Abstract
Due to a high evolutionary turnover many satellite DNAs are restricted to a group of closely related species. Here we demonstrate that the satellite DNA family PSUB, abundant in the beetle Palorus subdepressus, is distributed in a low number of copies among diverse taxa of Coleoptera (Insecta), some of them separated for an evolutionary period of up to 60 Myr. Comparison of PSUB cloned from the species Tribolium brevicornis with the PSUB family previously characterized in Palorus subdepressus revealed high sequence conservation and absence of fixed species-specific mutations. The most polymorphic sites are those with ancestral mutations shared among clones of both species. Since the ancestral mutations contribute significantly to overall diversity, it could be proposed that a similar mutational profile already existed in an ancestral species. The pattern of variability along the satellite monomer is characterized by the presence of conserved and variable regions. The nonrandom pattern of variability as well as the absence of sequence divergence is also discerned for PRAT satellite DNA, cloned previously from two Palorus species and a distantly related Pimelia elevata. Since PRAT and PSUB are present in parallel in diverse taxa of Coleoptera, we propose that their long evolutionary preservation suggests a possible functional significance. This indication is additionally supported not only by the high evolutionary conservation of the sequences, but also by the presence of significantly conserved and variable regions along the monomers.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
It is proposed that biologically important elements are evolutionarily conserved and cross-species sequence comparison is used as a method for identifying sequences that are likely to be functional. Satellite DNAs are usually preserved among a limited number of closely related species and are subjected to rapid changes in sequence and in copy number (Charlesworth et al. 1994; Ugarković and Plohl 2002). Despite their high turnover, satellite DNAs act as a main centromere building element and are proposed to drive evolution of centromeric histones (Henikoff et al. 2001). However, there are examples when preservation of satellite sequences can be observed between evolutionarily very distant species or species groups, indicating possible functionality. The most striking example is the sequence of human α satellite DNA, which is found highly conserved in chicken and zebrafish (Li and Kirby 2003). This sequence is transcribed during early embryogenesis of both species and it is proposed that it may serve as a control element in gene regulation. It has recently been shown that transcripts of satellite DNAs and other repetitive sequences are functional in the form of small interfering RNAs which act as signals necessary for establishment and/or maintenance of heterochromatin in different eukaryotes (Volpe et al. 2002; Aravin et al. 2003).
Preservation of satellite DNAs for long evolutionary periods, up to 90 Myr, has been observed in some species of fish (Garrido-Ramos et al. 1999; De la Herrán et al. 2001) as well as in whales, which retain the same satellite for almost 50 Myr (Grétarsdóttir and Arnason 1992). In these species groups, satellite DNA sequences evolve gradually, accumulating mutations very slowly. Due to the low rate of turnover mechanisms, the changes in the amount of satellite DNAs are also very moderate. The remarkable antiquity of these satellite DNAs is probably related to mutation rate, which is in general low for fish and whales (De la Herrán et al. 2001; Grétarsdóttir and Arnason 1992).
Insect species from the family Tenebrionidae (Coleoptera) are characterized by a high amount of satellite DNAs, constituting up to 50% of the genome and representing the main constituents of pericentromeric and centromeric heterochromatin. The satellites are affected by turnover mechanisms such as gene conversion and unequal crossing-over, usually acting with a high frequency and resulting in a rapid change of satellite profiles among species (Ugarković and Plohl 2002). The profiles are usually characterized by a single highly abundant satellite DNA which is, in closely related species, either present in low copy number or even absent, probably as the outcome of very efficient turnover. Despite such high evolutionary dynamics of tenebrionid satellites, we have observed that some satellite DNAs, like those belonging to the genus Palorus, are preserved among congeneric species in low copy number, exhibiting no sequence divergence (Meštrović et al. 1998). Further study of PRAT satellite DNA, which represents the major, abundant satellite in the species Palorus ratzeburgii, revealed its wide distribution far beyond the level of genus. The sequence of PRAT satellite DNA cloned from species that are separated by a significant evolutionary period of about 50–60 Myr showed high conservation without any fixed species-specific mutation (Mravinac et al. 2002).
In this paper we show that PSUB satellite DNA, previously characterized as a major satellite of P. subdepressus (Plohl et al. 1998), acts after the fashion of the PRAT satellite concerning the distribution and sequence conservation. In order to gain a deeper understanding of the phenomenon of evolutionarily conserved satellite sequences, we studied the genesis of mutational profiles of both satellites as well as the distribution of sequence variability along the sequences. The possible causes for the observed features such as wide distribution, evolutionary conservation of sequence, and uneven distribution of variability are discussed in light of recent data dealing with satellite DNA function.
Materials and Methods
Insect Species
Insect cultures of Palorus, Tribolium, Latheticus, and Tenebrio were purchased from the Central Science Laboratory (Sand Hutton, York, UK). Pimelia criba and Pimelia elevata specimens were collected by Dr. E. Petitpierre at Mallorca and Ibiza (Spain), respectively. Leptinotarsa decemlineata specimens were collected near Zagreb (Croatia).
PCR Amplification and Sequencing
Genomic DNA used for PCR amplification was isolated from a single insect specimen by the ROSE (rapid one-step extraction) procedure (Steiner et al. 1995). A primer pair, synthesized at GIBCO BRL Life Technologies (USA) and having the following sequence (5′-ATC AAG CCG ATT TAC AGC G-3′ and 5′-AAT CTG GCA AAT AAA CAG CG-3′), was used to amplify 126 bp of PSUB sequence (Fig. 3) from different insect species. The primer pair was selected according to the consensus sequence of major P. subdepressus satellite DNA (Plohl et al. 1998). Due to the high A + T content of PSUB (71.5%) and its complex substructure, it was not possible to find a suitable primer pair for amplification of the whole 144-bp monomer. The 25-μl reaction mixture contained 1× AmpliTaq buffer, 2.5 mM MgCl2, 0.2 mM dNTPs, 1 U of AmpliTaq DNA polymerase (Perkin Elmer), 20 ng of template DNA (or 1 pg of positive control) and a 0.4 μM concentration of each primer. The amplification was carried out in a GeneAmp PCR System 2 400 (Perkin Elmer) and the program started with predenaturation at 94°C for 3 min, followed by 33 cycles composed of denaturation at 94°C for 1 min, annealing at 54°C for 1 min, and extension at 72°C for 1 min. The final extension was performed at 72°C for 7 min. PCR products were separated by electrophoresis on a 1% agarose gel, and an ∼130-bp-long fragment obtained by amplification of T. brevicornis DNA was isolated from the gel and cloned using the pGEM-T Vector System I (Promega, USA). Positive recombinant clones were sequenced in both directions with an ALFexpress Sequencer (Amersham Pharmacia Biotech) using Amersham’s ThermoSequenase Cy5 Terminator Kit.
Hybridization
Genomic DNAs used for dot-blot and Southern hybridization were isolated using standard phenol extraction. For dot-blot analysis, 100 ng, 500 ng, and 1 μg of each genomic DNA was spotted on a nylon membrane. As a negative control, DNA from the lizard Podarcis sicula was used in the same quantities, while positive control P. subdepressus DNA was spotted in amounts of 10, 15, and 20 pg. Southern hybridization analysis of PCR products and dot-blotted genomic DNAs was performed under high-stringency conditions at 65°C, using as a probe a previously cloned satellite monomer, labeled with digoxigenin dUTP in a random priming reaction (DIG DNA Labeling and Detection Kit; Roche). The relative amount of PSUB satellite DNA in the examined species was determined by densitometry. For the genomic Southern blots, 4 μg of T. brevicornis DNA and 2 μg of P. subdepressus DNA were digested with appropriate restriction enzymes, blotted, and hybridized using as a probe a satellite monomer cloned previously from P. subdepressus, labeled with [α-32P]dATP in a random priming reaction. Hybridization was performed under high-stringency conditions at 65°C as well as at lower stringency at 60°C with a probe concentration of 106 cpm/ml. After high-stringency washes in 0.1× SSC and 0.1% SDS, the blot with P. subdepressus DNA was exposed on X-ray film for 1 h, while the T. brevicornis blot was exposed for 10 days.
Sequence Analysis
Nucleotide sequences were compared using the default parameters of the ClustalX v.1.81 program (Thompson et al. 1997). DNA polymorphism and the distribution of variability along the sequence were analyzed using DnaSP v.3.99 (Rozas and Rozas 1999). The conserved and variable segments were defined by sliding window analysis using a window size of 20 bp, a step size of 1, and the same program. Windows that exhibited a deviation of more than 2 SD from the average variability were considered. In the case of window clusters with significant deviation, the window with the highest deviation was taken as a center of the variable or conserved segments. The pattern of variation at each nucleotide position of satellite DNA was analyzed by the method described by Strachan et al. (1985) and Pons et al. (2002). Each position in the satellite was compared between the pair of species and classified in one of the six stages of homogenization in the process of concerted evolution. Class 1 represents complete homogeneity of all clones from a pair of species, while classes 2–4 are intermediate stages leading to the fixation of a mutation in all clones of one species (class 5). Class 6 shows the first stage of subsequent replacement of the mutation fixed in stage 5. Analysis of molecular variance was performed using the ARLEQUIN software (Schneider et al. 2000). Phylogenetic trees were obtained using the PAUP* version 4b10 program (Swofford 1998). Distance tree was built using the neighbor-joining (NJ) method and the uncorrected p distances. Maximum parsimony (MP) searches were performed using 1000 random additions with TBR branch swapping and the Multrees option. All nucleotide positions in the matrix were considered equivalent and weighted equally, while gaps were treated as missing data. The strict consensus tree of 495 trees is presented. Bootstrap values were obtained after 1000 replicates. Motifs homologous to the human CENP-B box (YTTCGTTGGAARCGGGA [Masumoto et al. 1989]), with Y = C or T and R=A or G, were searched using the MicroGenie (Beckmann) program package.
Results
Distribution and Organization of PSUB inT. brevicornis
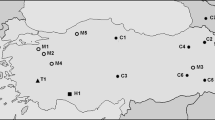
The distribution of the PSUB satellite sequence, which represents the most abundant satellite DNA in the insect species P. subdepressus (Plohl et al. 1998), was analyzed in eight species: seven from the family Tenebrionidae (subfamilies Tenebrioninae and Pimelinae) and Leptinotarsa decemlineata from family Chrysomelidae (Table 1). Dot-blot hybridization gave a positive signal in five species (Fig. 1, Table 1), three from the subfamily Tenebrioninae, and two from the subfamily Pimelinae. Tenebrio obscurus, Latheticus oryzae, and L. decemlineata DNAs did not react with the PSUB probe as well as DNA from the lizard Podarcis sicula, used as a negative control. The strongest hybridization signal was detected in genomic DNA of Tribolium brevicornis, estimated as 0.05% of the genomic DNA, and slightly less intense signal in Tribolium madens, constituting approximately 0.04% of the genome. Considering the genome size of T. brevicornis and T. madens (Alvarez-Fuster et al. 1991), PSUB is present in 1.3 × 103 and 7.0 × 102 copies per haploid genome, respectively. The contribution of PSUB to the genomes of other species was in the range of 0.002%, as determined in Tenebrio molitor, to 0.02%, in Pimelia elevata. The more sensitive PCR assay followed by Southern hybridization, using as probe a labeled PSUB monomer cloned from P. subdepressus, confirmed the results of dot blot and disclosed the presence of a PSUB-related sequence in the additional species L. oryzae (not shown; Table 1). A regular ladder-like pattern disclosed by Southern hybridization indicates tandem organization of PSUB in all six species. T. brevicornis was selected for cloning and sequencing of PSUB satellite since it exhibits the strongest hybridization signal in dot blot as well as in PCR.
Analysis of distribution of the PSUB sequence in eight species of Coleoptera by dot-blot hybridization using cloned PSUB as a probe. The amounts (μg) of DNAs blotted are indicated at the top. DNA from the lizard Podarcis sicula was used in the same amounts as a negative control, while P. subdepressus DNA served as a positive control in the amounts of 10, 15, and 20 pg.
The presence and organization of PSUB in the T. brevicornis genome were studied by genomic Southern hybridization and compared with those in P. subdepressus, where PSUB comprises 20% of the genome (Plohl et al. 1998). T. brevicornis and P. subdepressus genomic DNAs were digested with the restriction enzymes AluI and DraI, each of them having two recognition sites in the 144-bp-long PSUB sequence. The expected ladder, composed mostly of satellite monomer and low-size multimers, is observed in P. subdepressus DNA after hybridization with labeled PSUB monomer (Fig. 2A). However, hybridization of T. brevicornis DNA digested with AluI and DraI revealed a satellite DNA profile predominantly composed of longer fragments, starting from an ∼1000-bp-long fragment corresponding to the satellite heptamer (Fig. 2B). This pattern, which is not typical for enzymes cutting within a consensus sequence, might indicate that the frequency of restriction sites for AluI and DraI is lower in PSUB in the T. brevicornis genome relative to PSUB in P. subdepressus. The complete absence of satellite monomer and other small-size fragments on T. brevicornis genomic blots could be related to the lower efficiency of binding and hybridization of such fragments with respect to larger ones. The Southern hybridization of T. brevicornis DNA digested with HinfI, which does not cut the PSUB consensus sequence but whose recognition sites could be created by single mutation events, reveals an expected profile comparable to those obtained with the same enzyme on P. subdepressus genomic DNA (Figs. 2A, B). The profiles obtained by hybridization of PSUB on T. brevicornis DNA digested with all three enzymes are not composed of regular ladders as is the case in P. subdepressus. In P. subdepressus, PSUB is organized in long arrays encompassing the whole pericentromeric heterochromatin (Plohl et al. 1998). In T. brevicornis, with a diploid chromosome number of 18, there are on average 150 PSUB copies per chromosome. They are organized not as a single homogeneous domain, but as a number of short segments dispersed along heterochromatic block, as revealed by primed in situ labeling (not shown). Such dispersed arrays or even single-copy repeats could be responsible for the relatively irregular pattern of bands observed on genomic Southern blots.
The results of genomic Southern blots confirm the repetitive nature of the PSUB sequence and its presence in the T. brevicornis genome. The hybridization profile obtained under low-stringency conditions at 60°C (not shown) was identical to those at a high stringency shown in Fig. 2B, which indicates conservation and homogeneity of the PSUB sequence in the T. brevicornis genome. The fact that satellite monomers were not detected by this method indicates their low copy number, and therefore we decided to clone PSUB from T. brevicornis using PCR amplification.
Cloning and Sequence Variability
A primer pair specific for PSUB sequence was constructed, encompassing a 126-bp-long region (Fig. 3), and a PCR product corresponding to the satellite monomer was cloned, followed by sequencing of 14 positives. Cloning of fragments corresponding to satellite dimer was inefficient and clones were not stable, probably due to the internal repetitive substructure of the satellite monomer (Plohl et al. 1998). Sequences of PSUB satellite cloned from T. brevicornis (SINB clones) were aligned with seven previously cloned monomers (ps) of PSUB from P. subdepressus (Fig. 3) and the nucleotide diversity Pi of 0.05737 was determined. Of 144 sites, 50 were polymorphic due to 55 point mutations without any deletion or insertion. Mutations that appear in several clones at a particular position are counted only once since it is proposed that they result from a single mutation event and are spread among clones by gene conversion. The shared mutations can be classified into two groups: those shared among clones of both species assumed to be ancestral ones and those appearing in clones of a single species at a particular site. There are eight ancestral mutations at seven sites in the PSUB sequence (Fig. 3). Analysis of mutation spreading using the method introduced by Strachan (1985) showed that 93.7% of the sites can be classified in the proposed homogenization stages. Most of the sites (68.6%) fall into class 1, representing complete homogeneity of all clones from a pair of species, and the rest fall into class 2, represented by rare mutations spread among less than half of the clones of a particular species. This indicates a slow incidence of new mutations and supports our assumption that mutations which we consider as ancestral did not arise independently in both species, but more probably have a common origin. Statistical analysis of variation using the AMOVA test (Schneider et al. 2000) reveals that 95.47% of PSUB variation is intraspecific, while only 4.53% is interspecific. The phylogenetic tree calculated by either the NJ or the MP method does not separate the clones of the two species into different clusters, as illustrated by the MP tree in Fig. 4.
Alignment of PSUB satellite monomers cloned from P. subdepressus (ps clones [Plohl et al. 1998]) and from T. brevicornis (SINB; accession numbers AJ289024–AJ289037). The consensus sequence (PSUBcon) representing the most prevailing nucleotide at each position is shown. The positions of sequence identity to the consensus are indicated by dots, while missing parts are marked by a dashed line. Ancestral mutations spread among clones of both species at a particular position are indicated in boldface. Other mutations are shown in italics. Arrows indicate the positions of primers used to amplify SINB clones.
The midpoint rooted strict consensus tree of PSUB satellite monomers cloned from P. subdepressus (ps clones [Plohl et al. 1998]) and from T. brevicornis (SINB clones) obtained by maximum parsimony analysis. The interpolated bootstrap values represent the percentages of trees supporting the particular node, of 1000 bootstrap replicates.
Distribution of variability along the PSUB satellite monomer reveals the most variable positions in the sequence to be those with ancestral mutations, which contribute 29.3% to the overall diversity. Using sliding window analysis with a window size of 20 and a step size of 1, we were able to define variable and conserved segments in the PSUB satellite monomer (Fig. 5A). Two regions 13 bp long (positions 52–64 and 79–91) exhibit variability ≥ (average + 2 SD), each containing two ancient mutations, which contribute 37.2% and 44%, respectively, to their diversity. In addition to variable regions, a single 13-bp-long region (positions 34–46) that shows variability ≤ (average – 2 SD) is detected and is defined as a conserved region. A segment from nucleotide position 107 to the end of the sequence also exhibits variability significantly lower than the average, but since it overlaps with the position of the primer used for amplification of SINB clones, the observed conservation is probably highly affected by that.
Identification of conserved (light gray) and variable (dark gray) segments in PSUB (A) and PRAT (B) satellite monomer using sliding window analysis (window size, 20 bp; step size, 1). Pi represents nucleotide diversity. The average variability is indicated by the solid line, while the average variability ± 2 SD is marked by the dashed lines.
For the comparison, we have performed analysis of sequence variation along previously characterized 49 monomers of PRAT satellite cloned from three species: P. ratzeburgii, P. subdepressus, and P. elevata (Mravinac et al. 2002). Nucleotide diversity Pi calculated for PRAT monomers is 0.05290, very similar to that observed for PSUB clones. Among 145 sites, 67 are polymorphic due to 73 point mutations, 3 and 2 single-nucleotide insertions and deletions, respectively. Eighteen ancestral substitutions are found at 18 sites in the sequence, 9 of them shared among clones of all three species, 7 shared only among two Palorus species, and 2 among P. elevata and P. ratzeburgii clones. Analysis of transition stages of homogenization and mutation spreading, performed previously (Mravinac et al. 2002), revealed about 70% of the positions in class 1 and the remaining 30% in class 2, similar to the case in PSUB. Only in the case of PRAT clones from the P. elevata, P. subdepressus species pair is a single position found at a stage close to fixation. Analysis of DNA polymorphism along PRAT satellite monomers disclosed sites with ancestral mutations as the most variable ones, contributing 61.6% to the overall diversity. The highest proportion of ancestral mutation variability (70%) stems from the mutations shared among clones of all three species. The sliding window analysis performed in the same way as described for PSUB revealed the presence of two variable fragments, 26 bp long (positions 29–54) and 16 bp long (positions 118–133), and a single 19-bp conserved segment (positions 65–83) within the PRAT monomer (Fig. 5B). Ancestral mutations contribute 73.0% and 69.0%, respectively, to the diversity of variable segments. The conserved segment is, however, free of ancestral mutations.
Discussion
The results presented in this paper reveal that PSUB satellite DNA not only is distributed within congeneric Palorus species, as previously reported (Meštrović et al. 1998), but is widespread among members of two tenebrionid subfamilies: Tenebrioninae and Pimelinae. The separation of the two subfamilies is, according to the phylogeographic data, timed in the Paleocene, about 50–60 Myr ago (Kwieton 1977). The genera Tribolium and Palorus represent closely related taxa belonging to the subfamily Tenebrioninae. On the basis of the large number of morphological differences, Hinton (1948) proposed an ancient origin of the Tribolium species group, dating them as early as the Cretaceous. The preliminary phylogenetic analysis of Tribolium species based on two mitochondrial markers revealed unexpectedly high sequence divergence for closely related species, favoring the proposed ancient origin (unpublished results). Divergence of the mitochondrial (mt) COI gene among Palorus species also indicates that they are separated for a considerable amount of evolutionary time, roughly approximated to a minimum of 7 Myr for the two closest species (Meštrović et al. 2000). However, due to the lack of fossil and biogeographic data, it is not possible to calibrate the evolutionary rate of mt genes for either the Tribolium or the Palorus species group and to assess the time when the two genera last shared a common ancestor.
In addition to the PSUB satellite, the PRAT satellite has been found in all analyzed species, both present in a similar, low number of copies and interspersed within heterochromatic regions of all chromosomes (not shown; Mravinac et al. 2002). The wide distribution and preservation of PSUB and PRAT satellite sequences for long evolutionary periods might be an indication of potential function. Their parallel existence in the genomes could be related to possible cooperative activity, as suggested for some noncoding conserved elements in mammals (Frazer et al. 2004).
The restriction profiles of PSUB in T. brevicornis and P. subdepressus exhibit some differences which might be related to the organization of this sequence in two genomes. While in P. subdepressus, PSUB represents a highly abundant satellite forming long domains in the heterochromatic regions (Plohl et al. 1998), in T. brevicornis it is a low-copy-number satellite organized in the form of short regions dispersed among arrays of the major satellite. The dispersed type of organization is characteristic for Tribolium madens satellite II (Durajlija Žinić et al. 2000), while a combination of tandem and dispersed repetitive elements has been observed in subtelomeric satellite DNA of the plant Zamia paucijuga (Cafasso et al. 2003). It is possible that the hybridization profile of the PSUB satellite in T. brevicornis results from hybrid genomic fragments composed of satellite segments linked to unknown DNA sequences. Such a fragment detected in the hybridization pattern of Donax trunculus genomic DNA revealed a junction between the satellite monomer and adjacent sequences (Plohl and Cornudella 1996). In addition, hybridization bands that are digested with PSUB consensus restriction enzymes can be due to repeat variants with mutated DNA sequence. Monomer sequence variants characterized by a single point mutation affecting restriction site can be represented at various frequencies in a satellite family, as found in Tenebrio molitor satellite DNA (Plohl et al. 1992). In a low-copy-number satellite the frequencies of particular variants can depart from those detected in the high-copy P. subdepressus satellite. However, the existence of tandemly repeated low-copy satellite monomers that could not be distinguished from high-copy variants was corroborated by PCR analysis. Unfortunately, these repeats were not detected by genomic Southern hybridization, probably because of relatively inefficient hybridization to short, A+T-rich DNA fragments present in a low number of copies.
The PSUB satellite sequence cloned from P. subdepressus and T. brevicornis exhibits high sequence conservation without species-specific fixed mutation. The most polymorphic sites are those with mutations shared among clones of both species, considered to be ancestral ones. They contribute 29.3% to the overall diversity of PSUB. The PRAT satellite, previously cloned from three species, has diversity very similar to that of PSUB, based 61.6% on ancestral mutations. The distribution of variability along the PSUB and PRAT sequence reveals the nonrandom pattern: two variable 13-bp regions and the conserved one of the same size were detected in PSUB, while in PRAT a 19-bp-long conserved and two variable segments, of 26 and 16 bp, respectively, are found. The presence of conserved and variable regions has been reported previously for the satellite DNAs of Arabidopsis thaliana, human, and Tribolium beetles (Hall et al. 2003; Mravinac et al. 2004). The contribution of ancestral mutations to the diversity of variable regions is about 40% in PSUB, while in PRAT it reaches 70%. Based on the variability analysis and mutation spreading, it can be assumed that in both satellites there are some positions prone to mutation which probably acquired polymorphism in an ancestral species and have preserved it in the clones of the extant species. The mutations occurring after the species split are still in the initial phases of homogenization and fixation in both PRAT and PSUB despite the significant time since the separation of species. The low rate of occurrence and spreading of new mutations is responsible for the relatively high sequence conservation along the monomers, as well as for the lack of divergence of each satellite.
These observations further indicate that there could be pressure on the PSUB and PRAT sequences favoring restoration of the original mutation profile. At some positions in the sequence this pressure allows generation and limited spreading of particular mutations, while in the remainder of the sequence this process is suppressed. We think that the preservation of the mutation profile of PSUB and PRAT satellites speaks in favor of its functional significance. It could be hypothesized that conserved and variable regions might be of equal importance, both defining the periodicity of recognition by specific proteins along the satellite DNA array. The level of conservation could be correlated with the periodicity of protein-binding: conserved regions, or motifs within them might be recognized more often by certain proteins than the variable regions. Among satellite DNA binding proteins the best characterized is the CENP-B protein, which recognizes a 17-bp motif in human α-satellite DNA known as the CENP-B box (Masumoto et al. 1989). Homologs of CENP-B protein are widespread among eukaryotes and they bind relatively diverged DNA motifs (Kipling and Warburton 1997). Within centromeric satellite DNAs of insect species, motifs with 59–70% homology to the human CENP-B box are found in Chironomus palidivitattus (Lopez and Edstrom 1998), ants of the Messor genus (Lorite et al. 2002), and beetles of the genus Tribolium (Mravinac et al. 2004). In the PRAT and PSUB sequences a motif 70.6% homologous to the human CENP-B box is detected. It is located mostly within the variable region (positions 78–94) of PSUB and within the region of average variability (positions 51–68) of PRAT. In human α-satellite DNA the CENP-B box is found within the variable region since only about 23% of α-satellite monomers have a functional motif (Hall et al. 2003). This indicates that the CENP-B protein binds with a relatively low periodicity along the satellite array, but in spite of that it seems to be indispensable for the assembly of centromere-specific chromatin (Ohzeki et al. 2002).
Our preliminary experiments reveal that the PSUB and PRAT sequences are actively transcribed (not shown). Based on this finding it can be proposed that the long evolutionary preservation and high sequence conservation of both satellites could be related to a potential biological role for their transcripts. Small interfering RNAs cognate to satellite DNAs are abundant during Drosophila melanogaster embryogenesis and are involved in chromatin modification, probably acting through sequence-specific interaction with DNA (Aravin et al. 2003).
In conclusion, evolutionary preservation and the parallel presence of PRAT and PSUB satellite DNAs in different coleopteran species suggest the possible biological significance of these sequences. This indication is supported by unexpected sequence conservation concerning the period of species separation. The pattern of variability of both satellites is marked by the ancestral mutations which contribute significantly to the overall sequence diversity as well as by the absence of species-specific mutations that are close to the fixation. The antiquity of mutational profiles and the nonrandom pattern of evolution resulting in conserved and variable regions strongly indicate biological constraints on PSUB and PRAT satellite sequences.
References
Alvarez-Fuster A, Juan C, Petitpierre E (1991) Genome size in Tribolium flour-beetles: inter- and intraspecific variation. Genet Res 58:1–5
Aravin AA, Lagos-Quintana M, Yalcin A, Zavolan M, Marks D, Snyder B, Gaasterland T, Meyer J, Tuschl T (2003) The small RNA profile during Drosophila melanogaster development. Cell 5:337–350
Cafasso D, Cozzolino S, De Luca P, Chinali G (2003) An unusual satellite DNA from Zamia paucijuga (Cyclades) characterized by two different organizations of the repetitive units in the plant genome. Gene 311:71–79
Charlesworth B, Sniegowski P, Stephan W (1994) The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 371:215–220
De la Herrán R, Fontana F, Lanfredi M, Congiu L, Leis M, Rossi R, Ruiz Rejón C, Ruiz Rejón M, Garrido-Ramos MA (2001) Slow rates of evolution and sequence homogenization in an ancient satellite DNA family of sturgeons. Mol Biol Evol 18:432–436
Durajlija Žinić S, Ugarković Đ, Cornudella L, Plohl M (2000) A novel interspersed type of organization of satellite DNA in Tribolium madens heterochromatin. Chromosome Res 8:201–212
Frazer KA, Tao H, Osoegawa K, de Jong PJ, Chen X, Doherty MF, Cox DR (2004) Noncoding sequences conserved in a limited number of mammals in the SIM2 interval are frequently functional. Genome Res 14:367–372
Garrido-Ramos MA, de la Herrán R, Jamilena M, Lozano R, Ruiz Rejón C, Ruiz Rejón M (1999) Evolution of centromeric satellite DNA and its use in phylogenetic studies of Sparidae family (Pisces, Perciformes). Mol Phylogenet Evol 12:200–204
Grétarsdóttir G, Arnason U (1992) Evolution of the common cetacean highly repetitive DNA component and the systematic position of Orcaella brevirostris. J Mol Evol 34:201–208
Hall SE, Kettler G, Preuss D (2003) Centromere satellites from Arabidopsis populations: maintenance of conserved and variable domains. Genome Res 13:195–205
Henikoff S, Ahmad K, Malik HS (2001) The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293:1098–1102
Hinton HE, (1948) A synopsis of the genus Tribolium MacLey with some remarks on the evolution of its species groups. Bull Entomol Res 39:13–55
Kipling D, Warburton PE (1997) Centromeres, CENP-B and Tigger too. Trends Genet 13:141–145
Kwieton E, (1977) Esquisse phylogenetique du genre Pimelia F. Acta Ent Mus Nat Pragae 39:559–589
Li YX, Kirby ML (2003) Coordinated and conserved expression of alphoid repeat and alphoid repeat-tagged coding sequences. Dev Dynamics 228:72–81
Lopez CC, Edström JE (1998) Interdispersed centromeric element with a CENP-B box-like motif in Chironomus pallidivitatus. Nucleic Acids Res 26:4168–4172
Lorite P, Carrillo JA, Tinaut A, Palomeque T (2002) Comparative study of satellite DNAs in ants of the Messor genus. Gene 297:113–122
Masumoto H, Masukata H, Muro Y, Nozaki N, Okazaki T (1989) A human centromere antigen (CENP-B) interacts with a short specific sequence in alphoid DNA, a human centromere satellite. J Cell Biol 109:1963–1973
Meštrović N, Plohl M, Mravinac B, Ugarković Đ (1998) Evolution of satellite DNAs from the genus Palorus—experimental evidence for the library hypothesis. Mol Biol Evol 15:1062–1068
Meštrović N, Mravinac B, Juan C, Ugarković Đ, Plohl M (2000) Comparative study of satellite sequences and phylogeny of five species from the genus Palorus (insecta, Coleoptera). Genome 43:776–785
Mravinac B, Plohl M, Meštrović N, Ugarković Đ (2002) Sequence of PRAT satellite DNA “frozen” in some coleopteran species. J Mol Evol 54:774–783
Mravinac B, Plohl M, Ugarković Đ (2004) Conserved patterns in the evolution of Tribolium satellite DNAs. Gene 332:169–177
Ohzeki J, Nakano M, Okada T, Masumoto H (2002) CENP-B box is required for de novo centromere chromatin assembly on human alphoid DNA. J Cell Biol 159:765–775
Plohl M, Cornudella L (1996) Characterization of a complex satellite DNA in the mollusc Donax trunculus: analysis of sequence variation and divergence. Gene 169:157–164
Plohl M, Borštnik B, Lucijanić-Justić V, Ugarković Đ (1992) Evidence for random distribution of sequence variants in Tenebrio molitor satellite DNA. Genet Res 60:7–13
Plohl M, Meštrović N, Bruvo B, Ugarković Đ (1998) Similarity of structural features and evolution of satellite DNAs from Palorus subdepressus (Coleoptera) and related species. J Mol Evol 46:234–239
Pons J, Petitpierre E, Juan C (2002) Evolutionary dynamics of satellite DNA family PIM357 in species of the genus Pimelia (Tenebrionidae, Coleoptera). Mol Biol Evol 19:1329–1340
Rozas J, Rozas R (1999) DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15:174–175
Schneider S, Roessli D, Excoffier L (2000) ARLEQUIN: a software for population genetics data analysis, version 2.000. Genetics and Biometry Laboratory, University of Geneva, Switzerland
Steiner JJ, Poklemba CJ, Fjellstrom RG, Elliott LF (1995) A rapid one-tube genomic DNA extraction process for PCR and RAPD analyses. Nucleic Acids Res 23:2569–2570
Strachan T, Webb D, Dover GA (1985) Transition stages of molecular drive in multiple-copy DNA families in Drosophila. EMBO J 4:1701–1708
Swofford DL (1998) PAUP*: Phylogenetic Analysis Using Parsimony (*and other methods), version 4. Sinauer Associates, Sunderland, MA
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24:4876–4882
Ugarković Đ, Plohl M (2002) Variation in satellite DNA profiles—causes and effects. EMBO J 21:5955–5959
Volpe TA, Kidner C, Hall IM, Teng G, Grewal SIS, Martienssen RA (2002) Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297:1833–1837
Acknowledgments
We are grateful to Dr. Joan Pons for the critical reading of and valuable comments on the manuscript. We also thank Dr. Mary Sopta for suggestions and improvement of the English. This work was supported by the Research Fund of the Republic of Croatia, Projects 0098074 and 0098075.
Author information
Authors and Affiliations
Corresponding author
Additional information
[Reviewing Editor: Dr. Jerzy Jurka]
Rights and permissions
About this article
Cite this article
Mravinac, B., Plohl, M. & Ugarković, Ð. Preservation and High Sequence Conservation of Satellite DNAs Suggest Functional Constraints. J Mol Evol 61, 542–550 (2005). https://doi.org/10.1007/s00239-004-0342-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-004-0342-y