates interferon gamma. Blood 108:983–985
Premkumar DR, Jane EP, Agostino NR, Scialabba JL, Pollack IF (2010) Dasatinib synergizes with JSI-124 to inhibit growth and migration and induce apoptosis of malignant human glioma cells. J Carcinog 9:7
Rahaman SO, Harbor PC, Chernova O, Barnett GH, Vogelbaum MA, Haque SJ (2002) Inhibition of constitutively active Stat3 suppresses proliferation and induces apoptosis in glioblastoma multiforme cells. Oncogene 21:8404–8413
Rajan P, McKay RD (1998) Multiple routes to astrocytic differentiation in the CNS. J Neurosci 18:3620–3629
Ramana CV, Chatterjee-Kishore M, Nguyen H, Stark GR (2000) Complex roles of Stat1 in regulating gene expression. Oncogene 19:2619–2627
Raz R, Lee CK, Cannizzaro LA, D’Eustachio P, Levy DE (1999) Essential role of STAT3 for embryonic stem cell pluripotency. Proc Natl Acad Sci U S A 96:2846–2851
Reddy EP, Korapati A, Chaturvedi P, Rane S (2000) IL-3 signaling and the role of Src kinases, JAKs and STATs: a covert liaison unveiled. Oncogene 19:2532–2547
Schaefer LK, Ren Z, Fuller GN, Schaefer TS (2002) Constitutive activation of Stat3alpha in brain tumors: localization to tumor endothelial cells and activation by the endothelial tyrosine kinase receptor (VEGFR-2). Oncogene 21:2058–2065
Schiavone D, Avalle L, Dewilde S, Poli V (2011) The immediate early genes Fos and Egr1 become STAT1 transcriptional targets in the absence of STAT3. FEBS Lett 585:2455–2460
Schmidt D, Muller S (2003) PIAS/SUMO: new partners in transcriptional regulation. Cell Mol Life Sci 60:2561–2574
Senft C, Priester M, Polacin M, Schroder K, Seifert V, Kogel D, Weissenberger J (2010) Inhibition of the JAK-2/STAT3 signaling pathway impedes the migratory and invasive potential of human glioblastoma cells. J Neurooncol 101:393–403
Shen J, Li R, Li G (2009) Inhibitory effects of decoy-ODN targeting activated STAT3 on human glioma growth in vivo. In Vivo 23:237–243
Sherry MM, Reeves A, Wu JK, Cochran BH (2009) STAT3 is required for proliferation and maintenance of multipotency in glioblastoma stem cells. Stem Cells 27:2383–2392
Shuai K, Liu B (2005) Regulation of gene-activation pathways by PIAS proteins in the immune system. Nat Rev Immunol 5:593–605
Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, Dirks PB (2003) Identification of a cancer stem cell in human brain tumors. Cancer Res 63:5821–5828
Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB (2004) Identification of human brain tumour initiating cells. Nature 432:396–401
Socolovsky M, Fallon AE, Wang S, Brugnara C, Lodish HF (1999) Fetal anemia and apoptosis of red cell progenitors in Stat5a−/−5b−/− mice: a direct role for Stat5 in Bcl-X(L) induction. Cell 98:181–191
Stea B, Falsey R, Kislin K, Patel J, Glanzberg H, Carey S, Ambrad AA, Meuillet EJ, Martinez JD (2003) Time and dose-dependent radiosensitization of the glioblastoma multiforme U251 cells by the EGF receptor tyrosine kinase inhibitor ZD1839 (‘Iressa’). Cancer Lett 202:43–51
Stephanou A, Latchman DS (2005) Opposing actions of STAT-1 and STAT-3. Growth Factors 23:177–182
Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, Curschmann J, Janzer RC, Ludwin SK, Gorlia T, Allgeier A, Lacombe D, Cairncross JG, Eisenhauer E, Mirimanoff RO (2005) Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 352:987–996
Su Y, Li G, Zhang X, Gu J, Zhang C, Tian Z, Zhang J (2008) JSI-124 inhibits glioblastoma multiforme cell proliferation through G(2)/M cell cycle arrest and apoptosis augment. Cancer Biol Ther 7:1243–1249
Takeda K, Kaisho T, Yoshida N, Takeda J, Kishimoto T, Akira S (1998) Stat3 activation is responsible for IL-6-dependent T cell proliferation through preventing apoptosis: generation and characterization of T cell-specific Stat3-deficient mice. J Immunol 161:4652–4660
Thieu VT, Yu Q, Chang HC, Yeh N, Nguyen ET, Sehra S, Kaplan MH (2008) Signal transducer and activator of transcription 4 is required for the transcription factor T-bet to promote T helper 1 cell-fate determination. Immunity 29:679–690
Turkson J (2004) STAT proteins as novel targets for cancer drug discovery. Expert Opin Ther Targets 8:409–422
Turkson J, Bowman T, Garcia R, Caldenhoven E, De Groot RP, Jove R (1998) Stat3 activation by Src induces specific gene regulation and is required for cell transformation. Mol Cell Biol 18:2545–2552
Turkson J, Bowman T, Adnane J, Zhang Y, Djeu JY, Sekharam M, Frank DA, Holzman LB, Wu J, Sebti S, Jove R (1999) Requirement for Ras/Rac1-mediated p38 and c-Jun N-terminal kinase signaling in Stat3 transcriptional activity induced by the Src oncoprotein. Mol Cell Biol 19:7519–7528
Turkson J, Ryan D, Kim JS, Zhang Y, Chen Z, Haura E, Laudano A, Sebti S, Hamilton AD, Jove R (2001) Phosphotyrosyl peptides block Stat3-mediated DNA binding activity, gene regulation, and cell transformation. J Biol Chem 276:45443–45455
Turkson J, Kim JS, Zhang S, Yuan J, Huang M, Glenn M, Haura E, Sebti S, Hamilton AD, Jove R (2004) Novel peptidomimetic inhibitors of signal transducer and activator of transcription 3 dimerization and biological activity. Mol Cancer Ther 3:261–269
Valente AJ, Xie JF, Abramova MA, Wenzel UO, Abboud HE, Graves DT (1998) A complex element regulates IFN-gamma-stimulated monocyte chemoattractant protein-1 gene transcription. J Immunol 161:3719–3728
Vescovi AL, Galli R, Reynolds BA (2006) Brain tumour stem cells. Nat Rev Cancer 6:425–436
Vila-Coro AJ, Rodriguez-Frade JM, Martin De Ana A, Moreno-Ortiz MC, Martinez AC, Mellado M (1999) The chemokine SDF-1alpha triggers CXCR4 receptor dimerization and activates the JAK/STAT pathway. FASEB J 13:1699–1710
Villalva C, Martin-Lanneree S, Cortes U, Dkhissi F, Wager M, Le Corf A, Tourani JM, Dusanter-Fourt I, Turhan AG, Karayan-Tapon L (2010) STAT3 is essential for the maintenance of neurosphere-initiating tumor cells in patients with glioblastomas: a potential for targeted therapy? Int J Cancer 128:826–838
Wagner BJ, Hayes TE, Hoban CJ, Cochran BH (1990) The SIF binding element confers sis/PDGF inducibility onto the c-fos promoter. EMBO J 9:4477–4484
Wang H, Zhang W, Huang HJ, Liao WS, Fuller GN (2004) Analysis of the activation status of Akt, NFkappaB, and Stat3 in human diffuse gliomas. Lab Invest 84:941–951
Wang H, Lathia JD, Wu Q, Wang J, Li Z, Heddleston JM, Eyler CE, Elderbroom J, Gallagher J, Schuschu J, MacSwords J, Cao Y, McLendon RE, Wang XF, Hjelmeland AB, Rich JN (2009) Targeting interleukin 6 signaling suppresses glioma stem cell survival and tumor growth. Stem Cells 27:2393–2404
Wei J, Barr J, Kong LY, Wang Y, Wu A, Sharma AK, Gumin J, Henry V, Colman H, Sawaya R, Lang FF, Heimberger AB (2010) Glioma-associated cancer-initiating cells induce immunosuppression. Clin Cancer Res 16:461–473
Weissenberger J, Loeffler S, Kappeler A, Kopf M, Lukes A, Afanasieva TA, Aguzzi A, Weis J (2004) IL-6 is required for glioma development in a mouse model. Oncogene 23:3308–3316
Weissenberger J, Priester M, Bernreuther C, Rakel S, Glatzel M, Seifert V, Kogel D (2010) Dietary curcumin attenuates glioma growth in a syngeneic mouse model by inhibition of the JAK1,2/STAT3 signaling pathway. Clin Cancer Res 16:5781–5795
Wen Z, Zhong Z, Darnell JE Jr (1995) Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell 82:241–250
Wirnsberger G, Hebenstreit D, Posselt G, Horejs-Hoeck J, Duschl A (2006) IL-4 induces expression of TARC/CCL17 via two STAT6 binding sites. Eur J Immunol 36:1882–1891
Wong M, Fish EN (1998) RANTES and MIP-1alpha activate stats in T cells. J Biol Chem 273:309–314
Wormald S, Hilton DJ, Smyth GK, Speed TP (2006) Proximal genomic localization of STAT1 binding and regulated transcriptional activity. BMC Genomics 7:254
Yang F, Brown C, Buettner R, Hedvat M, Starr R, Scuto A, Schroeder A, Jensen M, Jove R (2010) Sorafenib induces growth arrest and apoptosis of human glioblastoma cells through the dephosphorylation of signal transducers and activators of transcription 3. Mol Cancer Ther 9:953–962
Yiin JJ, Hu B, Schornack PA, Sengar RS, Liu KW, Feng H, Lieberman FS, Chiou SH, Sarkaria JN, Wiener EC, Ma HI, Cheng SY (2010) ZD6474, a multitargeted inhibitor for receptor tyrosine kinases, suppresses growth of gliomas expressing an epidermal growth factor receptor mutant, EGFRvIII, in the brain. Mol Cancer Ther 9:929–941
Yoshimura A (2005) Negative regulation of cytokine signaling. Clin Rev Allergy Immunol 28:205–220
Yu H, Jove R (2004) The STATs of cancer–new molecular targets come of age. Nat Rev Cancer 4:97–105
Yu CL, Meyer DJ, Campbell GS, Larner AC, Carter-Su C, Schwartz J, Jove R (1995) Enhanced DNA-binding activity of a Stat3-related protein in cells transformed by the Src oncoprotein. Science 269:81–83
Yu H, Kortylewski M, Pardoll D (2007) Crosstalk between cancer and immune cells: role of STAT3 in the tumour microenvironment. Nat Rev Immunol 7:41–51
Yu H, Pardoll D, Jove R (2009) STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer 9:798–809
Yuan X, Curtin J, Xiong Y, Liu G, Waschsmann-Hogiu S, Farkas DL, Black KL, Yu JS (2004) Isolation of cancer stem cells from adult glioblastoma multiforme. Oncogene 23:9392–9400
Yue P, Turkson J (2009) Targeting STAT3 in cancer: how successful are we? Expert Opin Investig Drugs 18:45–56
Zhang F, Li C, Halfter H, Liu J (2003) Delineating an oncostatin M-activated STAT3 signaling pathway that coordinates the expression of genes involved in cell cycle regulation and extracellular matrix deposition of MCF-7 cells. Oncogene 22:894–905
Zhang Y, Cheng MB, Zhang YJ, Zhong X, Dai H, Yan L, Wu NH, Shen YF (2010) A switch from hBrm to Brg1 at IFNgamma-activated sequences mediates the activation of human genes. Cell Res 20:1345–1360
Zhou H, Miki R, Eeva M, Fike FM, Seligson D, Yang L, Yoshimura A, Teitell MA, Jamieson CA, Cacalano NA (2007) Reciprocal regulation of SOCS 1 and SOCS3 enhances resistance to ionizing radiation in glioblastoma multiforme. Clin Cancer Res 13:2344–2353
Acknowledgements
We thank Kavita Ramji for a critical reading of the manuscript. Studies were supported by a grant N N405621938 from the Ministry of Science and Higher Education.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Swiatek-Machado, K., Kaminska, B. (2020). STAT Signaling in Glioma Cells. In: Barańska, J. (eds) Glioma Signaling. Advances in Experimental Medicine and Biology, vol 1202. Springer, Cham. https://doi.org/10.1007/978-3-030-30651-9_10
Download citation
DOI: https://doi.org/10.1007/978-3-030-30651-9_10
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-30650-2
Online ISBN: 978-3-030-30651-9
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)