Abstract
The suppressor of cytokine signaling 3 (SOCS3) gene is one of eight structurally related genes of the SOCS family and has been suggested to function as a tumor suppressor by inhibition of the JAK/STAT signaling pathway. We investigated 60 human gliomas of different histological types for SOCS3 alterations and found frequent SOCS3 promoter hypermethylation and transcriptional downregulation. However, SOCS3 promoter hypermethylation was virtually absent in primary glioblastomas, which are characterized by frequent epidermal growth factor receptor (EGFR) amplification and overexpression. Assessment of the relationship between SOCS3 and EGFR aberrations revealed that SOCS3 promoter hypermethylation was inversely related to both the EGFR gene dosage as well as the EGFR protein expression, thus suggesting SOCS3 inactivation as a mechanism substituting for EGFR activation in a subset of gliomas. In support of this hypothesis, stable shRNA-mediated SOCS3 knock-down in U251 glioblastoma cells resulted in an activation of EGFR-related signaling pathways, i.e. an increase in the activation levels of STAT3, FAK and to a lesser extent MAPK, while the AKT phosphorylation levels remained unaffected. Functionally, SOCS3-depletion caused strongly increased tumor cell invasion with no obvious effect on tumor cell proliferation. In summary, our findings suggest that SOCS3 inactivation by promoter hypermethylation is mutually exclusive to EGFR activation in gliomas and preferentially promotes glioma cell invasion through STAT3 and FAK activation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
SOCS3 belongs to a family of eight structurally related proteins (CIS and SOCS1-7) that function as negative regulators of cytokine signaling [3, 8]. Cytokine-mediated activation of the JAK/STAT signal transduction pathway leads to a transcriptional increase of several genes, including SOCS3 that, in turn, acts as part of a negative feedback loop by inhibiting JAK activity through a mechanism involving receptor association. SOCS3 thus serves as a crucial regulator controlling for an excessive activation of cytokine-related JAK/STAT signaling [8].
In human cancers, JAK/STAT signaling is known to promote the oncogenic properties of human cancer cells [6, 32] and SOCS3 as a negative regulator of this signaling pathway has been suggested to bear tumor suppressor function [1]. In fact, inhibition of SOCS3 facilitates tumor formation and growth in multiple human cancers, including lung and liver carcinomas, breast and prostate cancer as well as squamous cell carcinomas [1, 16, 21, 28, 31]. Inactivation of SOCS3 is commonly caused by promoter hypermethylation in various types of cancer [4, 5, 24, 26, 28]. In human gliomas, SOCS3 promoter hypermethylation has been reported to associate with an unfavorable clinical outcome in a series of 46 glioblastoma patients [12].
Gliomas are the most common primary tumors of the central nervous system and they comprise a heterogeneous group of neoplasms, among which glioblastoma is the most frequent and most malignant tumor entity [11]. Two clinically and molecularly distinct subtypes of glioblastoma, namely primary and secondary glioblastoma, have been identified [7, 15]. While primary glioblastomas arise de novo and demonstrate frequent EGFR amplification and PTEN mutation, secondary glioblastomas develop by progression from a preexisting lower-grade precursor lesion and are molecularly characterized by frequent IDH1 and TP53 mutations, while EGFR amplification is virtually absent [18, 20].
As JAK/STAT signaling is initiated by EGFR signaling [17] and SOCS3 functions as a regulator within this signaling pathway, we aimed to investigate the role of SOCS3 in a panel of human gliomas comprising different histologically and molecularly defined glioma subtypes. We found that SOCS3 inactivation by promoter hypermethylation was significantly less frequent in primary glioblastomas than in all other investigated glioma entities. Moreover, SOCS3 promoter hypermethylation was significantly inversely related to both the EGFR gene dosage as well as the EGFR protein expression, thus suggesting SOCS3 inactivation as a surrogate mechanism in those types of gliomas that lack aberrant EGF receptor signaling. Subsequent functional in vitro analyses supported this hypothesis by indicating increased STAT3 and FAK activation levels as well as a strong enhancement of tumor cell invasion in SOCS3 knock-down glioma cells.
Materials and methods
Tumor samples and cell lines
Tumors were selected from the tumor tissue archive of the Department of Neuropathology, Heinrich-Heine-University, Düsseldorf, Germany and investigated according to protocols approved by the institutional review board. Tumors were classified according to the World Health Organization (WHO) 2007 classification of tumors of the central nervous system [11]. Parts of each tumor were snap-frozen directly after operation and stored at −80°C. Only tissue samples with a tumor cell content of 80% or more were used for molecular analyses. The tumor series consisted of 60 gliomas, including 25 primary glioblastomas and 8 secondary glioblastomas, WHO grade IV (GB); 4 anaplastic astrocytomas, WHO grade III (AA); 3 diffuse astrocytomas, WHO grade II (A); 6 anaplastic oligoastrocytomas, WHO grade III (AOA); 4 oligoastrocytomas, WHO grade II (OA); 7 anaplastic oligodendrogliomas, WHO grade III (AO); and 3 oligodendrogliomas, WHO grade II (O) (Fig. 1; Supplementary Table 1). We used ten non-neoplastic brain samples from different individuals (NB1-NB10) as reference samples (5 tissue specimens obtained by neurosurgical resection or open biopsy, two tissue specimens obtained at autopsy and three tissue specimens or nucleic acids obtained from commercial sources; Supplementary Table 1). As a positive control for the methylation studies we used commercially available hypermethylated DNA (CpG Genome™ Universal Methylated DNA, Cat.-No. S7821; Millipore, Billerica, MA). The human glioma cell line U251MG for stable transfection with shRNAs directed against SOCS3 (see below) was obtained from the American Type Culture Collection (Manassas, VA, USA).
Synopsis of molecular aberrations of the SOCS3 and the EGFR gene in human gliomas. Note that SOCS3 is frequently inactivated by promoter hypermethylation (blue) in a panel of 60 human gliomas, however, in the subgroup of primary glioblastomas (pGB) that are characterized by a high frequency of activating EGF receptor aberrations SOCS3 methylation is absent in all but one patient. Moreover, EGFR amplification (gene copy number > 5; red) is merely observed in tumors that lack SOCS3 promoter hypermethylation. Prot. EGFR protein expression, Ampl. relative EGFR gene copy number >5 (+) or ≤5 (−), mRNA relative SOCS3 transcript levels, Meth. SOCS3 methylation status, % Score % SOCS3 promoter methylation score. The SOCS3 promoter methylation pattern has been assessed by direct bisulfite sequencing and is depicted for each individual CpG site within the analyzed region spanning the transcription start site of SOCS3. na not analyzed
DNA and RNA extraction
Extraction of DNA and RNA from frozen tumor tissue was performed by ultracentrifugation over a caesium chloride gradient as described elsewhere [27]. Total RNA from cell cultures was isolated using the RNeasy mini kit (Qiagen, Hilden, Germany) following the manufacturer’s protocol.
Real-time (reverse transcription) PCR analysis
Three micrograms of total RNA extracted from tumor tissue or cell lines was reverse transcribed into cDNA using random hexanucleotide primers and SuperScript® Reverse Transcriptase (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. SOCS3 mRNA expression levels were determined by real-time PCR on the StepOnePlus™ sequence detection system (Applied Biosystems, Foster City, CA) by means of incorporation of SybrGreen fluorescent dye as reported before [2]. Fold expression changes (for the tumor tissues relative to non-neoplastic brain tissue and for shRNA-transfected cell lines relative to corresponding control cells) were calculated with the ΔΔCT method [10] using ARF1 (ADP-ribosylation factor 1) as the reference transcript. For the assessment of EGFR gene copy number gains, we used a real-time PCR-based assay with the EGFR gene copy number being normalized to the copy number of the genomic marker D2S1743 (WI3306.1) at chromosomal band 2q21.2. Copy number gains of >5 were designated as EGFR amplification. All primer sequences are listed in Supplementary Table 2.
Methylation analysis using sodium bisulfite sequencing
Sodium bisulfite treatment of 1 μg DNA was performed overnight (16 h) according to a standard protocol [13]. A fragment of the SOCS3 5′-CpG island located at 17q25.3 between nucleotides 73,867,448 and 73,867,855 (numbering according to the UCSC genome browser; http://www.genome.ucsc.edu) was PCR-amplified from sodium bisulfite-modified DNA using the primers as reported in Supplementary Table 2. The amplified fragment covers 43 CpG sites and spans the transcription start site of SOCS3 (Fig. 1). Direct bisulfite sequencing and semiquantitative calculation of a promoter methylation score were carried out as described [25]. In short, the methylation status at each of the analyzed CpG sites was semi-quantitatively rated using the following scale: 0, completely unmethylated; 1, a weak methylated signal detectable in the sequence; 2, methylated signal approximately equal to unmethylated signal; 3, methylated signal markedly stronger than unmethylated signal. Based on this rating, a promoter methylation score in percentage was calculated for each tumor as well as the non-neoplastic brain tissue controls by adding the figures determined at the individual CpG sites divided by the maximum possible methylation score at all analyzed CpG sites. Tumors with a methylation score exceeding that of non-neoplastic brain tissue were regarded as hypermethylated (methylation score > 18; Fig. 1). False-positive results due to incomplete bisulfite conversion were excluded by controlling for the conversion of cytosines entrapped in the respective sequences that were not part of CG dinucleotides. Only sequences with 100% conversion rates were used for evaluation.
Mutational analysis
Single strand conformation polymorphism (SSCP)/heteroduplex analysis was performed to detect mutations within the entire coding sequence of SOCS3 (exon 2) in all 60 tumors. The respective primer sequences are listed in Supplementary Table 2. PCR products were separated by electrophoresis on 10–12% polyacrylamide gels at room temperature and at 4°C overnight. Gels were then silver stained to visualize band patterns and PCR products sequenced in case of aberrant band patterns.
Immunohistochemistry
Immunohistochemistry was performed on formalin-fixed paraffin-embedded tissue sections using a commercially available EGFR-specific mouse monoclonal antibody (Dako, Hamburg, Germany; 1:25 dilution). Immunoreactivity was detected with the EnVision™ Detection System (Dako) using 3,3′-diaminobenzidine tetrahydrochloride (DAB) as a chromogen. Negative controls without primary antibodies were performed for all reactions. Protein expression levels were semi-quantitatively assessed by a composite numerical score, based on the percentage of positively stained tumor cells multiplied by staining intensity, potentially ranging from 0 to 12 (Fig. 2d). The percentage of labeled cells was scored as follows: 0 (no or minimal reactivity, similar to non-neoplastic brain tissue), 1 (<10%), 2 (10–50%), 3 (50–90%) and 4 (>90%). Staining intensity was graded as 0 (negative), 1 (weak), 2 (moderate) or 3 (strong).
Statistical analyses of SOCS3 promoter hypermethylation in human gliomas. a SOCS3 promoter hypermethylation is significantly less frequent in primary glioblastomas than in all other investigated histological glioma subtypes (astrocytic tumors other than pGB, mixed oligoastrocytomas and oligodendrogliomas). b Tumors with SOCS3 promoter hypermethylation show significantly lower SOCS3 transcript levels than tumors without SOCS3 promoter hypermethylation. c–d SOCS3 promoter hypermethylation is inversely correlated with the presence of EGFR aberrations. Note that both the relative gene copy number of EGFR (c) as well as the immunohistochemically assessed protein expression level of EGFR (d) are significantly lower in tumors with SOCS3 promoter hypermethylation than in tumors lacking this molecular alteration. Boxplots: boxes extend from the 25th to the 75th percentile with the middle line at the 50th percentile, whiskers extend down to the lowest and up to the highest values. Asterisks indicate significant differences (*P < 0.05, **P < 0.01, ***P < 0.001)
Generation of glioblastoma cell lines with stable SOCS3 knock-down
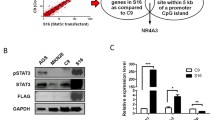
Glioblastoma cells with a stable SOCS3 knock-down were generated using vector-based shRNA-mediated RNAi. To control for potential off-target effects, we used two different predesigned shRNA oligonucleotides (Qiagen, Hilden, Germany) with the following sequences: sh1-sense, TGACGGTCTTCCGACAGAGAT; sh2-sense, TTTCTCATAGGAGTCCAGGTG. shRNA oligonucleotides were cloned into the pcDNA™ 6.2-GW/EmGFP-miR vector (Invitrogen, Carlsbad, CA) containing the human CMV promoter, EmGFP for fluorescent detection as well as a Blasticidin resistance gene for selection of stable cell lines. DNA sequencing was performed to confirm the proper orientation of the inserts into the vector and to exclude mutations. U251MG cells were then transfected with the shRNA oligonucleotide-containing vectors using GeneJuice transfection reagent (Merck, Darmstadt, Germany) according to the manufacturer’s recommendations. Cells were also transfected with the pcDNA™ 6.2-GW/EmGFP-miR-neg control plasmid containing an insert able to form a hairpin structure that is processed into mature miRNA, but is not predicted to target any known vertebrate gene. Stable transfectants were selected for by growing the cells under 20 μg/ml blasticidin. Blasticidin-resistant cells were then separated and individual cell clones were expanded for further analyses. Two different subclones of each of the two hairpins were investigated in the in vitro experiments to control for insertion-site-specific side effects (Fig. 3).
Generation of U251 glioblastoma cells with a stable shRNA-mediated SOCS3 knock-down. Two different hairpins (sh1 and sh2) were investigated to exclude off-target effects of the respective shRNAs. To correct for insertion-site-specific effects we investigated two different subclones of each hairpin (second digit within the respective designation). Note that in comparison to the scrambled negative control shRNA (scr) all four selected clones exhibit markedly reduced SOCS3 expression levels relative to the ARF1 reference gene (knock-down efficacy is provided in percentage on top of each band)
SDS-polyacrylamide gel electrophoresis (PAGE) and western blot analysis
Proteins (30 μg) extracted from U251MG transfected and control cells were denatured, electrophoretically separated on 10–12% gels and transferred to a nitrocellulose membrane (Whatman, Maidstone, UK). After blocking (5% BSA in TBS-T), the membrane was incubated overnight at 4°C with the following primary antibodies: phospho-STAT3 (phosphorylated on Tyr705; 1:500 dilution; #9138; Cell Signaling, Danvers, MA), phospho-MAPK (Thr202/Tyr204; 1:500; #9101; Cell Signaling), phospho-Akt (Ser473; 1:1,000; #4060; Cell Signaling), phospho-FAK (Tyr397; 1:500; #3283; Cell Signaling) and α-tubulin as a loading control (1:3000; #T5168; Sigma-Aldrich, Taufkirchen, Germany). Blots were then incubated for 2 h with HRP-linked anti-mouse or anti-rabbit antibodies, respectively (1:10,000 dilution; Thermo Fisher Scientific, Waltham, MA). To visualize primary antibody binding by chemiluminescence, the membranes were treated with Immobilon® Western HRP Substrate containing luminol and peroxide solution (Millipore, Billerica, MA) and recorded with the LAS-3000 mini system (Fujifilm Life Science, Stanford, CT). Results were reproduced in at least two independent experiments (Fig. 4).
Western blot analysis for the activation of EGFR downstream signaling targets in U251MG SOCS3 knock-down cells. Protein phosphorylation levels for both STAT3 and FAK are uniformly increased (+) in all four SOCS3 knock-down clones relative to U251MG scrambled negative control cells (scr). The SOCS3 knock-down effect on the activation levels of MAPK is inconsistently associated with an increase in protein phosphorylation (i.e. activation) in only one of the two investigated hairpins (clones sh1-1 and sh1-2). p-AKT expression levels appear completely unaffected by the modulation of SOCS3 expression in U251MG glioblastoma cells. α-Tubulin was employed as a control for equal protein loading. Results were reproduced in at least two independent experiments
Cell-based functional assays
The proliferation rate of SOCS3 knock-down clones was compared to control cells using a commercially available BrdU incorporation assay (Roche, Mannheim, Germany). After incubation with BrdU labeling solution for 24 h at 37°C, cells were fixed and absorption values of the colorimetric reaction were measured. Differences in apoptotic activity between SOCS3-depleted cells and controls were assessed by a fluorometric caspase-3/7 assay (Promega, Mannheim, Germany) according to the manufacturer’s protocol. Fluorimetric measurements were optimized for the U251MG cell line and performed after 150 min of incubation with the caspase substrate at room temperature. For investigating the impact of SOCS3 inactivation on the invasive properties of glioblastoma cells we used a 24-well modified Boyden chamber assay (BD Biosciences, San Jose, CA). After serum-starvation for 24 h, cells were seeded on top of a transwell insert containing a Matrigel™-coated fluorescence blocking membrane with 8.0 μm pores. Inserts were then transferred into a lower chamber filled with DMEM containing 10% fetal bovine serum as a chemoattractant and cells were allowed to invade along the serum gradient. Following incubation for 24 h, the cells on the membrane were fixed in paraformaldehyde (4% in PBS) and stained with 4′-6-diamidino-2-phenylindole (DAPI). Membranes were then sliced out from the inserts and transferred upside down on glass slides. Cells that had invaded were counted on the underside of the membrane by means of a fluorescent microscope. All cell-based functional assays were performed in at least six replicates (Fig. 5).
Cell-based assays for the functional effects of a reduction of SOCS3 expression levels in U251MG glioblastoma cells. Cell proliferation was measured using a BrdU incorporation assay (a), apoptosis was quantified using a fluorometric caspase-3/7 assay (b) and tumor cell invasion was assessed using a transwell assay with Matrigel-coated membranes (c). Note that inhibition of SOCS3 does not appear to affect proliferation, slightly inhibits apoptosis in a subset of the investigated clones, but reveals the most pronounced overall effects on tumor cell invasion. All four SOCS3-depleted clones show an uniform increase in tumor cell invasion that is significant in three out of the four investigated clones. The average result in the negative control cells (scr) has been set to 1. Error bars indicate standard deviations and asterisks significant differences. All assays were performed in at least six replicates
Transient siRNA-mediated silencing of SOCS3
To independently validate the SOCS3 effect on invasion observed in the stable U251MG knock-down clones, we performed siRNA-mediated silencing of SOCS3 in U251MG and A172 cells (Suppl. Fig. 1). Transfections were done with HiPerFect transfection reagent (Qiagen, Hilden, Germany) following the manufacturer’s protocol. For testing knock-down efficiency by quantitative real-time RT-PCR, transfections were performed in 24-well plates with 30,000 cells/well. The RNeasyPlus kit (Qiagen, Hilden, Germany) was used for RNA extraction. For invasion assays, cells were transfected in 6-well plates with 240,000 cells/well. All experiments were performed with 50 nM siRNA (Qiagen, Hilden, Germany). Sequences of siRNAs used were as follows: siRNA1-sense, CCAAGAACCUGCGCAUCCATT; siRNA2-sense, GAAGAGCCUAUUACAUCUATT; siRNA3-sense, GGGAGUUCCUGGACCAGUATT; siRNA4-sense, CGCUCAGCGUCAAGACCATT.
Statistical analyses
Fisher’s exact test was used to assess frequency differences of SOCS3 promoter hypermethylation between the different histological glioma entities (Fig. 2a). In this regard, we compared primary glioblastomas (pGB, n = 25) to other astrocytic gliomas (A + AA + sGB, n = 14), mixed oligoastrocytomas (OA + AOA, n = 11) and pure oligodendroglial neoplasms (O + OA, n = 10). P values were verified for significance by employing Benjamini-Hochberg corrections for multiple testing.
Mann–Whitney U test analyses were used to compare SOCS3 mRNA expression (Fig. 2b), EGFR gene copy number (Fig. 2c) and EGFR protein expression scores (Fig. 2d) between tumors with and without SOCS3 promoter hypermethylation. In the cell-based functional assays, two-sided Student’s t test analyses were utilized to compare results between transfected and control cells (Fig. 5). All statistical analyses were computed using the Graph Pad Prism Software (Version 5). The P value cut-off level for significance was set to 0.05 (*). (**) indicates a P value of <0.01 and (***) and a P value of <0.001.
Results
Promoter hypermethylation and transcriptional downregulation of SOCS3 in human gliomas
We assessed the methylation status of the SOCS3 promoter in a panel of 60 human gliomas of different histological subtypes and grades using direct bisulfite sequencing analysis (Fig. 1). Methylation of the SOCS3 promoter in the tumor samples exceeded that of non-neoplastic brain tissue in 17 out of 60 glioma patients (28%). There was a significant inverse correlation between SOCS3 promoter hypermethylation and SOCS3 transcript levels (Mann–Whitney U test, P = 0.001). SOCS3 mRNA expression levels were significantly lower in the group of gliomas with a hypermethylated SOCS3 promoter (median 0.3; interquartile range, IQR 0.5) when compared to the group of tumors with promoter methylation scores equal to or lower than the reference brain tissue samples (median 1.5, IQR 11.4) (Fig. 2b).
When comparing the SOCS3 methylation status between the different glioma entities represented in our panel, we found that SOCS3 promoter hypermethylation—with a single exception (pGB115)—was absent in primary glioblastomas (1 out of 25 cases, 4%). Furthermore, SOCS3 promoter hypermethylation was significantly less frequent in primary glioblastomas compared to all other glioma entities, i.e. diffusely infiltrating astrocytic (A + AA + sGB: 4 out 14, 29%; Fisher’s exact test, P = 0.047), mixed oligoastrocytic (OA + AOA: 6 out of 11, 55%; P = 0.001) and oligodendroglial (O + AO: 6 out of 10, 60%; P = 0.0008) neoplasms (Fig. 2a).
To screen for potential alternative SOCS3 inactivation mechanisms, we performed an additional mutation analysis of the entire coding sequence (exon 2) of SOCS3 by means of SSCP/heteroduplex analysis but did not detect any tumor-associated (somatic) mutations in the 60 gliomas investigated.
SOCS3 promoter hypermethylation is inversely correlated with EGFR gene dosage and EGFR protein expression
Following up on our observation of a significantly less frequent SOCS3 promoter hypermethylation in primary glioblastomas, which usually contain a high fraction of activating EGFR aberrations, we assessed the EGFR gene dosage by real-time PCR analysis as well as the EGFR protein expression by immunohistochemistry in our tumor panel and correlated the results with the SOCS3 promoter methylation status. A total of 15 tumors (13 glioblastomas and 2 anaplastic oligoastrocytomas) showed a more than fivefold increase in the EGFR gene dosage, consistent with EGFR amplification (Fig. 1) and none of these tumors exhibited concomitant SOCS3 promoter hypermethylation. Statistical analysis underscored this observation and revealed significantly lower EGFR gene copy numbers in tumors containing a hypermethylated SOCS3 promoter (median 0.9, IQR 0.4) when compared to tumors without SOCS3 promoter hypermethylation (median 1.7, IQR 19.0, Mann–Whitney U test, P = 0.0006; Fig. 2c).
We also compared the semiquantitatively determined EGFR protein expression levels between tumors with and without SOCS3 promoter hypermethylation. The median EGFR protein expression score was significantly lower in tumors with SOCS3 promoter hypermethylation (median 1.5, IQR 3.0) compared to tumors without this aberration (median 8.5, IQR 6.0, Mann–Whitney U test, P = 0.0004; Fig. 2c). Of note, the single primary glioblastoma (pGB115) with SOCS3 promoter hypermethylation did show neither a relevant increase in EGFR gene copy number (fold change: 1.2) nor EGFR protein expression (Score: 0).
Stable shRNA-mediated SOCS3 knock-down increases STAT3 and FAK activation levels in glioblastoma cells
To further assess the impact of SOCS3 on potential downstream targets within the EGFR signaling pathway, we used vector-based shRNA-mediated RNAi to generate U251 glioblastoma cells with a stable SOCS3 knock-down. Two different hairpins (sh1 and sh2) were used to exclude off-target effects of the respective shRNAs and two different subclones of each hairpin (shX-1 and shX-2) to exclude insertion-site-specific side effects. Transfection with both hairpins resulted in a consistent SOCS3 knock-down in comparison to the negative scrambled (scr) control with a subtotal depletion of SOCS3 expression in glioblastoma cells transfected with hairpin 1 (86–99%) and a somewhat lesser reduction of SOCS3 expression in cells transfected with hairpin 2 (62–65%; Fig. 3).
Employing phosphorylation-specific antibodies, we studied the SOCS3 effect on protein phosphorylation, i.e. activation levels of STAT3 (Tyr705), FAK (Tyr397), MAPK (Thr202/Tyr204) and Akt (Ser473). Of note, SOCS3 depletion resulted in a strong and unequivocal increase in the phosphorylation levels of STAT3 and FAK in all investigated tumor cell subclones relative to the respective negative control cells (Fig. 4). The phosphorylation pattern of MAPK in dependence on SOCS3 reduction was less uniform with an increased expression of p-MAPK only in hairpin 1 and a reduction of p-MAPK levels in U251 glioblastoma cells transfected with hairpin 2. For AKT, we did not detect any relevant changes in the protein phosphorylation levels comparing SOCS3 knock-down and control cells (Fig. 4).
Cell-based functional assays reveal a major effect of SOCS3 depletion on tumor cell invasion
To evaluate the functional consequences of SOCS3 inactivation in U251 glioblastoma cells we performed cell-based assays for tumor cell proliferation, apoptosis and invasion. Tumor cell proliferation measured by BrdU incorporation exhibited an uneven either minor increase or reduction in individual SOCS3 knock-down clones in comparison to control cells but none of these changes was statistically significant (Fig. 5a). Using a caspase-3/7 assay we observed a moderate inhibition of apoptotic activity in the majority (3 out of 4) of the investigated SOCS3 knock-down clones with statistically significant results for two of the investigated subclones (Students’s t test; sh1-2, P = 0.0476; sh2-2, P = 0.0001; Fig. 5b).
The predominant functional effect of SOCS3 reduction, however, was detected when investigating the differential invasive capacities of U251 glioblastoma cells using a modified Matrigel™-coated Boyden chamber assay. All four investigated SOCS3-depleted tumor cell clones showed an unequivocal increase in their invasive capacities relative to the respective control cells with the highest, up to nearly fivefold increase in subclone sh2-2. Using Student’s t test analyses this increase proved highly significant in three out of the four investigated SOCS3 knock-down clones (sh1-1, P = 0.005; sh2-1, P = 0.004; sh2-2, P = 0.002; Fig. 5c). To independently corroborate the SOCS3 effect on tumor cell invasion, we employed siRNA-mediated silencing of SOCS3 in U251MG and A172 glioblastoma cells. In line with our findings in the stably transfected SOCS3 knock-down U251MG clones, we observed an uniform and significant increase of glioma cell invasion after SOCS3 depletion in both cell lines (Suppl. Fig. 1).
Discussion
SOCS3 functions as a negative regulator of the JAK/STAT signal transduction pathway and has been described as a tumor suppressor in human cancers. In diffusely infiltrating human gliomas, the role of SOCS3 was less clear, particularly in regard to the fact that these tumors consist of a number of different subentities that are characterized by distinct patterns of molecular aberrations [11]. As such, primary glioblastomas contain a high frequency of activating EGFR aberrations, while secondary glioblastomas and their lower-grade precursor lesions in their majority lack EGFR aberrations and are instead characterized by frequent mutations in the IDH1 and TP53 genes [15]. These different molecular preconditions may imply to the distribution of SOCS3 alterations between the different subtypes of gliomas, particularly in regard to the fact that SOCS3 and EGFR receptor signaling share common downstream signaling targets. In a preceding publication, Martini and colleagues [12] reported on the negative prognostic effect of SOCS3 promoter hypermethylation in gliomas but restricted their study to primary glioblastomas and did not refer to the EGFR status in their patients.
We investigated a panel of 60 human gliomas of different histological subtypes and WHO grades for SOCS3 alterations and made the striking observation that SOCS3 inactivation by promoter hypermethylation was observed with the lowest frequency in primary glioblastomas (4%), i.e. exactly in those tumors that contain the highest rate of EGFR aberrations. We then more closely assessed the correlation between EGF receptor aberrations and SOCS3 promoter hypermethylation in our tumor panel. Indeed, tumors with SOCS3 promoter hypermethylation exhibited both significantly lower relative EGFR gene copy numbers as well as significantly lower EGFR protein expression scores than tumors without this epigenetic aberration, suggesting that SOCS3 inactivation by promoter hypermethylation in gliomas might be an alternative molecular alteration to EGFR amplification/overexpression. Interestingly, two other recent publications add further proof to our hypothesis of an inverse relation between SOCS3 protein function and EGFR signaling. In human hepatocytes and in human embryonic kidney 293 cells SOCS3 was found to impair the phosphorylation of STAT3 in response to EGF [22, 29], thus indicating that SOCS3 differentially regulates EGF receptor signaling. Also, in line with this hypothesis, the single primary glioblastoma (pGB115) in our panel with a hypermethylated SOCS3 promoter did show neither elevated EGFR gene copy numbers nor EGFR protein overexpression. On histological review, the tumor exhibited the classical morphological features of glioblastoma and molecularly the tumor was wild-type for IDH1 and IDH2, as typical for the majority of primary glioblastomas. Thus, our results do not exclude that primary glioblastomas may require a hypermethylated SOCS3 promoter, nevertheless the low frequency of SOCS3 hypermethylation (4%) is contrary to the one described by Martini and colleagues [12] who report on up to 35% of primary glioblastomas with SOCS3 promoter hypermethylation. While in the 25 primary glioblastomas in our panel we had a nearly equal proportion of tumors with (13 cases) and without (12 cases) EGF receptor amplifications, the higher SOCS3 methylation frequency in the paper by Martini et al. might be due to an overrepresentation of EGFR wildtype tumors in their tumor panel that are potentially more susceptible to require SOCS3 hypermethylation. On the other hand, closer to the low SOCS3 methylation frequencies observed in our panel, an alternative study by Zhou and colleagues [34] did detect neither SOCS3 hypermethylation nor reduced SOCS3 expression levels in 10 glioblastoma cell lines and 12 primary glioblastoma tissues.
Further investigation of the downstream signaling effects of SOCS3 within the EGF receptor signaling pathway revealed a strong and unequivocal increase in the phosphorylation levels of STAT3 and FAK in all investigated SOCS3 knock-down clones relative to respective U251MG control cells. The positive effect of SOCS3 depletion on STAT3 activation is in line with the functional role of SOCS3 as a negative regulator of JAK/STAT signaling [3, 8] and also our finding of an increased FAK activation after SOCS3 knock-down had been concordantly reported in progenitor B lymphocytes and human hepatocellular carcinoma cells. The question on how exactly SOCS3 depletion increases FAK Y397 phosphorylation remains a matter for further investigation. One study reported that SOCS3 regulates CXCL12-induced FAK phosphorylation through the ubiquitin–proteasome pathway during B lymphopoiesis. Another report in human hepatocellular carcinoma points to a direct physical interaction of SOCS3 with phosphorylated FAK and this interaction mediating polyubiquitination of FAK, resulting in proteasome-dependent degradation of FAK [9, 14].
As both STAT3 and FAK can be alternatively phosphorylated by activated growth factor receptors, such as the epidermal growth factor receptor (EGFR), these findings may add further proof to our hypothesis that SOCS3 inactivation may substitute for the activation of identical intracellular signaling pathways in those types of gliomas that do not bear activated EGF receptors. By means of STAT3 activation SOCS3-depleted cancer cells may transactivate multiple target genes impacting such diverse cell conditions as proliferation, differentiation or migration and invasion [6, 32]. The activation of FAK has been described to impact tumor cell growth, cytoskeletal organization and microtubule organization and is functionally even more directly linked to an increase of invasive properties in a variety of human tumors, including gliomas [19].
As two further downstream EGFR signaling intermediates, p44/p42 MAPK (mitogen-activated protein kinase, ERK1/2) and AKT (protein kinase B), had been described to be regulated by SOCS3 [16, 23, 33], we investigated the activation levels of both molecules in our SOCS3 knock-down glioma cells. For p44/p42 MAPK we obtained non-uniform results with a moderate increase in activation levels for only one of the transfected hairpins (sh1). This partial lack of a MAPK activation increase may be potentially due to the weaker SOCS3 knock-down efficiency of hairpin 2 (sh2) but may also argue for a less consistent impact of SOCS3 on the p44/p42 MAPK activation levels in glioma cells. For AKT there was no increase of phosphorylation levels after SOCS3 depletion. Thus, our results are not indicative of a close interrelationship between these two molecules in human gliomas.
As to the functional implications of SOCS3 inactivation in human glioma cells, we found a predominant impact on tumor cell invasion with uniformly and significantly increased invasive capacities in SOCS3-depleted glioma cells. Our findings are in line with reports on an increased migratory and invasive potential after SOCS3 knock-down in tumor entities other than gliomas [14, 16, 30]. Moreover, the above-described specific activation pattern of STAT3 and particularly FAK after SOCS3 knock-down is compatible with a predominant functional impact on tumor cell invasion.
We observed an only moderate reduction of apoptotic cell properties that, however, was statistically significant in two out of the four investigated SOCS3 knock-down clones. The only moderate and uneven effect of SOCS3 reduction on apoptosis may be due to the circumstance that we measured basal apoptosis without stimulation. Also, the detection of a significant dysregulation in two subclones, each from different hairpins, argues against an off-target and for a true anti-apoptotic effect of impaired SOCS3 in glioma cells. We did not observe consistent or significant effects on tumor cell proliferation after SOCS3 inactivation, arguing for a negligible effect of SOCS3 on tumor cell proliferation in glioblastoma cells. Recapitulating that we found a more frequent SOCS3 promoter hypermethylation and transcriptional downregulation in lower-grade astrocytic and oligodendroglial tumors than in glioblastomas and that these tumors share the diffuse infiltrative pattern but not the high proliferative activity with glioblastomas, it is thus tempting to speculate that SOCS3 inactivation in these tumors may mainly substitute for the stimulating effect of EGFR on invasion but not on proliferation. Nevertheless, the intracellular signaling relationships are certainly more complex and will need further clarification exceeding the one provided in this manuscript.
In summary, our study molecularly dissects the inactivation mechanisms of SOCS3 in different histological subtypes of human gliomas and functionally investigates the interrelationships of SOCS3 with EGF receptor signaling in gliomas. We conclude that SOCS3 inactivation by promoter hypermethylation is significantly less frequent in primary glioblastomas than in all other investigated glioma entities and mutually exclusive to EGFR amplification/overexpression. Functionally, SOCS3 silencing preferentially promotes glioma cell invasion through STAT3 and FAK activation.
References
Baltayiannis G, Baltayiannis N, Tsianos EV (2008) Suppressors of cytokine signaling as tumor repressors. Silencing of SOCS3 facilitates tumor formation and growth in lung and liver. J Buon 13:263–265
Barski D, Wolter M, Reifenberger G, Riemenschneider MJ (2010) Hypermethylation and transcriptional downregulation of the TIMP3 gene is associated with allelic loss on 22q12.3 and malignancy in meningiomas. Brain Pathol 20:623–631
Cooney RN (2002) Suppressors of cytokine signaling (SOCS): inhibitors of the JAK/STAT pathway. Shock 17:83–90
Fourouclas N, Li J, Gilby DC, Campbell PJ, Beer PA, Boyd EM, Goodeve AC, Bareford D, Harrison CN, Reilly JT, Green AR, Bench AJ (2008) Methylation of the suppressor of cytokine signaling 3 gene (SOCS3) in myeloproliferative disorders. Haematologica 93:1635–1644
He B, You L, Uematsu K, Zang K, Xu Z, Lee AY, Costello JF, McCormick F, Jablons DM (2003) SOCS-3 is frequently silenced by hypermethylation and suppresses cell growth in human lung cancer. Proc Natl Acad Sci USA 100:14133–14138
Imada K, Leonard WJ (2000) The Jak-STAT pathway. Mol Immunol 37:1–11
Kleihues P, Ohgaki H (1999) Primary and secondary glioblastomas: from concept to clinical diagnosis. Neurooncology 1:44–51
Krebs DL, Hilton DJ (2000) SOCS: physiological suppressors of cytokine signaling. J Cell Sci 113(Pt 16):2813–2819
Le Y, Zhu BM, Harley B, Park SY, Kobayashi T, Manis JP, Luo HR, Yoshimura A, Hennighausen L, Silberstein LE (2007) SOCS3 protein developmentally regulates the chemokine receptor CXCR4-FAK signaling pathway during B lymphopoiesis. Immunity 27:811–823
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25:402–408
Louis DN, Ohgaki H, Wiestler OD, CaveneeWK (eds) (2007) WHO Classification of tumours of the central nervous system, 4th edn. IARC Press, Lyon
Martini M, Pallini R, Luongo G, Cenci T, Lucantoni C, Larocca LM (2008) Prognostic relevance of SOCS3 hypermethylation in patients with glioblastoma multiforme. Int J Cancer 123:2955–2960
Mueller W, Nutt CL, Ehrich M, Riemenschneider MJ, von Deimling A, van den Boom D, Louis DN (2007) Downregulation of RUNX3 and TES by hypermethylation in glioblastoma. Oncogene 26:583–593
Niwa Y, Kanda H, Shikauchi Y, Saiura A, Matsubara K, Kitagawa T, Yamamoto J, Kubo T, Yoshikawa H (2005) Methylation silencing of SOCS-3 promotes cell growth and migration by enhancing JAK/STAT and FAK signalings in human hepatocellular carcinoma. Oncogene 24:6406–6417
Ohgaki H, Kleihues P (2007) Genetic pathways to primary and secondary glioblastoma. Am J Pathol 170:1445–1453
Puhr M, Santer FR, Neuwirt H, Marcias G, Hobisch A, Culig Z (2010) SOCS-3 antagonises the proliferative and migratory effects of fibroblast growth factor-2 in prostate cancer by inhibition of p44/p42 MAPK signalling. Endocr Relat Cancer 17:525–538
Quesnelle KM, Boehm AL, Grandis JR (2007) STAT-mediated EGFR signaling in cancer. J Cell Biochem 102:311–319
Riemenschneider MJ, Jeuken JW, Wesseling P, Reifenberger G (2010) Molecular diagnostics of gliomas: state of the art. Acta Neuropathol 120:567–584
Riemenschneider MJ, Mueller W, Betensky RA, Mohapatra G, Louis DN (2005) In situ analysis of integrin and growth factor receptor signaling pathways in human glioblastomas suggests overlapping relationships with focal adhesion kinase activation. Am J Pathol 167:1379–1387
Riemenschneider MJ, Reifenberger G (2009) Molecular neuropathology of gliomas. Int J Mol Sci 10:184–212
Sasi W, Jiang WG, Sharma A, Mokbel K (2010) Higher expression levels of SOCS 1, 3, 4, 7 are associated with earlier tumour stage and better clinical outcome in human breast cancer. BMC Cancer 10:178
Seki E, Kondo Y, Iimuro Y, Naka T, Son G, Kishimoto T, Fujimoto J, Tsutsui H, Nakanishi K (2008) Demonstration of cooperative contribution of MET- and EGFR-mediated STAT3 phosphorylation to liver regeneration by exogenous suppressor of cytokine signalings. J Hepatol 48:237–245
Senn JJ, Klover PJ, Nowak IA, Zimmers TA, Koniaris LG, Furlanetto RW, Mooney RA (2003) Suppressor of cytokine signaling-3 (SOCS-3), a potential mediator of interleukin-6-dependent insulin resistance in hepatocytes. J Biol Chem 278:13740–13746
Sutherland KD, Lindeman GJ, Choong DY, Wittlin S, Brentzell L, Phillips W, Campbell IG, Visvader JE (2004) Differential hypermethylation of SOCS genes in ovarian and breast carcinomas. Oncogene 23:7726–7733
Tepel M, Roerig P, Wolter M, Gutmann DH, Perry A, Reifenberger G, Riemenschneider MJ (2008) Frequent promoter hypermethylation and transcriptional downregulation of the NDRG2 gene at 14q11.2 in primary glioblastoma. Int J Cancer 123:2080–2086
Tokita T, Maesawa C, Kimura T, Kotani K, Takahashi K, Akasaka T, Masuda T (2007) Methylation status of the SOCS3 gene in human malignant melanomas. Int J Oncol 30:689–694
van den Boom J, Wolter M, Kuick R, Misek DE, Youkilis AS, Wechsler DS, Sommer C, Reifenberger G, Hanash SM (2003) Characterization of gene expression profiles associated with glioma progression using oligonucleotide-based microarray analysis and real-time reverse transcription-polymerase chain reaction. Am J Pathol 163:1033–1043
Weber A, Hengge UR, Bardenheuer W, Tischoff I, Sommerer F, Markwarth A, Dietz A, Wittekind C, Tannapfel A (2005) SOCS-3 is frequently methylated in head and neck squamous cell carcinoma and its precursor lesions and causes growth inhibition. Oncogene 24:6699–6708
Xia L, Wang L, Chung AS, Ivanov SS, Ling MY, Dragoi AM, Platt A, Gilmer TM, Fu XY, Chin YE (2002) Identification of both positive and negative domains within the epidermal growth factor receptor COOH-terminal region for signal transducer and activator of transcription (STAT) activation. J Biol Chem 277:30716–30723
Yang SF, Yeh YT, Wang SN, Hung SC, Chen WT, Huang CH, Chai CY (2008) SOCS-3 is associated with vascular invasion and overall survival in hepatocellular carcinoma. Pathology 40:558–563
Ying M, Li D, Yang L, Wang M, Wang N, Chen Y, He M, Wang Y (2010) Loss of SOCS3 expression is associated with an increased risk of recurrent disease in breast carcinoma. J Cancer Res Clin Oncol 136:1617–1626
Yu H, Pardoll D, Jove R (2009) STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer 9:798–809
Yu ZB, Bai L, Qian P, Xiao YB, Wang GS, Qian GS, Bai CX, Min JX (2009) Restoration of SOCS3 suppresses human lung adenocarcinoma cell growth by downregulating activation of Erk1/2, Akt apart from STAT3. Cell Biol Int 33:995–1001
Zhou H, Miki R, Eeva M, Fike FM, Seligson D, Yang L, Yoshimura A, Teitell MA, Jamieson CA, Cacalano NA (2007) Reciprocal regulation of SOCS 1 and SOCS3 enhances resistance to ionizing radiation in glioblastoma multiforme. Clin Cancer Res 13:2344–2353
Acknowledgments
The authors would like to thank Nadine Lottmann (Department of Neuropathology, Düsseldorf) for skillful technical assistance. This work was financially supported by grants from the German Cancer Aid (Max-Eder Junior Research Group Program, Grant no. 107709 and 109426) and the Academy of Sciences of North Rhine-Westphalia/Mercator foundation (“Junges Kolleg”) (both to Markus J. Riemenschneider).
Author information
Authors and Affiliations
Corresponding author
Additional information
C. Lindemann and O. Hackmann contributed equally to this paper.
Electronic supplementary material
Below is the link to the electronic supplementary material.
401_2011_832_MOESM1_ESM.jpg
Suppl. Fig. 1. SOCS3 effect on tumor cell invasion after siRNA-mediated knock-down in U251 and A172 gioblastoma cells. (a) In comparison to the transfection with a scrambled negative control siRNA (CTRL), two different siRNAs per cell line were used to significantly downregulate SOCS3 expression levels relative to the ARF1 reference gene. (b) Note that tumor cell invasion significantly increases in both cell lines after siRNA-mediated SOCS3 knock-down in comparison to the transfection with respective negative control siRNAs. These findings corroborate the strong SOCS3 effect on glioma cell invasion observed in the stably transfected U251 knock-downclones. (JPEG 259 kb)
Rights and permissions
About this article
Cite this article
Lindemann, C., Hackmann, O., Delic, S. et al. SOCS3 promoter methylation is mutually exclusive to EGFR amplification in gliomas and promotes glioma cell invasion through STAT3 and FAK activation. Acta Neuropathol 122, 241–251 (2011). https://doi.org/10.1007/s00401-011-0832-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00401-011-0832-0