Abstract
In eukaryotes, the haploid DNA content (C-value) varies widely across lineages without an apparent correlation with the complexity of organisms. This incongruity has been called the C-value paradox and has been solved by demonstrating that not all DNA is constituted by genes but, on the contrary, most of it is made up of repetitive DNA. In vertebrates, the increasing number of sequenced genomes has shown that differences in genome size between lineages are ascribable to a variation in transposon content. These mobile elements, previously perceived as “junk DNA” or “selfish DNA,” are now recognized as the major players in shaping genomes. During vertebrate evolution, transposable elements have been repeatedly co-opted and exapted to generate regulatory sequences, coding exons, or entirely new genes that lead to evolutionary advantages for the host. Moreover, transposable elements are also responsible for substantial rearrangements such as insertions, deletions, inversions, and duplications potentially associated with, or following, speciation events.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
1 Genome Size Variation: A Fascinating Enigma
In 1948, Vendrely and Vendrely (1948) reported a “remarkable constancy in the nuclear DNA content of all the cells in all the individuals within a given animal species.” This constancy, referred to the haploid nuclear DNA content, was defined as the C-value (Swift 1950).
Comparing the haploid DNA content or C-value in eukaryotes, there is a notable lack of correlation between DNA content and organism complexity (Thomas 1971). Indeed, DNA is the stuff of genes and the more complex the organism is, the more genes it should have, and thus more DNA. However, it has been demonstrated that simple organisms such as some amoebas have hundreds of times more DNA than humans (Thomas 1971). The lack of correlation between the size and complexity of eukaryotic genomes is known as the C-value paradox and remained a mystery for almost half a century. Research in this field revealed that this incongruity is only apparent since not all DNA is made up of genes but, on the contrary, most of it is constituted by non-coding DNA and often repetitive DNA. The comparison between the genomes of prokaryotes and multicellular eukaryotes has shown an increase in genome size that is associated not only with a proliferation of repeated elements but also with an increased number of genes. Moreover, an expansion in the size and number of introns has also been related to the gigantism of genomes (Lynch and Conery 2003).
The presence of repetitive DNA in the genome of eukaryotes opens up a number of questions regarding, first of all, why some species possess a great amount of repetitive DNA and others present compact genomes. It would be interesting to reveal the mechanisms by which repetitive DNA spreads or is deleted from genomes during evolution, together with the effects and functions that it might have on chromosomes, nuclei, cells, and organisms. It is also intriguing to investigate whether this DNA has an adaptive role, and if this is not the case, it would be interesting to understand why natural selection has tolerated so much extra DNA.
According to the adaptive theory, this DNA, given its amount, influences phenotype. Indeed, the amount of DNA can directly or indirectly increase nuclear and cell size, the duration of mitosis and meiosis, the rate of basal metabolism, as well as embryonic development time and body size (Bennett 1971; Cavalier-Smith 1978; Vinogradov 1995, 1997). It has been pointed out that for birds, it is advantageous to have a smaller genome size because this implies smaller cells and thus a higher surface-to-volume ratio which provides a consequently more efficient transfer of oxygen through the cell membrane (Olmo 1983; Hughes and Hughes 1995). This hypothesis is also supported by the reduced genome size of bats compared with other mammals (Burton et al. 1989; Van den Bussche et al. 1995).
In 2002, Petrov (2002) suggested the hypothesis of the “mutation equilibrium model” according to which genome size is determined as the equilibrium between the rate of sequence loss by the deletion bias of small indels and the rate of sequence gain by long insertions.
The extra DNA could also have a protective function since mutations can statistically occur more frequently in the non-coding repetitive fraction (Vinogradov 1998).
Repetitive DNA also includes mobile elements that, given their ability to replicate themselves, have been defined as “selfish DNA” (Doolittle and Sapienza 1980). The proliferation of such elements in the genome depends on the strength of natural selection so that the final genome size is the highest tolerable value. However, smaller genomes seem to be favored by natural selection also in relation to the population size: species that have experienced a population reduction present larger genomes and are more prone to extinction (Vinogradov 2003, 2004; Kraaijeveld 2010).
Alternatively, DNA accumulation might be non-adaptive and thus useless. For this reason, repetitive DNA was initially labeled as “junk DNA,” fixed by random drift and carried passively in the genome (Ohno 1972). On the contrary, an increasing number of studies are now supporting an unexpected dynamicity of repetitive DNA which was originally thought to be silent and inert (Biscotti et al. 2015a, b; Biemont 2010).
Several approaches have been adopted to estimate the nuclear DNA content. In the 1970s and 1980s, it became relatively common to use reassociation kinetics to assess the composition and size of genomes. This method consists in the extraction of DNA from cells and denaturation by heating. The solution of denaturated genomic DNA is placed in an environment conducive to renaturation. The rates of reassociation of the DNA strands are proportional to the number of times that specific sequences are found in the genome, providing information on the repetitive and low-copy components. Moreover, they can be calibrated against a standard to give an estimate of absolute DNA content. DNA reassociation of a eukaryotic genome is described by a Cot curve where Co is the starting concentration of nucleotides and t is the reassociation time; low values correspond to highly repetitive DNA while high values indicate single and low-copy DNA sequences (Britten et al. 1974). Thus, Cot analysis provides considerable information on the size and structure of eukaryotic genomes. However, reassociation kinetics for large size genomes are very slow and not particularly accurate and consequently, alternative methods have been adopted. Flow cytometry is a technique used in several applications including genome sizing. It consists in isolating the nuclei and staining them with a fluorescent dye that binds stoichiometrically to DNA. The amount of fluorescent light emitted by each nucleus is converted into a digital signal and compared with a known amount of DNA in order to determine the absolute DNA content in the species of interest. Using this method, the sources of error are the choice of fluorochrome and the presence of inhibitors or cytoplasmatic constituents released during nuclei isolation. Feulgen microdensitometry is a method based on staining nuclei with the Feulgen technique and then measuring the amount of light absorbed by the stain. However, this method can be influenced by chromatin condensation since histones, in particular, restrict the accessibility of DNA to fluorochromes and thus, the stoichiometry of DNA staining is affected. This depends on cell types but also on the fluorochromes.
The use of next-generation genome sequencing techniques has provided information on genome size, organization, and composition in an increased number of species. However, GC-rich regions or arrays of repetitive DNA are under-represented in assemblies (Peona et al. 2018). Until sequencing technologies will not allow obtaining scaffolds which span the entire length of individual chromosomes, genome assemblies will continue to be far from complete. Moreover, organisms such as lamprey are characterized by the physical restructuring of the genome during development that consists in the elimination of about 0.5 Gb of DNA from the 2.3 Gb genome. As a consequence, somatic cell types possess a smaller gene complement compared to germ cells possessing a full complement (Smith et al. 2018).
An understanding of the C-value paradox will only be achieved through studies on the non-coding portion of the genome; the so-called dark matter, which currently, given the technical difficulties in identifying and understanding its function, is a subject of interest for many research groups (Blaxter 2010; Kapranov and Laurent 2012).
1.1 Types of Repetitive DNA Sequences
Repetitive DNA includes sequences present in multiple copies in the genome and can account for up to 90% of the genome size in some species (Biscotti et al. 2015a; Lopez-Flores and Garrido-Ramos 2012).
This DNA, together with sequences removed from mRNA before translation (5′ and 3′ untranslated regions and introns) and pseudogenes, is part of non-coding DNA (Fig. 12.1).
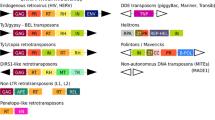
Repetitive DNA is constituted by sequences repeated thousands of times that can be grouped into two main types: transposable elements (TEs) and tandem repeats (Fig. 12.1).
Among repetitive sequences, TEs are mostly responsible for the pronounced differences in genome size (Garrido-Ramos 2017). They are genetic elements characterized by their ability to insert themselves in novel genome locations of the host and to increase in number by replication. On the basis of their transposition mechanism, TEs can be further distinguished into (i) Class I retroelements or retrotransposons and (ii) Class II DNA transposons (Goerner-Potvin and Bourque 2018; Bourque et al. 2018).
Retroelements (Class I) are provided by an RNA intermediate that is then reverse transcribed into complementary DNA using a copy and paste mechanism. In Class I, long terminal repeat (LTR) retrotransposons and non-LTR (non-LTR) retrotransposons can both be found.
LTR retrotransposons are characterized by direct LTR-flanking sequences of about 250–600 bp, necessary for the transcription and consequent insertion into the host genome. LTR retrotransposons are structurally very similar to retroviruses with the exception of the envelope gene that is only present in retroviruses (Naville et al. 2016). Moreover, these elements, unlike retroviruses, are not able to move between cells and to infect them (Malik et al. 2000; Ribet et al. 2008). Besides direct LTR-flanking sequences, LTR retrotransposons are constituted by some genes, essential for the complete synthesis of all the components of reverse transcriptase machinery: gag protein, reverse transcriptase (RT), protease (prt), RNAse H, and integrase (int). After the RT-mediated cDNA synthesis, integrase inserts the cDNA into a new position of the genome. The subclassification of LTR retrotransposons includes three main TE superfamilies in vertebrates: Ty1/Copia (Pseudoviridae), Ty3-gypsy-like (Metaviridae), and BEL/Pao (Chalopin et al. 2015).
Non-LTR retroelements are defined as autonomous retrotransposons and are mainly represented by long interspersed nuclear elements (LINEs) and short interspersed nuclear elements (SINEs). LINEs are non-LTR retrotransposons that may contain one or two open reading frames (ORFs) (Wicker et al. 2007). One of the two ORFs encodes for a reverse transcriptase (RT) and an endonuclease (en) domain encoding for a protein responsible for integration of the TE copy in a different region of the genome. On the contrary, SINEs are non-autonomous retrotransposons and do not code for a RT, thus they need LINE reverse transcriptase in order to be transposed (Kramerov and Vassetzsky 2011). The origin of SINEs can be ascribed to the reverse transcription of Pol III transcripts (Lopez-Flores and Garrido-Ramos 2012).
Class II DNA transposons are transposed by moving their genomic DNA copies from one chromosomal location to another without any RNA intermediates (Goerner-Potvin and Bourque 2018; Bourque et al. 2018) and can be divided into subclasses I and II. In subclass I, Crypton elements and terminal inverted repeat (TIR) transposons can be found. For these elements, both DNA strands are cleaved and transposed following the canonical cut and paste mechanism of transposition. In this case, the number of these elements remains unchanged. In subclass II, the major representatives are Helitrons and Maverick/Polinton elements in which the transposition follows the copy and paste mechanism (Wicker et al. 2007; Kapitonov and Jurka 2008). Class II also comprises MITEs, non-autonomous transposons originated from DNA transposons, not encoding for a transposase and therefore unable to copy themselves autonomously. Thus, they exploit transposase encoded by autonomous elements in order to transpose (Feschotte et al. 2003).
Beside TEs, repetitive DNA includes tandem repeats that are constituted by satellite DNAs (satDNAs), minisatellites, and microsatellites (Fig. 12.1). The former are highly repetitive DNA sequences contributing considerably to the C-value (Biscotti et al. 2015a) while the latter show a lower number of repeats and are classified as moderately repeated sequences. However, there are some exceptions, as found in snakes which are characterized by a great number of minisatellites and microsatellites (Adams et al. 2016; Pasquesi et al. 2018). SatDNAs are organized as long arrays of head-to-tail linked repeats and are mainly localized at telomeric, centromeric, or pericentromeric level. Their preferential localization may suggest a possible involvement in biological or functional processes (Biscotti et al. 2015b) such as centromeric DNA packaging (Heslop-Harrison and Schwarzacher 2011; Levitsky et al. 2013), chromosome segregation, and kinetochore formation (Schueler et al. 2001). These sequences may be characterized by a high dynamism so as to be species or genus-specific (Garrido-Ramos 2015) but may also be conserved in some lineages for long evolutionary periods (Chaves et al. 2017; Mravinac et al. 2005; Petraccioli et al. 2015; Biscotti et al. 2018).
2 Genome Size in Vertebrates: An Overview
The genome size database (http://www.genomesize.com/) contains information about the DNA content of 6222 species of which 3793 are vertebrates (Gregory 2019). Currently, the genome size of the smallest animal is estimated as 0.02 pg/N and belongs to the plant-parasitic nematode Pratylenchus coffeae while the largest animal genome is found in the marbled lungfish Protopterus aethiopicus and is estimated as 132.83 pg/N. Changes in genome size are mainly ascribable to the gain or loss of repetitive DNA. The advent of high throughput sequencing technologies has led to an increase in the number of sequenced genomes and this has been extremely important for extending our knowledge on the amount and the landscape of TEs in several species.
Although data available on primitive metazoans are limited, they seem to suggest a low genome size and probably a low percentage of transposons at the origin of metazoans. In protostomes, the variability in genome size is mainly related to the expansion of various classes of transposons (Canapa et al. 2015). Indeed, a positive correlation exists between genome size and the percentage of transposons. Most of the data on protostomes derives from species belonging to the Arthropoda phylum. Within insects, genome size is mainly comprised between 0.09 and 4 pg/N, with the exception of orthopterans that reach 16.93 pg/N. These differences are attributable to the percentage of transposons showing variable rates of amplification in the different groups of insects (Canapa et al. 2015). Although in invertebrates, genome size rarely exceeds 20 pg/N, in crustaceans, some species have genomes of over 50 pg/N. It has been noted that such values are restricted to species adapted to extreme environments, such as polar regions or hydrothermal vents (Bonnivard et al. 2009; Dufresne and Jeffery 2011).
Among deuterostomes, primitive chordates (urochordates and cephalochordates) present smaller genomes than vertebrates. This seems to be related to the whole-genome duplication events (WGDs) that have affected vertebrates during evolution. These events determined the appearance of new features leading to an increase in complexity. However, the variation in genome size does not follow a common trend among the different vertebrate lineages. Indeed, comparing the estimated genome size among vertebrates, some classes experienced expansions while others experienced strong contractions (Fig. 12.2). For example, the genome of the teleost fish Takifugu rubripes is one-eighth of that of Homo sapiens even if both contain a comparable number of protein-coding genes (Aparicio et al. 2002); amphibians and lungfish show the widest range of genome size variation (Fig. 12.2) and both taxa have organisms with the largest genomes among vertebrates.
DNA content in vertebrate lineages. On the left: the evolutionary relationships between vertebrate lineages. On the right: bars indicating the range of DNA amount for each lineage. Orange branches represent jawless fishes; green branch represents Chondrichthyes; blue branches represent Osteichthyes. The scale indicates the amount of DNA in pg/nucleus. Note that for coelacanths, only one species has been analyzed but with different methods. This justifies the presence of a range of C-values in the figure
Regarding agnathes, the genome size data are available for seven species of Myxiniformes and for 10 species of Petromyzontiformes (Gregory 2019). These organisms have moderate genomes ranging from 1.29 to 4.59 pg/N, thus ranking after birds among vertebrates (Fig. 12.2). Data on transposon contribution are available only for the sea lamprey Petromyzon marinus for which the analysis of the sequenced genome revealed that 34.7% is made up of mobile elements (Smith et al. 2013). More than 20% of these mobile elements are unknown while the remaining portion (about 15%) is constituted by LINEs, LTRs, and DNA transposons (Chalopin et al. 2015).
Considerable genomic dimensions are reported for some species of cartilaginous fish, reaching 17 pg/N. The analysis of the sequenced genome of the elephant shark Callorhinchus milii demonstrated that more than 40% is composed of transposons with a major contribution of LINE retroelements (Chalopin et al. 2015; Venkatesh et al. 2014).
Actinopterygians comprise of Polypteriformes, Acipenseriformes, holostei, and teleosts. The latter is one of the most successful groups, representing more than 99.8% of ray-finned fishes. The genome size of bony fish is comprised between 0.34 and 9.32 pg/N, including organisms with the most compact genome among vertebrates, such as some species belonging to the Tetraodontidae family (Gregory 2019). In this lineage, the link between genome size variation and transposon contribution is particularly evident. Indeed, within actinopterygians, the content of mobile elements is extremely variable, ranging between a very low amount in puffer fish (about 6%) to more than 50% in zebrafish. Moreover, ray-finned fish genomes present a higher mobile element diversity than other vertebrate lineages which is also maintained in the most compact genomes. Overall, in teleosts, the major contribution is provided by DNA transposons (Chalopin and Volff 2017) while compact genomes do not show a prevalence of any transposon type. In non-teleost species, such as the spotted gar Lepisosteus oculatus, a predominance of non-LTR retrotransposons is evident (Chalopin et al. 2015; Chalopin and Volff 2017).
Lobe-finned fish includes two species of coelacanths (Latimeria chalumnae and Latimeria menadoensis) and six species of lungfish (Protopterus annectens, Protopterus dolloi, Protopterus aethiopicus, Protopterus amphibius, Lepidosiren paradoxa, and Neoceratodus forsteri). While the former presents a moderate amount of DNA of about 3.5 pg/N, the latter have a genome size ranging from 40 to 132.83 pg/N. The contribution of transposons has been well evaluated in L. chalumnae for which genome sequencing is available (Amemiya et al. 2013). Analyses performed on the genome of this taxon revealed that 20% is made up of transposons with about one-third of SINEs (Chalopin et al. 2015). In lungfish, the huge genome size represents a drawback for current sequencing techniques and assembly procedures. However, a study performed on a small portion of the N. forsteri genome estimated that 40% of the genome is made up of transposons and suggested that CR1 and L2 (non-LTR) are predominant (Metcalfe et al. 2012). These data obtained in basal sarcopterygians at genome level reflect the results obtained by analyzing the activity of mobile elements in the transcriptomes of the Indonesian coelacanth L. menadoensis (Forconi et al. 2014) and the West African lungfish P. annectens (Biscotti et al. 2016).
Among vertebrates, the Amphibia class shows the widest range of genome size from 0.95 to 120.60 pg/N (Fig. 12.2). Most of the analyzed species belong to the Anura and Urodela orders, while only three species have been investigated for the Gymnophiona order (Gregory 2019). In Anura and Gymnophiona, the genome size does not exceed 14 pg/N unlike Urodela in which the values range from 10.12 to 120.60 pg/N in species belonging to the Proteidae family (Gregory 2019). Mobile elements constitute from 20% to over 40% of the genome with a predominance of DNA transposons in Xenopus tropicalis (Chalopin et al. 2015; Sun et al. 2012, 2015) and of LTR in urodeles (Canapa et al. 2015; Nowoshilow et al. 2018) and in the Tibetan frog Nanorana parkeri (Sun et al. 2015). However, in amphibians, the genomic gigantism observed is not only due to the higher amount of repetitive DNA but also to longer introns, as found mainly in salamanders (Sun et al. 2012; Voss et al. 2013; Nowoshilow et al. 2018).
The genome size of the 420 non-bird reptile species analyzed to date ranges from 1.05 to 5.44 pg/N. In Squamata and Crocodylia, values are comprised between 1.05 and 3.95 pg/N while in Testudines and in the unique analyzed species of Sphenodontia, the genome size exceeds 4.00 pg/N reaching the value of 5.44 pg/N in Testudo graeca. In Squamata and Crocodylia, about 30% of the genome is TE-derived and the major contribution is ascribable to non-LTR and DNA transposons (Alföldi et al. 2011; Green et al. 2014; Castoe et al. 2011, 2013). The genomes of turtles sequenced to date indicate that around 10% is represented by TEs and that non-LTR retrotransposons constitute the predominant part of the mobilome, as is the case for the other two orders (Shaffer et al. 2013; Wang et al. 2013).
Birds present the smallest genomes among vertebrates (Fig. 12.2). Indeed, the 898 species included in the genome size database show values comprised between 0.91 and 2.16 pg/N (Gregory 2019). The compressed genomes of birds are characterized by a very low number of mobile elements with the loss of certain TE families; however, their mobilome is mainly constituted by retroelements (Chalopin et al. 2015). Studying fossil cell size in dinosaurs, the contraction of the genome size can be dated at 230–250 Mya in saurischians, the lineage from which birds originated. Comparative genomic analyses on the current descendants of this evolutionary lineage showed that the reduction observed in saurischians might be due to a strong reduction in non-LTR elements (Organ et al. 2007). It has been proposed that the genome size between 2.5 and 5.0 pg/N represents the ancestral condition of the sarcopterygian lineage and consequently, the large genomes of lungfish and salamanders, together with the contracted genomes of birds, are secondarily derived (Organ et al. 2011). Moreover, it is worth noting that the increase in the amount of DNA experienced in dipnoans and amphibians accompanied their transition from water to land life which, being one of the most important steps during vertebrate evolution, probably required significant changes at genome level.
Finally, the genome size of mammals ranges from 1.63 to 8.40 pg/N (Gregory 2019) and the TE content accounts for more than 30% of the mammalian genomes sequenced to date. Non-LTR elements are the most prominent type even if in therians (Metatheria and Eutheria) there is a prevalence of L1 LINE retroelements while in monotremes there is a predominance of L2 LINE retroelements (Chalopin et al. 2015). Moreover, in mammals, most species having small genomes (less than 2.5 pg/N of DNA) belong to the Chiroptera order. These organisms are the only group of mammals to have evolved powered flight and, interestingly, they are characterized by reduced genomes, similar to birds. However, while in the latter, there is a prevalence of retroelements, and in bats, there is an accumulation of DNA transposons (Pagán et al. 2012; Ray et al. 2007).
In conclusion, the general trend that can be extrapolated is that retroelements have shaped the genomes of jawless fish, cartilaginous fish, coelacanths, lungfish, birds, and mammals while DNA transposons have played a key role in ray-finned fish and Xenopus genome size.
Moreover, if some lineages are characterized by high transposon diversity others have experienced a reduction in retroelement diversity with the complete extinction of some families in certain lineages. Indeed, in mammals, only three families of retrotransposons are present: the non-LTR retrotransposons L1 (LINE1), L2, and L3/CR1 while in birds, L1 and L2 have been completely lost (Wicker et al. 2005). The extinction of ancient families of TEs has also been identified in teleosts, in which the non-LTR retrotransposon Rex3 is widespread, but not in salmonids (Volff et al. 2001; Carducci et al. 2018).
It is also interesting to note that the impact of the same TE family can be very different: the L1 family is highly dispersed in mammalian genomes while a much lower copy number is present in fish genomes (Volff et al. 2003, Furano et al. 2004); the L3/CR1 family is the major group of TEs in birds with 96,000 copies compared to the larger genome of placental mammals with only 8000 copies (Wicker et al. 2005).
Finally, some TEs were not present in the common ancestor of vertebrates but were introduced/originated ex novo in some lineages. This is the case of Alu elements derived from 7SL RNA, or SVA elements originated from Alu and SINE-R. Both these elements are non-autonomous retroelements specific of primates.
Overall, the TE content in a given species could be the result of an equilibrium between TE transposition, defense mechanisms of the genome, and natural selection constraints allowing genome functionality to be maintained.
The determination of the TE landscape in a genome depends on the methods used to identify and annotate TEs. This issue requires considerable efforts due to the great variability of TEs and to the accumulation of mutations in old and inactive TE sequences. Currently, there is no reliable strategy to overcome this problem. However, three approaches are commonly used: library-based methods, signature-based methods, and de novo consensus methods (Goerner-Potvin and Bourque 2018; Lerat 2010; Girgis 2015; Tempel 2012). The library-based methods use Repeat-Masker program, usually in association with Repbase. The signature-based methods identify specific traits such as long terminal repeats. The methods based on de novo consensus such as the REPET package combine both the previous strategies. Each of these approaches presents advantages and disadvantages and thus, different strategies or pipelines have been developed to improve TE annotation (Guizard et al. 2016; Su et al. 2019).
3 The Impact of Transposable Elements on Genome Evolution: Adaptations and Speciation
Vertebrates represent a highly successful lineage that has adapted to a wide range of different environments from arid deserts to cold polar regions or from high altitudes to deep oceans. They originated during the Cambrian explosion and the appearance of relevant innovations allowed the colonization of a broad range of biotopes.
The comprehension of the evolution of organisms is based on knowledge of the functional and structural complexity of their genomes. Given the significant impact of TEs in genome plasticity, it is mandatory to get information on content, diversity, distribution, and the family abundance of mobile elements.
In this context the sequenced genomes available allowed comparative analyses to be performed and the evolution of vertebrates (Fig. 12.3), the origin, and the successful diversification of these organisms to be traced.
TEs are widely present in the genomes of mammals, non-bird reptiles, sharks, lamprey, and some fish but are poorly represented in the genomes of birds and some fish, such as puffer fish. In fact, mammals contain 10 times more TEs than birds; the zebrafish genome is composed of 55% TEs while Tetraodon has less than 6%. Although all types of TEs have been identified in vertebrates, the composition, the copy number and the age of the mobile elements are extremely variable (Chalopin et al. 2015; Warren et al. 2015). TE diversity decreases from basal sarcopterygians to mammals. Indeed, few TE superfamilies are present in the genomes of mammals and birds, unlike reptiles and amphibians.
Within superfamilies, the human genome contains 20% of L1 while the zebrafish genome harbors more than 30 different L1 families even if with a lower copy number (Furano et al. 2004). The prevalence of a specific TE family could be due to competition, rate of transposition, rate of DNA elimination, population size, mode of reproduction, and host defense mechanisms. Moreover, horizontal transfer can also affect TE diversity, leading to the insertion of mobile elements from distant species into a new genome.
Overall, these observations indicate that TEs might have had a different impact on genome evolution in various lineages. Indeed, it is well-known that TEs are responsible for the origin of key adaptations leading to evolutionary advantages and the success of host species (Chalopin et al. 2015; Warren et al. 2015) and thus could be among the main drivers of speciation and major evolutionary transitions. Notably, one of the most important events in the evolution of vertebrates was the transition from water to land life that was accompanied by drastic changes in genome size and in the percentage of TEs, as observed in lungfish and salamanders.
Given their activity, TEs play a key role in genome organization through chromosomal rearrangements such as deletions, inversions, translocations, and duplication events (Fig. 12.4) that have provoked a rapid evolution of a specific lineage followed by reproductive isolation, thereby, determining species diversification (Rebollo et al. 2010).
Main effects of TE transposition activity. Positive effects are included in the green box while negative effect in blue box. On the upper right side, evolutionary advantages due to positive effects are indicated; on the lower right side, the defense mechanisms adopted by the host against the negative effect of TE transposition are listed
Moreover, mobile elements have significantly contributed to the complexity of vertebrate transcriptome and proteome (Horie et al. 2007). In fact, several reports have discussed the ability of TEs to generate regulatory elements, genetic novelties, and functional innovations (Fig. 12.4). In humans, 4% of genes contains coding sequences derived from TEs as well as 25% of promoters (Nekrutenko and Li 2001; van de Lagemaat et al. 2003).
TEs can insert near promoter regions and can be coopted to alter the gene expression of the nearby genes (Thornburg et al. 2006). Among the TE-derived regulatory sequences, the involvement of ERV elements is well-documented in the emergence of the placenta in mammals, which was one of the most important innovations in vertebrate evolution (Chuong et al. 2013). Indeed, promoters derived from these mobile elements trigger the expression of placenta-specific genes.
The neuronal enhancer responsible for the expression of the proopiomelanocortin gene (POMC) is responsible for encoding the prohormone of the adenocorticotropic hormone, the melanocyte-stimulating hormone, and endorphin derived from a SINE retroelement in mammals. The absence of this element in other non-mammalian vertebrates suggests that this event occurred in the common ancestor of placentals, marsupials, and monotremes (Santangelo et al. 2007). Another example of lineage-specific recruitment of regulatory sequences from TEs is the enhancer derived from a LF-SINE that controls the expression of the neurodevelopmental gene ISL1 encoding a LIM homeobox transcription factor required for motor neuron differentiation. This regulatory element has been found in mammals, chicken, and frogs suggesting that the co-option event occurred in the common ancestor of tetrapods (Bejerano et al. 2006).
In addition, TEs contribute to the occurrence of new exons in an existing host-coding protein through a process called exonization (Sorek 2007) and this seems to be very frequent in humans in which this process is mainly due to Alu elements (Sela et al. 2010). The insertion of these elements occurs preferentially at the beginning of the coding sequence in both human and mouse genes. Moreover, the analysis of SNPs has highlighted a population-specific pattern indicating that exonization may enhance divergence and thus speciation (Sela et al. 2010). Similarly, in primates, the histone methyltransferase SETMAR presents an exon derived from a mariner-like DNA transposon (Cordaux et al. 2006).
TEs can generate not only new exons but also entire new coding genes through a process called molecular domestication or exaptation. The emergence of new genes enriches the gene repertoire in genomes and thus represents an important contribution to the evolution of organisms. The immune system adaptability of jawed vertebrates took advantage of the emergence of the genes RAG1 and RAG2 derived from a transposon about 500 Mya. Indeed, these genes encode the recombinase which catalyzes the V(D)J recombination responsible for the generation of a wide repertoire of antibodies (Kapitonov and Jurka 2005; Schatz and Swanson 2011). A key event in the evolution of mammals is certainly represented by the emergence of the placenta, a specialized organ whose purpose is to guarantee the exchange of water, nutrients, and gasses between the mother and the developing fetus. Several genes expressed in this structure are derived from retrotransposons (Henke et al. 2015). One example is represented by the genes syncytin-1 and syncytin-2 that derived from the exaptation of retroviral env genes and are involved in cell–cell fusion and in the differentiation of the trophoblast layer in the placenta (Vernochet et al. 2014). Moreover, the mammalian protein CENP-B that binds the centromeric 17 base-pair CENP-B box derived from a pogo-like transposase before the divergence of placental mammals, marsupials, and monotremes (Casola et al. 2008).
Polyploidization represents a drastic event that is accompanied by substantial rearrangements useful for bringing the genome back to a state of diploidy. Although the mechanisms involved in this reinstatement are not completely understood, large scale movements, in particular, due to TEs, have been hypothesized as playing a major role in shaping genomes. Therefore, these observations suggest that polyploidization is associated with bursts of TE activity (Matzke and Matzke 1998). Moreover, these events have led to an increase in gene redundancy and less selective constraints against insertional mutagenesis resulting in an increase in TE content. In vertebrates, two rounds of whole-genome duplication are known to have occurred in the agnathe-gnathostome ancestor after the divergence from urochordates and cephalochordates, a third event occurred in teleost ancestor and a fourth in salmonids (Allendorf and Thorgaard 1984) (Fig. 12.3). Analyses have shown that bursts of transposon activity took place after genome duplication in salmonids and coincide with speciation events in this lineage (de Boer et al. 2007).
Mobile elements have also been proposed as responsible for the rapid adaptation of invasive species to new environments despite the reduction in genetic variation characterizing these species as a result of a genetic bottleneck. In fact, adaptation to novel habitats represents a stress condition that induces changes in the epigenetic control of TEs; consequently, TE transposition is altered and mobile elements contribute to increase genetic diversity (Stapley et al. 2015).
Despite the positive effects that TEs may have for the host genome, their movement could have strongly deleterious consequences and therefore, organisms have developed various mechanisms to control TE activity (Fig. 12.4). Mobile elements can be inactivated by methylation and/or interference of small RNAs such as piwi-interacting RNAs (Malone and Hannon 2009; Biscotti et al. 2017). However, under stress conditions, these mechanisms can be neutralized leading to an increase in TE activity (Piacentini et al. 2014).
The resolution of the C-value paradox is an ambitious challenge that many research groups are addressing in order to unravel why a parsimonious energy system such as the cellular one can tolerate a great amount of repetitive DNA. The advent of next-generation sequencing technologies has certainly provided a greater availability of genomic data that, as discussed in the present chapter, have strongly contributed to gaining insight into the functional, structural, and evolutionary meaning of repetitive DNA.
The ever-increasing number of available transcriptomes, together with more accurate annotations of TEs, will also allow information to be obtained on the transcriptional activity of the mobilome.
Abbreviations
- 7SL RNA:
-
eukaryotic small cytoplasmic RNA
- Alu :
-
Arthrobacter luteus restriction endonuclease
- CR1:
-
Chicken Repeat 1
- en:
-
endonuclease
- env:
-
envelope
- ISL-1:
-
Insulin gene enhancer protein ISL-1
- L1:
-
LINE1
- L2:
-
LINE2
- L3:
-
LINE3
- LINE:
-
Long Interspersed Nuclear Elements
- LTR:
-
Long Terminal Repeat
- MITEs:
-
Miniature Inverted-repeated Transposable Elements
- pg/N:
-
picograms/Nucleus
- POMC:
-
ProopioMelanoCortin gene
- RAG1 :
-
Recombination-ActivatinG Protein 1
- RAG2 :
-
Recombination-ActivatinG Protein 2
- RT:
-
Reverse Transcriptase
- SatDNA:
-
Satellite DNA
- SINE:
-
Short Interspersed Nuclear Elements
- SINE-R:
-
Short Interspersed Nuclear Elements-R, where R indicates a sequence of Retroviral origin
- SNPs:
-
Single Nucleotide Polymorphisms
- SETMAR:
-
SET domain and Mariner transposase fusion gene
- SVA:
-
SINE-VNTR-Alu
- TEs:
-
Transposable Elements
- TIR:
-
Terminal Inverted Repeat
- UTRs:
-
UnTranslated Regions
- VDJ:
-
Variable Diversity Joining
- VNTR:
-
Variable Number of Tandem Repeats
References
Adams RH, Blackmon H, Reyes-Velasco J et al (2016) Microsatellite landscape evolutionary dynamics across 450 million years of vertebrate genome evolution. Genome 59:295–310
Alföldi J, Di Palma F, Grabherr M et al (2011) The genome of the green anole lizard and a comparative analysis with birds and mammals. Nature 477:587–591
Allendorf FW, Thorgaard GH (1984) Tetraploidy and the evolution of salmonid fishes. In: Turner BJ (eds) Evolutionary genetics of fishes. Monographs in evolutionary biology. Springer, Boston, MA
Amemiya CT, Alfoldi J, Lee AP et al (2013) The African coelacanth genome provides insight into tetrapod evolution. Nature 496:311–316
Aparicio S, Chapman J, Stupka E et al (2002) Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science 297:1301–1310
Bejerano G, Lowe CB, Ahituv N et al (2006) A distal enhancer and an ultraconserved exon are derived from a novel retroposon. Nature 441:87–90
Bennett MD (1971) The duration of meiosis. Proc R Soc Lond B Biol Sci 178:259–275
Biemont (2010) A brief history of the status of transposable elements: from junk DNA to major players in evolution. Genetics 186:1085–1093
Biscotti MA, Olmo E, Heslop-Harrison JS (2015a) Repetitive DNA in eukaryotic genomes. Chromosome Res 23:415–420
Biscotti MA, Canapa A, Forconi M, Olmo E, Barucca M (2015b) Transcription of tandemly repetitive DNA: functional roles. Chromosome Res 23:463–477
Biscotti MA, Gerdol M, Canapa A et al (2016) The lungfish transcriptome: a glimpse into molecular evolution events at the transition from water to land. Sci Rep 6:21571
Biscotti MA, Canapa A, Forconi M et al (2017) The small noncoding RNA processing machinery of two living fossil species, lungfish and coelacanth, gives new insights into the evolution of the argonaute protein family. Genome Biol Evol 9:438–453
Biscotti MA, Barucca M, Canapa A (2018) New insights into the genome repetitive fraction of the Antarctic bivalve Adamussium colbecki. PLoS ONE 13(3):e0194502
Blaxter M (2010) Genetics. Revealing the dark matter of the genome. Science 330:1758–1759
Bonnivard E, Catrice O, Ravaux J et al (2009) Survey of genome size in 28 hydrothermal vent species covering 10 families. Genome 52:524–536
Bourque G, Burns KH, Gehring M et al (2018) Ten things you should know about transposable elements. Genome Biol 19:199
Britten RJ, Graham DE, Neufeld BR (1974) Analysis of repeating DNA sequences by reassociation. Method Enzymol 29:363–405
Burton DW, Bickham JW, Genoways HH (1989) Flow-cytometric analyses of nuclear DNA content in four families of neotropical bats. Evolution 43:756–765
Canapa A, Barucca M, Biscotti MA et al (2015) Transposons, genome size, and evolutionary insights in animals. Cytogenet Genome Res 147:217–239
Carducci F, Barucca M, Canapa A et al (2018) Rex retroelements and teleost genomes: an overview. Int J Mol Sci 19:11
Casola C, Hucks D, Freschotte C et al (2008) Convergent domestication of pogo-like transposases into centromere-binding proteins in fission yeast and mammals. Mol Biol Evol 25:29–41
Castoe TA, de Konig AP, Hall KT et al (2013) The Burmese python genome reveals the molecular basis for extreme adaptation in snakes. Proc Natl Acad Sci USA 110:20645–20650
Castoe TA, Hall KT, Guibotsy Mboulas ML et al (2011) Discovery of highly divergent repeat landscapes in snake genomes using high-throughput sequencing. Genome Biol Evol 3:641–653
Cavalier-Smith T (1978) Nuclear volume control by nucleoskeletal DNA, selection for cell volume and cell growth rate, and the solution of the DNA C-value paradox. J Cell Sci 43:247–278
Chalopin D, Naville M, Plard F et al (2015) Comparative analysis of transposable elements highlights mobilome diversity and evolution in vertebrates. Genome Biol Evol 7:567–580
Chalopin D, Volff JN (2017) Analysis of the spotted gar genome suggests absence of causative link between ancestral genome duplication and transposable element diversification in teleost fish. J Exp Zool B Mol Dev Evol 328:629–637
Chaves R, Ferreira D, Mendes-da-Silva et al (2017) FA-SAT is an old satellite DNA frozen in several bilateria genomes. Genome Biol Evol 9:3073–3087
Chuong EB, Rumi MAK, Soares MJ et al (2013) Endogenous retroviruses function as species-specific enhancer elements in the placenta. Nat Genet 45:325–329
Cordaux R, Udit S, Batzer MA et al (2006) Birth of a chimeric primate gene by capture of the transposase gene from a mobile element. Proc Natl Acad Sci USA 103:8101–8106
de Boer JG, Yazawa R, Davidson WS et al (2007) Bursts and horizontal evolution of DNA transposons in the speciation of pseudotetraploid salmonids. BMC Genom 8:422
Doolittle WF, Sapienza C (1980) Selfish genes, the phenotype paradigm and genome evolution. Nature 284:601–603
Dufresne F, Jeffery N (2011) A guided tour of large genome size in animals: what we know and where we are heading. Chromosome Res 19:925–938
Feschotte C, Swamy L, Wessler SR (2003) Genome-wide analysis of mariner-like transposable elements in rice reveals complex relationships with stowaway miniature inverted repeat transposable elements (MITEs). Genetics 163:747–758
Forconi M, Chalopin D, Barucca M et al (2014) Transcriptional activity of transposable elements in coelacanth. J Exp Zool B Mol Dev Evol 322:379–389
Furano AV, Duvernell DD, Boissinot S (2004) L1 (LINE-1) retrotransposon diversity differs dramatically between mammals and fish. Trends Genet 20:9–14
Garrido-Ramos MA (2015) Satellite DNA in plants: more than just rubbish. Cytogenet Genome Res 146:153–170
Garrido-Ramos MA (2017) Satellite DNA: an evolving topic. Genes (Basel) 8(9)
Girgis HZ (2015) Red: an intelligent, rapid, accurate tool for detecting repeats de-novo on the genomic scale. BMC Bioinf 16:227
Goerner-Potvin P, Bourque G (2018) Computational tools to unmask transposable elements. Nat Rev Genet 19:688–704
Green RE, Braun EL, Armstrong J et al (2014) Three crocodilian genomes reveal ancestral patterns of evolution among archosaurs. Science 346:1254449
Gregory TR (2019) Animal genome size database. http://www.genomesize.com
Guizard S, Piégu B, Arensburger P et al (2016) Deep landscape update of dispersed and tandem repeats in the genome model of the red jungle fowl, Gallus gallus, using a series of de novo investigating tools. BMC Genom 17(1):659
Henke C, Strissel PL, Schubert MT et al (2015) Selective expression of sense and antisense transcripts of the sushi-ichi-related retrotransposon-derived family during mouse placentogenesis. Retrovirology 12:9
Heslop-Harrison JS, Schwarzacher T (2011) Organisation of the plant genome in chromosomes. Plant J 66:18–33
Horie K, Saito ES, Keng VW et al (2007) Retrotransposons influence the mouse transcriptome: implication for the divergence of genetic traits. Genetics 176:815–827
Hughes AL, Hughes MK (1995) Small genomes for better fliers. Nature 377:391
Kapitonov VV, Jurka J (2005) RAG1 core and V(D)J recombination signal sequences were derived from Transib transposons. PLoS Biol 3:e181
Kapitonov VV, Jurka J (2008) A universal classification of eukaryotic transposable elements implemented in Repbase. Nat Rev Genet 9:411–412
Kapranov P, Laurent GS (2012) Genomic ‘dark matter’: implications for understanding human disease mechanisms, diagnostics, and cures. Front Genet 3:95
Kraaijeveld K (2010) Genome size and species diversification. Evol Biol 37:227–233
Kramerov DA, Vassetzky NS (2011) Origin and evolution of SINEs in eukaryotic genomes. Heredity 107:487–495
Lerat E (2010) Identifying repeats and transposable elements in sequenced genomes: how to find your way through the dense forest of programs. Heredity 104:520–533
Levitsky VG, Babenko VN, Vershinin AV (2013) The roles of the monomer length and nucleotide context of plant tandem repeats in nucleosome positioning. J Biomol Struct Dyn 32:115–126
López-Flores I, Garrido-Ramos MA (2012) The repetitive DNA content of eukaryotic genomes. In: Garrido-Ramos MA (ed) Repetitive DNA. Genome Dyn 7:1–28
Lynch M, Conery JS (2003) The origins of genome complexity. Science 302:1401–1404
Malik HS, Henikoff S, Eickbush TH (2000) Poised for contagion: evolutionary origins of the infectious abilities of invertebrate retroviruses. Genome Res 10:1307–1318
Malone CD, Hannon GJ (2009) Small RNAs as guardians of the genome. Cell 136:656–668
Matzke MA, Matzke AJ (1998) Polyploidy and transposons. Trends Ecol Evol 13:241
Metcalfe CJ, Filée J, Germon I et al (2012) Evolution of the Australian lungfish (Neoceratodus forsteri) genome: a major role for CR60 and L2 LINE elements. Mol Biol Evol 29:3529–3539
Mravinac B, Plohl M, Ugarković D (2005) Preservation and high sequence conservation of satellite DNAs suggest functional constraints. J Mol Evol 61:542–550
Naville M, Warren IA, Haftek-Terreau Z et al (2016) Not so bad after all: retroviruses and long terminal repeat retrotransposons as a source of new genes in vertebrates. Clin Microbiol Infect 22:312–323
Nekrutenko A, Li WH (2001) Transposable elements are found in a large number of human protein-coding genes. Trends Genet 17:619–621
Nowoshilow S, Schloissnig S, Fei JF et al (2018) The axolotl genome and the evolution of key tissue formation regulators. Nature 554:50–55
Ohno S (1972) So much ‘junk’ DNA in our genome. In: Smith HH (ed) Evolution of genetic systems. Gordon and Breach, New York
Olmo E (1983) Nucleotype and cell size in vertebrates: a review. Basic Appl Histochem 27:227–256
Organ CL, Shedlock AM, Meade A et al (2007) Origin of avian genome size and structure in non-avian dinosaurs. Nature 446:180–184
Organ CL, Canoville A, Reisz RR et al (2011) Paleogenomic data suggest mammal-like genome size in the ancestral amniote and derived large genome size in amphibians. J Evol Biol 24:372–380
Pagán HJ, Macas J, Novák P et al (2012) Survey sequencing reveals elevated DNA transposon activity, novel elements, and variation in repetitive landscapes among vesper bats. Genome Biol Evol 4:575–585
Pasquesi GIM, Adams RH, Card DC et al (2018) Squamate reptiles challenge paradigms of genomic repeat element evolution set by birds and mammals. Nat Commun 9:2774
Peona V, Weissensteiner MH, Suh A (2018) How complete are “complete” genome assemblies?-An avian perspective. Mol Ecol Resour 18(6):1188–1195
Petraccioli A, Odierna G, Capriglione T et al (2015) A novel satellite DNA isolated in Pecten jacobaeus shows high sequence similarity among molluscs. Mol Genet Genomics 290:1717–1725
Petrov DA (2002) Mutational equilibrium model of genome size evolution. Theor Popul Biol 61:531–544
Piacentini L, Fanti L, Specchia V et al (2014) Transposons, environmental changes, and heritable induced phenotypic variability. Chromosoma 123:345–354
Ray DA, Pagan HJ, Thompson ML et al (2007) Bats with hATs: evidence for recent DNA transposon activity in genus Myotis. Mol Biol Evol 24:632–639
Ribet D, Harper F, Esnault C et al (2008) The GLN family of murine endogenous retroviruses contains an element competent for infectious viral particle formation. J Virol 82:4413–4419
Rebollo R, Horard B, Hubert B et al (2010) Jumping genes and epigenetics: towards new species. Gene 454:1–7
Santangelo AM, de Souza FSJ, Franchini LF et al (2007) Ancient exaptation of a CORE–SINE retroposon into a highly conserved mammalian neuronal enhancer of the proopiomelanocortin gene. PLoS Genet 3:1813–1826
Schatz DG, Swanson PC (2011) V(D)J recombination: mechanisms of initiation. Annu Rev Genet 45:167–202
Schueler MG, Higgins AW, Rudd MK, Gustashaw K, Willard HF (2001) Genomic and genetic definition of a functional human centromere. Science 294:109–115
Sela N, Mersch B, Hotz-Wagenblatt A et al (2010) Characteristics of transposable element exonization within human and mouse. PLoS ONE 5:e10907
Shaffer HB, Minx P, Warren DE et al (2013) The western painted turtle genome, a model for the evolution of extreme physiological adaptations in a slowly evolving lineage. Genome Biol 14:R28
Smith JJ, Kuraku S, Holt C et al (2013) Sequencing of the sea lamprey (Petromyzon marinus) genome provides insights into vertebrate evolution. Nat Genet 45:415–421
Smith JJ, Timoshevskaya N, Ye C et al (2018) The sea lamprey germline genome provides insights into programmed genome rearrangement and vertebrate evolution. Nat Genet 50(2):270–277
Sorek R (2007) The birth of new exons: mechanisms and evolutionary consequences. RNA 13:1603–1608
Stapley J, Santure AW, Dennis SR (2015) Transposable elements as agents of rapid adaptation may explain the genetic paradox of invasive species. Mol Ecol 24:2241–2252
Su W, Gu X, Peterson T (2019) TIR-learner, a new ensemble method for TIR transposable element annotation, provides evidence for abundant new transposable elements in the maize genome. Mol Plant 12:447–460
Sun C, Lopez Arriaza JR, Mueller RL (2012) Slow DNA loss in the gigantic genomes of salamanders. Genome Biol Evol 4:1340–1348
Sun YB, Xiong ZJ, Xiang XY et al (2015) Whole–genome sequence of the Tibetan frog Nanorana parkeri and the comparative evolution of tetrapod genome. Proc Natl Acad Sci USA 112:1257–1262
Swift H (1950) The constancy of desoxyribose nucleic acid in plant nuclei. Proc Natl Acad Sci USA 36:643–654
Tempel S (2012) Using and understanding RepeatMasker. Methods Mol Biol 859:29–51
Thomas CA Jr (1971) The genetic organization of chromosomes. Annu Rev Genet 5:237–256
Thornburg BG, Gotea V, Makalowski W (2006) Transposable elements as a significant source of transcription regulating signals. Gene 365:104–110
Van den Bussche RA, Longmire JL, Baker RJ (1995) How bats achieve a small C-value: frequency of repetitive DNA in Macrotus. Mamm Genome 6:521–525
van de Lagemaat LN, Landry JR, Mager DL et al (2003) Transposable elements in mammals promote regulatory variation and diversification of genes with specialized functions. Trends Genet 19:530–536
Vendrely R, Vendrely C (1948) La teneur du noyau cellulaire en acide désoxyribonucléique à travers les organes, les individus et les espèces animales: techniques et premiers résultats. Experientia 4:434–436
Venkatesh B, Lee AP, Ravi V et al (2014) Elephant shark genome provides unique insights into gnathostome evolution. Nature 505:174–179
Vernochet C, Redelsperger F, Harper F et al (2014) The captured retroviral envelope syncytin-A and syncytin-B genes are conserved in the Spalacidae together with hemotrichorial placentation. Biol Reprod 91:148
Vinogradov AE (1995) Nucleotypic effect in homeotherms: body-mass-corrected basal metabolic rate of mammals is related to genome size. Evolution 49:1249–1259
Vinogradov AE (1997) Nucleotypic effect in homeotherms: body-mass independent resting metabolic rate of passerine birds is related to genome size. Evolution 51:220–225
Vinogradov AE (1998) Buffering: a possible passive-homeostasis role for redundant DNA. J Theor Biol 193:197–199
Vinogradov AE (2003) Selfish DNA is maladaptive: evidence from the plant red list. Trends Genet 19:609–614
Vinogradov AE (2004) Testing genome complexity. Science 304:389–390
Volff JN, Körting C, Froschauer A et al (2001) Non-LTR retrotransposons encoding a restriction enzyme-like endonuclease in vertebrates. J Mol Evol 52:351–360
Volff JN, Bouneau L, Ozouf-Costaz C et al (2003) Diversity of retrotransposable elements in compact pufferfish genomes. Trends Genet 19:674–678
Voss SR, Putta S, Walker JA et al (2013) Salamander Hox clusters contain repetitive DNA and expanded non-coding regions: a typical Hox structure for non-mammalian tetrapod vertebrates? Hum Genomics 7:9
Wang Z, Pascual-Anaya J, Zadissa A et al (2013) The draft genomes of soft-shell turtle and green sea turtle yield insights into the development and evolution of the turtle specific body plan. Nat Genet 45:701–706
Warren IA, Naville M, Chalopin D et al (2015) Evolutionary impact of transposable elements on genomic diversity and lineage-specific innovation in vertebrates. Chromosome Res 23:505–531
Wicker T, Sabot F, Hua-Van A et al (2007) A unified classification system for eukaryotic transposable elements. Nat Rev Genet 8:973–982
Wicker T, Robertson JS, Schulze SR et al (2005) The repetitive landscape of the chicken genome. Genome Res 15:126–136
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Biscotti, M.A., Carducci, F., Olmo, E., Canapa, A. (2019). Vertebrate Genome Size and the Impact of Transposable Elements in Genome Evolution. In: Pontarotti, P. (eds) Evolution, Origin of Life, Concepts and Methods. Springer, Cham. https://doi.org/10.1007/978-3-030-30363-1_12
Download citation
DOI: https://doi.org/10.1007/978-3-030-30363-1_12
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-30362-4
Online ISBN: 978-3-030-30363-1
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)