Abstract
Self-splicing group II introns are large ribozymes and mobile retroelements initially identified in the mitochondrial and chloroplast genomes of lower eukaryotes and plants and subsequently found in bacteria and archaea. Group II introns display structural, functional and mechanistic similarities to eukaryotic pre-mRNA nuclear introns, which may have evolved from mobile group II introns. As in spliceosomal introns, the ribozyme of group II introns excises the intron as a branched, lariat structure, through two sequential transesterification reactions. The movement of group II introns is mediated by a ribonucleoprotein (RNP) complex consisting of the IEP encoded by the ORF and the spliced intron lariat RNA, which remains associated with the IEP. These RNP complexes recognize intron targets through both the IEP and the intron lariat RNA. New possibilities for the use of these introns as biotechnological tools are emerging, due to the small number and flexibility of interactions between IEPs and target sites, through the modification of the intron RNA motifs that recognize DNA target sites by base pairing.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Group II intron s
- Reverse transcriptase
- Maturase
- Splicing
- Self-catalytic RNA
- Ribozymes
- Retroelements
- Targetrons
- Retrohoming
- Mobile genetic elements
Characteristics of Group II Introns
Group II intron s are mobile metalloribozymes that self-splice from precursor RNA to generate excised intron lariat RNA forms, which invade new DNA genomic locations by reverse splicing . These retroelements also encode a reverse transcriptase that stabilizes the RNA structure for forward and reverse splicing and finally converts the inserted intron RNA back to DNA. For these reasons, group II introns initially identified in the mitochondrial and chloroplast genomes of lower eukaryotes and plants, and subsequently found in bacteria and archaea are thought to be the ancestors of nuclear spliceosomal introns and non-long terminal repeat (non-LTR) retrotransposons [1–4]. Recently identified structural and functional similarities between group II introns and spliceosomal nuclear RNAs have suggested that group II introns may have played an important role at the very start of eukaryote evolution. It is now thought that their invasion of pre-eukaryotic genomes and their proliferation in those genomes may have driven the evolutionary separation of nucleus and cytoplasm [5].
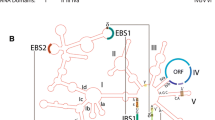
Typically, group II introns consist of a conserved RNA structure organized into six domains. Domain V is the most conserved of these domains and is considered to be an essential part of the catalytic core (Fig. 8.1) [6, 7]. In mobile introns, a protein (the intron-encoded protein) is commonly encoded by domain IV, which contains a specific subdomain (subdomain DIVa) responsible for facilitating the canonical interaction generating the ribonucleoprotein (RNP) particles involved in the invasion of new DNA targets [8]. The IEP has two conserved domains: an N-terminal RT domain and a maturase/thumb domain (also known as the X-domain). Some IEPs also have a C-terminal DNA-binding (D) region followed by a DNA endonuclease (En) domain (Fig. 8.1).
Secondary structure of group II intron RNAs and domain structure of the intron-encoded proteins (IEP). Structure of a representative ribozyme (not to scale): domains (DI–DVI), the EBS/IBS elements, the bulged adenosine within DVI, and the sequences involved in the tertiary interactions (Greek letters) occurring during splicing , as described in the text. Major differences between group IIA and IIB introns are indicated in brackets. Note that group IIC introns lack the EBS2 element. The loop of DIV, which encodes the IEP, is depicted by dashed lines, with a box showing the location and structure of DIVa, a high-affinity binding site for the IEP. Diagrams (drawn to scale) of the Ll.LtrB (IIA), RmInt1 (IIB) and GBSi1 (IIC) IEPs encoded within intron DIV are boxed on the right, with the predicted reverse transcriptase (RT) , maturase (X), variable DNA-binding region (D) and conserved DNA endonuclease (En) domains indicated. The C-tail denotes a C-terminal extension of 20 amino-acid residues conserved in the RmInt1 IEP and involved in maturase activity and DNA target recognition. The numbers above the RT domain identify conserved amino-acid sequence blocks characteristic of RTs
Group II Intron Ribozyme Sequences
Group II intron ribozymes are characterized by a conserved secondary structure, which varies in size from 100 nt up to about 3000 nt [9]. The first model was established on the basis of a phylogenetic data comparison, looking for potential base pairings that had been preserved by evolution despite primary sequence divergence [6, 10]. The only sequences of group II intron ribozymes that are strongly conserved are the intron boundaries (GUGYG at the 5′ exon junction and AY at the 3′ junction), which resemble those of spliceosomal introns (GU…AG) and few nucleotides dispersed throughout the rest of the structure [11].
Group II intron ribozymes are organized into six domains, DI–DVI, radiating from a central core (Fig. 8.1). They form a structure consisting of a set of double helices resulting from Watson-Crick and Crick wobble base-pairing [12]. The six domains fold into a catalytically active tertiary structure with the assistance of a series of conserved motifs involved in long-range tertiary interactions surrounding a catalytic four-metal-ion center [13]. Some interactions involve Watson-Crick base pairs (α-α′, β-β′, γ-γ′, δ-δ′, ε-ε′, IBS1-EBS1, IBS2-EBS2 and IBS3-EBS3), whereas other are tetraloop-receptor interactions of known geometries (ζ-ζ′, η-η′ and θ-θ′), or other types of less well defined non-Watson-Crick interactions (λ-λ′, κ-κ′ and μ-μ′) [14, 15]. Two of the six domains are essential for catalysis: the largest domain (DI), and DV. Recent crystallographic studies have revealed that the shape of the RNA molecule is dictated by a set of tertiary interaction networks within domains I and III that creates the scaffold responsible for binding and activating catalytic domain V [16]. DI is essential for exon recognition in forward and reverse splicing reactions. DV contains the catalytic triad, AGC at its base, the G residue being invariant and critical for splicing. Another important catalytic motif is the AC bulge of DV. Tertiary contacts between conserved nucleotides in the linker region J2/3 and the fifth nucleotide of the intron (λ position) have been described and reported to bring together these nucleotides to form a metal ion-binding platform directly involved in catalysis [10, 17–20]. Domain VI (DVI) contains a highly conserved bulged adenosine residue that acts as the branch point for lariat formation during splicing [7]. DVI also takes part in the long-range η-η′ interaction underlying the organization of the terminal loop of DVI and an internal helix of DII, which has been reported to be important for transesterification at the 3′-splice site [9].
Group II intron s can be classified into three structural subclasses, according to the recognition of their flanking exons. Subclass IIA and IIB introns, which display strong DNA target specificity, recognize their two exons via three base-pair interactions (IBS1/EBS1 and IBS2/EBS2 for the 5′ exon and δ-δ′ (IIA) or IBS3/EBS3 (IIB) for the 3′ exon), whereas subclass IIC introns use only two of these interactions (IBS1/EBS1 and IBS3/IBS3) and require a stem-loop structure in ssDNA that is generally derived from a transcription terminator by an as yet unknown recognition mechanism [21]. Additional subclasses have also been defined on the basis of specific structural differences: A1, A2, B1, B2 and B3 [10, 22].
Group II Intron-Encoded Proteins
The IEP component is a multifunctional protein containing a reverse transcriptase (RT) domain with subdomains conserved across other RT families (subdomains 0, 1, 2, 2a, 3, 4, 5, 6, 7) [8, 23]. Downstream from the RT domain is domain X, which functions as the thumb domain of the RT, and has a sequence conserved among group II introns but not between group II introns and other types of RTs [4, 24]. Domain X is immediately followed by a DNA-binding domain (D), defined on the basis of its function, but displaying no sequence conservation. Finally, many group II IEPs have an endonuclease domain (En) at their C-terminus that is required for the retromobility of these introns (Fig. 8.1). However, most bacterial IEPs have no En domain. Instead, they have a C-terminal portion that constitutes a distinctive, characteristic signature of this ORF class and has probably been conserved throughout evolution. This region of the protein is a group-specific functionally important protein region participating in both maturase function and intron mobility [25]. Recent studies on the consensus amino-acid sequences of the maturase and C-terminal domains have expanded our knowledge of subclasses of intron ORFs with no recognizable D/En region [26].
The classification and phylogenetic analysis of group II introns on the basis of their IEPs have resulted in the definition of several main groups: A, B, C, D, E, F, CL1 (chloroplast-like 1), CL2 (chloroplast-like 2) and ML (mitochondrion-like) [4, 26, 27], although additional types of intron ORF have recently been identified [26]. The introns of classes A, C, D, E and F and the newly identified g1 introns encode proteins with no En domain [26]. The En domain seems to have been acquired only once in recent phylogenetic lineages of group II introns, by the common ancestor of classes B, CL and ML [26]. Intron RNA structures are generally congruent with ORF phylogeny [27].
Group II Intron Splicing
Group II intron s have developed a series of splicing mechanisms to ensure their removal from the pre-mRNA and, hence, their survival in the host genome [28]. Organellar group II introns mostly interrupt essential genes, whereas prokaryotic group II introns are found in mobile genetic element s or intergenic regions [29]. Some group II introns can self-splice in vitro [30], but excision in vivo is generally assisted by protein factors that facilitate either ribozyme folding or the splicing reaction itself [31].
Excision Mechanisms of Group II Introns
Several excision mechanisms have been described for group II introns, generating different intron splicing products and ligated exons [15, 32]. These mechanisms generally co-exist within the same host [33–36], but some group II introns have been reported to display excision exclusively via a specific pathway [37–39]. Major advances have been made towards understanding the mechanism and kinetics of group II intron splicing in studies based on self-splicing assays, in which the intron precursor is incubated in non-physiological conditions (high temperatures and salt concentrations) in vitro [30].
Group II intron catalysis generally involves two sequential transesterification reactions (Fig. 8.2a). Mg2+ ions are involved in ensuring the correct folding of the catalytic core of the intron ribozyme and they also orchestrate the rearrangement of the single active site between the first and second splicing reactions [19, 40, 41]. The branching reaction is the most common excision pathway among group II introns [15, 42, 43] (Fig. 8.2a [1]). The 2′-OH of a bulged adenosine located in ribozyme domain VI initiates a nucleophilic attack on the phosphate bond at the intron-5′ exon junction, generating an intron intermediate in which the adenosine is covalently linked to the first intron nucleotide. In a second step, the free 3′-OH of the 5′ exon triggers a nucleophilic attack on the 3′ splice site, leading to exon ligation and intron lariat release. Alternatively, the first transesterification reaction can be initiated by a hydroxyl group from a water molecule, generating a linear excised intron and ligated exons [44–46] (Fig. 8.2a [2]). This excision pathway has classically been associated with group II introns that have lost the bulged adenosine [37–39]. Both steps in the branching reaction are reversible [47], but the first reaction of the hydrolytic pathway seems to be irreversible [33, 48]. The most recently discovered intron excision mechanism mediates the release of the intron as a true circle [35, 36, 49–51] (Fig. 8.2a [3]). The mechanism underlying this reaction remains unknown, but it has been suggested that the 3′-OH of a free 5′ exon could attack the 3′ splice site, releasing the 5′ exon-intron linear intermediate and linked exons. The hydroxyl group at the end of the intron thus reacts with the first intron residue and forms the circular intron. The maturase activity of the IEP modulates the balance between intron lariats and circles in vivo [52]. It has recently been suggested that the splicing of wheat mitochondrial group II introns under cold stress plays a regulatory role [53]. Introns with a conventional branchpoint structure display classical lariat-type splicing regardless of germination temperature. By contrast, the excision of non-conventional introns shifts from a predominantly hydrolytic pathway at room temperature to the production of circular molecules in the cold.
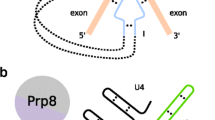
Mechanisms of group II intron excision. (a) Intron lariat, linear and circular molecules may be generated, depending on the excision pathway: branching [1], hydrolysis [2] and circle formation [3], respectively. The intron RNA seq uence is indicated by a dark blue line, and the exon sequences are represented by an empty blue box (5′ exon) or a light blue box (3′ exon). Orange bars correspond to the EBSs/IBSs interaction. The bulged adenosine in intron domain VI is represented as an A. Dotted red lines indicate the nucleophilic attacks occurring during each step of the reaction. (b) Structural similarities between the catalytic core of group II introns and activated spliceosomes. Dark blue lines represent the intron RNA, whereas light blue boxes and lines correspond to exon sequences (dotted lines indicating connecting regions of variable length omitted from the diagram for simplification). snRNA segments are shown in black or dark gray, and pink spheres represent Mg2+ ions. The catalytic triad is shown in red; the unpaired CA region in intron domain V and the equivalent segment in the U6 snRNA are shown in green and the bulged adenosine (A) region is shown in violet. The nucleophilic attack of the bulged adenosine on the 5′ exon-intron junction is indicated by a red arrow. Orange bars correspond to the EBSs/IBSs interaction. Other long-range contacts are identified by yellow and light gray bars
The 5′ and 3′ splice sites are recognized principally by base pairing. The EBS1-IBS1 interaction is crucial to 5′ splice site recognition during the splicing reaction. By contrast, the EBS2-IBS2 duplex is entirely dispensable for catalysis and splice site fidelity [36, 54]. Indeed, group IIC introns naturally lack the EBS2-IBS2 interaction, instead requiring the recognition of an upstream transcription terminator stem-loop [21]. Similarly, recognition of the 3′ splice site involves base pairing between the first nucleotide of the 3′ exon and the nucleotide preceding the EBS1 sequence (δ) in group IIA introns, or the EBS3 residue located in the coordination loop in group IIB and IIC introns [55, 56]. In addition to the long-range interactions underlying the catalytic conformation, tertiary contacts and structural constraints determine the efficiency and fidelity of identification, for both splice sites [11, 36, 57] (Fig. 8.2b, left panel).
Degeneration is common in organellar group II introns, for both RNA structures and ORF motifs. Thus, the intron ribozyme may be encoded in two or more pieces located at different positions in the genome , with disruptions often observed within dIV [58]. The intron and the flanking exons are transcribed separately, in a manner similar to that for cis-splicing introns, making the ligation of two distant exons possible. Trans-splicing is generally reported in higher plants, and this process requires the assistance of host-encoded protein factors [59]. A more dramatic process, intron fragmentation, has been observed in the chloroplast genome of Euglena gracilis, in which 155 small group II intron fragments are found [60].
Alternative splicing reactions have also been reported for bacterial and organellar group II introns [61–63]. These reactions occur at low frequency and result from misrecognition of the 3′ or 5′ splice site, inducing ORF truncations or small insertions. Alternative excision was initially thought to result in unproductive processing, but a recent study has revealed that this constitutive regulated process involving an unknown mechanism generates four functional surface-layer protein isoforms in the human pathogen Clostridium tetani [64].
Proteins Assisting Intron Excision
The efficiency of group II intron splicing in vivo is dependent on two groups of proteins [8]: those encoded by intron domain IV (IEPs), which are involved in cis-splicing reactions and found mostly in bacteria, and a group of host-encoded proteins with diverse functions, mediating the trans-splicing of organellar group II introns in yeasts, algae and higher plants [31, 65]. IEPs promote the splicing of group II introns in the maturase domain. They are highly specific splicing factors, playing little or no role in the excision of any intron other than the intron that encodes them [66–68]. They are usually expressed in cis, but they can promote the splicing of genomic copies of ORF-less introns [68, 69].
The organellar introns have diverged considerably from their bacterial ancestors, through a decrease in the number of maturase-encoding genes. A few intron-encoded ORFs are found in the mitochondria of lower eukaryotes (i.e., Marchantia polymorpha), but only one organelle-encoded protein has been reported to assist in the splicing of about 20 group II introns in vascular plants. This protein is called MatR in mitochondria, and MatK in chloroplasts [70, 71]. Moreover, a series of maturase-related proteins (nMat1a, nMat1b, nMat2a, and nMat2b) have been identified in the nucleus of angiosperms. After translation, these proteins are imported into mitochondria and chloroplasts to mediate the excision of group II introns [72–74]. An extensive search of the genomic sequences of recently sequenced green algae and land-plant mitochondria identified a number of new genes potentially encoding maturases, the role of which in the maturation of group II introns remains to be elucidated [75].
A plethora of nuclear-encoded proteins from various families, with diverse functions, has been reported to contribute to the correct folding and excision of organellar group II introns [9, 31, 65, 76]. One of the most numerous and well characterized groups of proteins identified is the DEAD-box proteins, which act as ATP-dependent RNA chaperones, ensuring the correct folding of the ribozyme or resolving inactive kinetic traps and then triggering productive RNA folding (the yeast factor MSS116, CYT-19 in N. crassa; Ded1 in S. cerevisiae; SrmB in E. coli; PMH2 in A. thaliana) [77–80]. Only a small number of these proteins (MSS116, CYT-19 and Ded1) can stimulate group II intron splicing in vitro in near-physiological conditions, suggesting that additional cofactors must be required to trigger splicing in vivo. Genetic and biochemical data have shown that organellar group II intron RNAs and multiple protein factors form functional high-molecular weight, spliceosome-like complexes [81, 82] (Fig. 8.2b).
Group II Intron Mobility
In addition to acting as catalytic RNAs, some group II introns are also target-specific mobile genetic element s (recently reviewed in [8, 83]). Mobile group II introns can insert site-specifically into a DNA sequence identical to the splice site (homing), at a frequency of up to 100 % [84–86], or more randomly, at low frequency, into ectopic sites (transposition) [87–89]. These mechanisms occur through an RNA intermediate and are referred to as retrohoming and retrotransposition, respectively [90, 91]. Both events occur via full reverse splicing , mediated by a ribonucleoprotein particle (RNP) formed by the association of the IEP with DIVa and the catalytic core regions of the excised intron RNA [92]. The IEP is essential for maintenance of the active intron RNA structure, to ensure that the intron can reverse splice into the DNA target site. Group II intron mobility mechanisms were first studied for the yeast mtDNA introns aI1 and aI2 [93–96], and have since been investigated in bacteria [66, 90, 97]. The main difference between yeast and bacterial mobile introns is that one or both exons may accompany the intron (co-conversion of flanking exons) in yeast, but not in bacteria [98].
The retrohoming of group II introns is highly site-specific, because a ≈20–25-bp DNA target sequence is recognized by the RNP via domain D or other regions. The IEP recognizes the upstream (positions −23 to −1) and downstream (positions +4 to +9) exon DNA sequences (Fig. 8.3a). Thirteen nucleotides of the DNA target are recognized by base pairing between the intron RNA and exon sequences, through EBS2/IBS2, EBS1/IBS1 and either δ-δ′ (IIA introns) or EBS3/IBS3 (IIB, IIC introns) interactions (Fig. 8.3b; [12, 32, 99, 100]). The essential role of each of the base-pairing interactions has been demonstrated by mutating the DNA target site and observing the inhibition of reverse splicing in vitro or of intron mobility in vivo. These mutations can be rescued by compensatory mutations in the intron RNA. DNA target specificity is mostly controlled by the intron RNA, as the IEP can recognize only a few nucleotide positions. Initial recognition appears to involve interactions in the major groove between the IEP and key bases in the distal region of the 5′ exon, in the chain into which the intron subsequently reverse splices [101]. These interactions, enhanced by contact between the phosphate backbone and the IEP, involve unwinding of the DNA, allowing the reverse splicing of the intron RNA by pairing with IBS and/or δ sequences. DNA target sites have been defined experimentally for the yeast introns coxI-I1 [102] and coxI-I2 [103], the L. lactis Ll.LtrB intron [99], the S. meliloti RmInt1 intron [104], the B. halodurans B.h.I1 intron [21], the E. coli E.c.I5 intron [105], the T. elongatus T.e.I4h intron [106] and the Enterobacter cloacae group IIC E.cl.GOC intron [107]. Group IIA, IIB and IIC introns differ in terms of DNA target site recognition, and these differences affect design and performance in the biotechnological context (Fig. 8.3b).
DNA target site recognition: (a) RNP complexes recognize the target site on double- or single-stranded DNA primarily through EBS-IBS pairing (and by δ–δ′ interactions in subgroup IIA), whereas the IEP also binds specifically to key nucleotide residues in distal 5′ and 3′ exon regions indicated by dotted lines in the diagram. (b) Comparison between the base-pairing interactions used by group IIA, IIB and IIC introns for DNA target site recognition. EBS exon binding site, IBS intron binding site
Group II Intron Mobility Pathways
After reverse splicing of the RNA into the DNA target site, at least three different mechanisms may complete the mobility of yeast group II introns [96, 108]. One of these mechanisms involves a minor pathway through which a small proportion of the intron mobility events occur in natural conditions without the coconversion of exons, probably through the synthesis of a full-length intron cDNA, which is joined by DNA repair . A second RT -independent pathway (≈40 %), in which intron integration occurs by homologous recombination of both the 5′ and 3′ exon sequences, involves the repair of the nicks generated by RNPs in the DNA target, by the double-strand break reaction (DSBR) mechanism (Fig. 8.4c). Finally, the major pathway (≈60 %) entails the coconversion of the 5′ exon only, and involves cDNA synthesis by target-primed reverse transcription (TPRT) and the integration of the intron by homologous recombination (DSBR) (Fig. 8.4a, b).
Group II intron mobility pathways: Mechanisms a, b and c have been described only in yeast; all these mechanisms are dependent on homologous recombination. a and b are the major retrohoming pathways in yeast, whereas c is the minor pathway and is RT -independent. d is the major pathway in Ll.ltrB and a minor pathway in yeasts. This mechanism is independent of homologous recombination. e is the retrohoming pathway for introns lacking the En domain and the mechanism associated with retrotransposition. f is the retrohoming pathway for linear group II introns
The bacterial group II intron mobility pathway was first described for the L. lactis Ll.LtrB IIA intron, in studies involving in vivo plasmid-based genetic assays in both L. lactis and E. coli [66, 90]. Retrohoming was subsequently characterized by analyzing the biochemical characteristics of RNPs reconstituted from the purified IEP (LtrA) and in vivo excised lariat RNA [67, 109]. In vivo, the RNPs bind the DNA nonspecifically and then scan for the accurate target site by facilitated diffusion [109]. Ll.LtrB RNPs recognize a relatively long target region through three sequence motifs in DI of the RNA (EBS1, EBS2 and δ), and they base pair to complementary sequences in the DNA target site (IBS1, IBS2 and δ′) and through interactions of the IEP with nucleotides located in positions −25 to +9 of the insertion site (Fig. 8.3a) [83, 99, 100, 110, 111]. Once the target has been recognized, retrohoming occurs by TPRT (Fig. 8.4d). The RNA cleaves the sense strand of the double-stranded DNA at the exon junctions, and the intron RNA integrates into the target site. At the same time, LtrA cleaves the antisense strand at position +9, through its En activity. The 3′ end of the antisense strand is used by the RT domain of the IEP for the reverse transcription of the inserted RNA intron. The resulting cDNA is then integrated into the host DNA by homologous recombination-independent repair mechanisms [90].
Some mobile group IIB introns have an IEP with no En domain. Their IEPs have RT activity, and their RNPs can mediate reverse splicing into double- or single-stranded DNA substrates but cannot carry out site-specific second-strand cleavage; they therefore require a variant of the TPRT retrohoming pathway (Fig. 8.4e) [91, 105, 112, 113]. Most En− group II introns are inserted into the strand used as a template for synthesis of the lagging strand during replication [114]. They can therefore insert into single-stranded DNA only when the replication fork has overtaken the insertion site. The IEP thus uses the nascent lagging strand to prime reverse transcription [91]. Other mechanisms have also been suggested for the initiation of the cDNA synthesis, including random nicks in the antisense strand (Schizosaccharomyces pombe cob-I1 intron, [115]), or de novo initiation (RT encoded by the Mauriceville plasmid in Neurospora, [116]). The best studied IIB-like intron is RmInt1 [69, 91, 112], which recognizes a DNA target site extending 20 nt into the 5′ exon and 5 nt into the 3′ exon. Target recognition occurs primarily by base-pairing between the EBS1, EBS2 and EBS3 of the intron RNA and the corresponding IBS sequences in the DNA target. The RmInt1 RT recognizes two critical nucleotide residues, possibly with the contribution of additional sequences [104, 112].
Unlike group IIA and IIB introns, group IIC introns have limited specificity due to the recognition of short IBS1 and IBS3 sequences (Fig. 8.3b). Moreover, the IBS2/EBS2 pairing seems to be replaced by the recognition of a palindromic Rho-independent transcription terminator motif or phage attachment site (attC sites), through an as yet unidentified mechanism [21, 37, 116–118]. Group IIC intron-encoded proteins also lack the En domain, but retain both domain Z (DNA binding) and domain X (maturase activity) [107]. Introns of this kind are found after non-identical terminators, inserted into the top or bottom strand, with a leading or inverse orientation, respectively [12, 107]. The integration of these introns resembles that of IIB introns, as it occurs through reverse splicing into single-stranded DNA at the replication fork or transcription bubble, with the nascent lagging strand preferentially used to prime reverse transcription of the intron.
It was thought that linear introns could not undergo reverse splicing [119–121], but recent studies have shown that yeast and bacterial linear group II introns reverse splice efficiently (Fig. 8.4f) [122–124]. The retrohoming of linear Ll.LtrB intron was demonstrated in eukaryotes, by the microinjection of RNPs into Xenopus laevis oocyte nuclei or Drosophila melanogaster embryos [123, 124]. The linear RNA undergoes the first reverse splicing reaction, becoming attached to the 3′ exon but not to the 5′ exon. The IEP then reverse transcribes the RNA, and the cDNA is ligated to the 5′ exon by the non-homologous end-joining (NHEJ) factor Lig 4 and the DNA repair polymerase θ (polQ). Other DNA ligases and polymerases can also perform this function, but at lower efficiency [124]. This mechanism may also mediate the retrohoming of linear RNAs, not only in eukaryotes, but also in many prokaryotes with homologous NHEJ machinery [8, 125].
Group II intron s can also retrotranspose to ectopic DNA target sites, albeit at low frequency (10−4–10−5) [88, 114, 126–128]. The pattern of spread of Ll.LtrB within the L. lactis genome is consistent with intron retrotransposition into double- or single-stranded DNA through a homologous recombination-independent mechanism [114], similar to that described for the mitochondrial and bacterial RmInt1 introns [32]. In L. lactis, the retrotransposition of the Ll.ltrB intron is biased towards reverse splicing into transiently single-stranded DNA, with priming by the nascent lagging strand (Fig. 8.4e). By contrast, the retrotransposition of Ll.LtrB in E. coli is characterized by the preferential use of double-stranded DNA targets, with or without En cleavage of the opposite strand [129], indicating a role of the host cell, in addition to the intron, in pathway selection [130].
Host Factors Influencing the Retrohoming Pathway of Group II Introns
Mobile group II introns are genetic elements with specific molecular characteristics favoring their retention and spread in the genome . However, their mobility depends on the genetic background of the host, and retrohoming is dependent on the completion of cell functions [111]. The replication machinery of the cell is required in the early stages, but the host repair machinery is essential during late stages of retrohoming. The first experiments performed with the group II intron Ll.LtrB from L. lactis in the heterologous host E. coli [131] led to the formulation of a model of retrohoming involving host factors that either increased or decreased the efficiency of mobility. Thus, exonucleases (Recj, MutD, and PolI) cutting the ends of the DNA, RNases (RNase H) degrading the RNA template after cDNA synthesis, DNA and repair polymerase complexes (PolII, PolIII, PolIV and PolV) ensuring correct synthesis of the second DNA chain and DNA ligases all facilitate intron mobility. By contrast, degradative enzymes may decrease retrohoming levels. For example, RNase I and E, may eliminate the intron RNA, and exonuclease III (XthA) may degrade the newly synthesized cDNA or top strand in the upstream exon. Further studies revealed that some enzymes from the degradosome (RNase E) may affect retrohoming levels, depending on the physiological status of the cell [132]. It was subsequently shown that Ll.LtrB mobility was influenced by cell interactions and responses to cellular or environmental stresses, through global regulators [133–136]. One recent study [137] confirmed previous findings and revealed, through genetic and biochemical analyses, a possible role for replication restart proteins in the retrohoming mechanism.
Use of Group II Introns in Biotechnology
Group II intron s have a number of characteristics that render them suitable for use as biotechnological tools: (1) they integrate into their DNA targets highly efficiently, in a homologous recombination-independent manner; (2) they can mobilize foreign DNA inserted within the intron; (3) minimal host functions, in the form of common cellular DNA repair mechanisms, are required for intron integration and (4) group II introns recognize the target DNA mostly through base pairing with the intron RNA . This last characteristic makes it possible to change intron specificity simply by changing the EBS/δ sequences. Currently, Ll.LtrB [138] a group IIA intron from Lactoccocus lactis, and the group IIB introns EcI5 [105] from Escherichia coli and RmInt1 from Sinorhizobium meliloti [139] are used as biotechnological tools. A chimeric intron based on the TeI3c ribozyme and the TeI4c IEP from Thermosynechococcus elongatus have also been used for gene targeting in thermophilic bacteria [140].
Introns were initially modified for the recognition of new target sites by identifying target sites matching the requirements of the IEP and then modifying the EBS/δ sequences to ensure base pairing with the new IBS/δ′ sequences. The retargeted introns are known as targetrons . Several algorithms have been developed for the retargeting of the Ll.LtrB [138], EcI5 [105] and RmInt1 [141] introns. These algorithms are based on the observed nucleotides frequencies obtained in invasion experiments using both randomized EBSs/δ-intron donor libraries and randomized IBSs/δ′-intron target site libraries. The algorithm scores a DNA sequence across a sliding window with 1 bp increments. The length of the sliding window depends on the intron: 45 bp for Ll.LtrB, 36 bp for EcI5 and 25 bp for RmInt1. A score is assigned to the potential target sites identified, with higher scores associated with a greater probability of a high invasion frequency. For each algorithm, a threshold value has been defined, above which the retargeted intron insertion frequency is high enough for the identification of intron insertion into the selected new target site in a simply assay, such as colony PCR. Once the best potential target site has been identified, the EBSs/δ sequences of the introns are modified to ensure base pairing with the new target site and are inserted into the intron donor plasmid, in which the IBSs/δ′ regions of the flanking exons are also modified to provide complementarity with the modified EBS/δ regions, for efficient RNA splicing . Intron donor plasmids also contain the IEP sequence, together with the corresponding intron from a position outside the DIV of the ribozyme (ΔORF) and downstream from the intron RNA. This conformation has been shown to be more efficient for retrohoming than the wild-type conformation with the IEP within DIV of the intron RNA [69, 105, 110]. Different promoters have been used for expression of the targetron and the associated IEP: constitutive promoters, such as the Km promoter used for the RmInt1 targetron [69], the T7 promoter recognized by the T7 RNA polymerase used in the expression of EcI5 and Ll.LtrB targetrons in E. coli, inducible promoters, such as the m-toluic acid-inducible promoter or tac promoter [142, 143] and endogenous promoters from the bacterial strain in which the targetron is used [144–146].
Intron integration can be detected by colony PCR or through the use of a selectable marker such as an antibiotic resistance gene. A retrotransposition-activated marker (RAM) has been developed for this purpose [145, 147]. The RAM cassette is based on a selectable marker with its own promoter inserted in reverse orientation into group II intron domain IV of the intron RNA . The marker is interrupted by the td group I intron in the forward orientation. The selectable marker is thus expressed only if retrohoming occurs. Subsequent modifications, with the selectable marker flanked by FRT sites recognized by the site-specific recombinase Flp, made it possible to remove the marker gene and led to the adaptation of the system for multiple gene disruptions.
Retargeted introns have been used in various species of the genera Agrobacterium [142], Azospirillum [148], Bacillus [149], Clostridium [145], Ehrlichia [150], Escherichia [105], Francisella [146], Lactococcus [151], Listeria [152], Paenibacillus [153], Pasteurella [154], Proteus [155], Pseudomonas [142], Ralstonia [143], Salmonella [111], Shewanella [156], Shigella [111], Sinorhizobium [139], Sodalis [157], Staphylococcus [144], Vibrio [158], and Yersinia [159]. In bacteria, targetrons are used primarily to obtain knockout mutants. In Clostridium, a genus in which transformation is difficult, retargeted intron derivatives of Ll.LtrB, known as ClosTron [145], have proved useful in several studies of the biology of the various species. When retargeted ΔORF introns insert into the sense strand, a conditional disruption is obtained as splicing can take place if the IEP is expressed, even in trans. However, intron targeting to the antisense strand leads to an unconditional mutation.
It is also possible to use targetrons to deliver foreign DNA into specific sequences [151, 160, 161]. The cargo gene is transported into the deleted region of DIV. For Ll.LtrB, fragments of less than 100 bp in length have a slight effect on intron insertion, but mobility efficiency is greatly reduced by fragments of more than 1 kb [162]. The secondary structure of the cargo sequences also affects intron mobility [156].
A method for bacterial genome editing using both targetrons and the Cre/lox system has recently been used [156]. This system has been used for insertions of 12 kb and deletions of up to 120 kb in E. coli and S. aureus, inversions in E. coli and Bacillus subtilis and one-step cut-and-paste manipulations for the translocation of 120 kb of genomic sequence to a site 1.5 Mb away.
Group II intron s (Ll.LtrB) have also been used in eukaryotic cells [163]. This approach is less well developed in eukaryotes than in prokaryotes and several hurdles have yet to be overcome. The principal problem concerns the concentration of Mg2+, which is below that required for the movement of Ll.LtrB in eukaryotic cells. Furthermore, the chromatinization of cellular DNA strongly inhibits intron integration. In eukaryotic cells, group II introns are microinjected into the cell nucleus as in vitro reconstituted ribonucleoproteins (RNPs). Xenopus laevis oocytes, and embryos of Drosophila melanogaster and zebrafish have been used for this purpose. RNPs have been reconstituted with both lariat and linear RNAs. In addition to the RNPs, a mixture of 500 mM Mg2+ and 17–20 mM each of dATP, dCTP, dGTP and dTTP is also injected into the nucleus, to optimize intron insertion. The RNPs and Mg2+ must be injected separately, because RNPs precipitate at this Mg2+ concentration. In these conditions, lariat RNPs injected into the X. laevis oocyte nuclei can both insert into an injected plasmid target at high frequency and stimulate DNA integration by homologous recombination, by producing target-site double-strand breaks. In D. melanogaster embryos, intron integration into the yellow gene has been achieved with introns retargeted against this gene. More knowledge is required about the behaviour of group II introns in eukaryotic cells, for the development of tools for use in eukaryotes.
Evolutionary Aspects of Bacterial Group II Introns
Group II intron s display structural, functional and mechanistic similarities to eukaryotic pre-mRNA nuclear introns [164–168]. Nuclear pre-messenger RNA introns [11] and non-long terminal repeat retrotransposons may have evolved from mobile group II introns [169]. It has been suggested [2, 168, 170] that, at an early stage in the evolution of eukaryotes, the ancestral group II intron structure was split into the non-catalytic spliceosomal introns and the catalytically active RNA component of the spliceosome. This transition was accompanied by the degradation of the reverse transcriptase ORF. Maturases may have persisted in plants, during evolution, through the acquisition of a targeting signal enabling them to function within the organelles, to support the splicing of organellar group II introns [75, 171]. The evolution of eukaryotic cell organization may also have been a defensive response to the deleterious effect of group II intron proliferation in the host genome [172, 173]. Nevertheless, a recent report [174] suggests that the compartmentalization of eukaryotic cells into nucleus and cytoplasm does not prevent group II intron invasion of the host genome, although it may control proliferation of the intron, through transient or stable nucleolar sequestration. Strikingly, when the IEP loses its maturase activity, the protein becomes localized in nuclear speckles, domains of the nucleus enriched in pre-mRNA splicing factors [175], including small nuclear ribonucleoproteins (snRNPs) and serine-arginine (SR) proteins located in the interchromatin regions of the nucleoplasm. This is consistent with the hypothesis that eukaryotic spliceosomal introns may have evolved from group II introns.
Bacterial group II introns are tending to evolve towards an inactive form by fragmentation, with the loss of the 3′ terminus, including the IEP [176, 177]. The significance of fragmented introns within a particular genome remains unclear. It has recently been suggested that, as for transposable element s (TEs), the dispersal and dynamics of group II intron spread within a bacterial genome follows a selection-driven extinction model, predicting the removal of highly colonized genomes from the population by purifying selection [178]. Only 25 % of the bacterial genomes sequenced to date [8] harbor recognizable group II introns. This suggests that these introns did not act as a major force with a broad effect in the promotion of evolutionary change, but caution is required in the interpretation of these observations, because the 5′ end of fragmented intron sequences lacking the encoded ORF is unlikely to have been detected in sequenced bacterial genomes.
It is generally accepted that the “selfish” features of mobile elements underlie their acquisition and maintenance in bacterial genomes, but these elements may also be beneficial to the host. In bacteria, group II introns are thought to be tolerated to some extent because they self-splice and preferentially home to sites outside key functional genes, generally within intergenic regions or other mobile genetic element s [179]. Other studies have suggested that group II introns are beneficial to the host because they control other potentially harmful mobile genetic elements [180], and contribute to the generation of diversity and the remodeling of genomes in times of stress [135]. These features may decrease negative effects on the host organism, resulting in the maintenance of these retroelements for longer periods in bacterial populations. It also seems likely that the gradual eradication of group II introns by the host during evolution would not result in the complete elimination of intron sequences, with some intron fragments remaining and continuing to evolve in the genome . It thus remains possible that these fragments provide sequence variation on which selection can act, leading to their persistence and continuing evolution in the genomes of some bacterial lineages [181].
References
Sharp PA (1991) Five easy pieces. Science 254:663
Martin W, Koonin EV (2006) Introns and the origin of nucleus-cytosol compartmentalization. Nature 440:41–45
Capy P, Vitalis R, Langin T, Higuet D, Bazin C (1996) Relationships between transposable elements based upon the integrase-transposase domains: is there a common ancestor? J Mol Evol 42:359–368
Simon DM, Kelchner SA, Zimmerly S (2009) A broadscale phylogenetic analysis of group II intron RNAs and intron-encoded reverse transcriptases. Mol Biol Evol 26:2795–2808
Doolittle WF (2014) The trouble with (group II) introns. Proc Natl Acad Sci U S A 111:6536–6537
Michel F, Jacquier A, Dujon B (1982) Comparison of fungal mitochondrial introns reveals extensive homologies in RNA secondary structure. Biochimie 64:867–881
Qin PZ, Pyle AM (1998) The architectural organization and mechanistic function of group II intron structural elements. Curr Opinion Struct Biol 8:301–308
Lambowitz AM, Zimmerly S (2010) Group II introns: mobile ribozymes that invade DNA. Cold Spring Harb Perspect Biol 3:a003616. doi:10.1101/cshperspect.a003616
Lehmann K, Schmidt U (2003) Group II introns: structure and catalytic versatility of large natural ribozymes. Crit Rev Biochem Mol Biol 38:249–303
Michel F, Umesono K, Ozeki H (1989) Comparative and functional anatomy of group II catalytic introns--a review. Gene 82:5–30
Michel F, Ferat JL (1995) Structure and activities of group II introns. Ann Rev Biochem 64:435–461
Lambowitz AM, Zimmerly S (2004) Mobile group II introns. Ann Rev Genet 38:1–35
Marcia M, Pyle AM (2014) Principles of ion recognition in RNA: insights from the group II intron structures. RNA 20:516–527
Dai L, Chai D, Gu SQ, Gabel J, Noskov SY, Blocker FJ et al (2008) A three-dimensional model of a group II intron RNA and its interaction with the intron-encoded reverse transcriptase. Mol Cell 30:472–485
Pyle AM (2010) The tertiary structure of group II introns: implications for biological function and evolution. Crit Rev Biochem Mol Biol 45:215–232
Toor N, Keating KS, Fedorova O, Rajashankar K, Wang J, Pyle AM (2010) Tertiary architecture of the Oceanobacillus iheyensis group II intron. RNA 16:57–69
Pyle AM (2002) Metal ions in the structure and function of RNA. J Biol Inorg Chem 7:679–690
Toor N, Keating KS, Pyle AM (2009) Structural insights into RNA splicing. Curr Opinion Struct Biol 19:260–266
de Lencastre A, Hamill S, Pyle AM (2005) A single active-site region for a group II intron. Nat Struct Mol Biol 12:626–627
de Lencastre A, Pyle AM (2008) Three essential and conserved regions of the group II intron are proximal to the 5′-splice site. RNA 14:11–24
Robart AR, Seo W, Zimmerly S (2007) Insertion of group II intron retroelements after intrinsic transcriptional terminators. Proc Natl Acad Sci U S A 104:6620–6625
Toor N, Hausner G, Zimmerly S (2001) Coevolution of group II intron RNA structures with their intron-encoded reverser transcriptases. RNA 7:1142–1152
Zimmerly S, Hausner G, Wu X (2001) Phylogenetic relationships among group II intron ORFs. Nucleic Acids Res 29:1238–1250
Blocker FJ, Mohr G, Conlan LH, Qi L, Belfort M, Lambowitz AM (2005) Domain structure and three-dimensional model of a group II intron-encoded reverse transcriptase. RNA 11:14–28
Molina-Sánchez MD, Martínez-Abarca F, Toro N (2010) Structural features in the C-terminal region of the Sinorhizobium meliloti RmInt1 group II intron-encoded protein contribute to its maturase and intron DNA-insertion function. FEBS J 277:244–254
Toro N, Martínez-Abarca F (2013) Comprehensive phylogenetic analysis of bacterial group II intron-encoded ORFs lacking the DNA endonuclease domain reveals new varieties. PLoS One 8:e55102
Toro N (2003) Bacteria and Archaea Group II introns: additional mobile genetic elements in the environment. Environ Microbiol 5:143–151
Pyle AM, Lambowitz AM (2006) Group II introns: ribozymes that splice RNA and invade DNA. In: Getsland RF, Cech TR, Atkins JF (eds) RNA World, 3rd edn. Cold Spring Harbor Laboratory Press, New York, pp 469–505
Robart AR, Zimmerly S (2005) Group II intron retroelements: function and diversity. Cytogenet Genome Res 110:589–597
Fedorova O, Zingler N (2007) Group II introns: structure, folding and splicing mechanism. Biol Chem 388:665–678
Solem A, Zingler N, Pyle AM, Li-Pook-Than J (2009) Group II introns and their protein collaborators. In: Walter NG, Woodson SA, Batey RT (eds) Non-protein coding RNAs. Springer, Berlin/Heidelberg, pp 167–182
Toro N, Jiménez-Zurdo JI, García-Rodríguez FM (2007) Bacterial group II introns: not just splicing. FEMS Microbiol Rev 31:342–358
Daniels D, Michels WJ, Pyle AM (1996) Two competing pathways for self-splicing by group II introns; a quantitative analysis of in vitro reaction rates and products. J Mol Biol 256:31–49
Costa M, Michel F, Molina-Sanchez MD, Martinez-Abarca F, Toro N (2006) An alternative intron-exon pairing scheme implied by unexpected in vitro activities of group II intron RmInt1 from Sinorhizobium meliloti. Biochimie 88:711–717
Molina-Sánchez MD, Martínez-Abarca F, Toro N (2006) Excision of the Sinorhizobium meliloti group II intron RmInt1 as circles in vivo. J Biol Chem 281:28737–28744
Nagy V, Pirakitikulr N, Zhou KI, Chillón I, Luo J, Pyle AM (2013) Predicted group II intron lineages E and F comprise catalytically active ribozymes. RNA 19:1266–1278
Granlund M, Michel F, Norgren M (2001) Mutually exclusive distribution of IS1548 and GBSi1, an active group II intron identified in human isolates of group B streptococci. J Bacteriol 183:2560–2569
Vogel J, Börner T (2002) Lariat formation and a hydrolytic pathway in plant chloroplast group II intron splicing. EMBO J 21:3794–3803
Toor N, Robart AR, Christianson J, Zimmerly S (2006) Self-splicing of a group IIC intron: 50 exon recognition and alternative 50 splicing events implicate the stem-loop motif of a transcriptional terminator. Nucleic Acids Res 34:6561–6573
Marcia M, Somarowthu S, Pyle AM (2013) Now on display: a gallery of group II intron structures at different stages of catalysis. Mob DNA 4:14
Kruschel D, Skilandat M, Sigel RKO (2014) NMR structure of the 5′ splice site in the group IIB intron Sc.ai5γ-conformational requirements for exon-intron recognition. RNA 20:295–307
Peebles CL, Perlman PS, Mecklenburg KL, Petrillo ML, Tabor JH, Jarrell KA, Cheng HL (1986) A self-splicing RNA excises an intron lariat. Cell 44:213–223
van der Veen R, Arnberg AC, van der Horst G, Bonen L, Tabak HF, Grivell LA (1986) Excised group II introns in yeast mitochondria are lariats and can be formed by self-splicing in vitro. Cell 44:225–234
van der Veen R, Kwakman JH, Grivell LA (1987) Mutations at the lariat acceptor site allow self-splicing of a group II intron without lariat formation. EMBO J 6:3827–3831
Jarrell KA, Peebles CL, Dietrich RC, Romiti SL, Perlman PS (1988) Group II intron self-splicing: alternative reaction conditions yield novel products. J Biol Chem 263:3432–3439
Podar M, Chu VT, Pyle AM, Perlman PS (1998) Group II intron splicing in vivo by first-step hydrolysis. Nature 391:915–918
Chin K, Pyle AM (1995) Branch-point attack in group II introns is a highly reversible transesterification, providing a potential proofreading mechanism for 5′-splice site selection. RNA 1:391–406
Chu VT, Liu Q, Podar M, Perlman PS, Pyle AM (1998) More than one way to splice an RNA: branching without a bulge and splicing without branching in group II introns. RNA 4:1186–1202
Murray HL, Mikheeva S, Coljee VW, Turczyk BM, Donahue WF, Bar-Shalom A, Jarrel KA (2001) Excision of group II introns as circles. Mol Cell 8:201–211
Li-Pook-Than J, Bonen L (2006) Multiple physical forms of excised group II intron RNAs in wheat mitochondria. Nucleic Acids Res 34:2782–2790
Chee GJ, Takami H (2011) Alternative splicing by participation of the group II intron ORF in extremely halotolerant and alkaliphilic Oceanobacillus iheyensis. Microbes Environ 26:54–60
Molina-Sánchez MD, Barrientos-Durán A, Toro N (2011) Relevance of the branch point adenosine, coordination loop, and 3′ exon binding site for in vivo excision of the Sinorhizobium meliloti group II intron RmInt1. J Biol Chem 286:21154–21163
Dalby SJ, Bonen L (2013) Impact of low temperature on splicing of atypical group II introns in wheat mitochondria. Mitochondrion 13:647–655
Barrientos-Durán A, Chillón I, Martínez-Abarca F, Toro N (2011) Exon sequence requirements for excision in vivo of the bacterial group II intron RmInt1. BMC Mol Biol 12:24. doi:10.1186/1471-2199-12-24
Costa M, Michel F, Westhof E (2000) A three-dimensional perspective on exon binding by a group II self-splicing intron. EMBO J 19:5007–5018
Hamill S, Pyle AM (2006) The receptor for branch-site docking within a group II intron active site. Mol Cell 23:831–840
Su L, Qin P, Michels W, Pyle A (2001) Guiding ribozyme cleavage through motif recognition: the mechanism of cleavage site selection by a group II intron ribozyme. J Mol Biol 306:665–668
Bonen L (2008) Cis- and trans-splicing of group II introns in plant mitochondria. Mitochondrion 8:26–34
Glanz S, Kück U (2009) Trans-splicing of organelle introns – a detour to continuous RNAs. Bioessays 31:921–934
Copertino DW, Hallick RB (1993) Group II and group III introns of twintrons: potential relationship with nuclear pre-mRNA introns. Trends Biochem Sci 18:467–471
Jenkins K, Hong L, Hallick R (1995) Alternative splicing of the Euglena gracilis chloroplast roaA transcript. RNA 1:624–633
Robart AR, Montgomery NK, Smith KL, Zimmerly S (2004) Principles of 3′ splice site selection and alternative splicing for an unusual group II intron from Bacillus anthracis. RNA 10:854–862
Costa M, Michel F, Toro N (2006) Potential for alternative intron-exon pairings in group II intron RmInt1 from Sinorhizobium meliloti and its relatives. RNA 12:338–341
McNeil BA, Simon DM, Zimmerly S (2014) Alternative splicing of a group II intron in a surface layer protein gene in Clostridium tetani. Nucleic Acids Res 42:1959–1969
Brown GG, Colas des Francs-Small C, Ostersetzer-Biran O (2014) Group II intron splicing factors in plant mitochondria. Front Plant Sci 5:35. doi:10.3389/fpls.2014.00035
Matsuura M, Saldanha R, Ma H, Wank H, Yang J, Mohr G et al (1997) A bacterial group II intron encoding reverse transcriptase, maturase, and DNA endonuclease activities: biochemical demonstration of maturase activity and insertion of new genetic information within the intron. Genes Dev 11:2910–2924
Saldanha R, Chen B, Wank H, Matsuura M, Edwards J, Lambowitz AM (1999) RNA and protein catalysis in group II intron splicing and mobility reactions using purified components. Biochemistry 38:9069–9083
Cui X, Matsuura M, Wang Q, Ma H, Lambowitz AM (2004) A group II intron-encoded maturase functions preferentially in cis and requires both the reverse transcriptase and X domains to promote RNA splicing. J Mol Biol 340:211–231
Nisa-Martínez R, Jiménez-Zurdo JI, Martínez-Abarca F, Muñoz-Adelantado E, Toro N (2007) Dispersion of the RmInt1 group II intron in the Sinorhizobium meliloti genome upon acquisition by conjugative transfer. Nucleic Acids Res 35:214–222
Wahleithner JA, MacFarlane JL, Wolstenholme DR (1990) A sequence encoding a maturase-related protein in a group II intron of a plant mitochondrial nad1 gene. Proc Natl Acad Sci U S A 87:548–552. doi:10.1073/pnas.87.2.548
Zoschke R, Nakamura M, Liere K, Sugiura M, Börner T, Schmitz-Linneweber C (2010) An organellar maturase associates with multiple group II introns. Proc Natl Acad Sci U S A 107:3245–3250. doi:10.1073/pnas.0909400107
Nakagawa N, Sakurai N (2006) A mutation in At-nMat1a, which encodes a nuclear gene having high similarity to group II intron maturase, causes impaired splicing of mitochondrial nad4 transcript and altered carbon metabolism in Arabidopsis thaliana. Plant Cell Physiol 47:772–783. doi:10.1093/pcp/pcj051
Keren I, Bezawork-Geleta A, Kolton M, Maayan I, Belausov E, Levy M et al (2009) AtnMat2, a nuclear-encoded maturase required for splicing of group-II introns in Arabidopsis mitochondria. RNA 15:2299–2311. doi:10.1261/rna.1776409
Cohen S, Zmudjak M, Colas des Francs-Small C, Malik S, Shaya F, Keren I et al (2014) nMAT4, a maturase factor required for nad1 pre-mRNA processing and maturation, is essential for holocomplex I biogenesis in Arabidopsis mitochondria. Plant J 78:253–268
Guo W, Mower J (2013) Evolution of plant mitochondrial intron-encoded maturases: frequent lineage-specific loss and recurrent intracellular transfer to the nucleus. J Mol Evol 77:43–54. doi:10.1007/s00239-013-9579-7
de Longevialle AF, Small ID, Lurin C (2010) Nuclearly encoded splicing factors implicated in RNA splicing in higher plant organelles. Mol Plant 3:691–705. doi:10.1093/mp/ssq025
Huang HR, Rowe CE, Mohr S, Jiang Y, Lambowitz AM, Perlman PS (2005) The splicing of yeast mitochondrial group I and group II introns requires a DEAD-box protein with RNA chaperone function. Proc Natl Acad Sci U S A 102:163–168
Halls C, Mohr S, del Campo M, Yang Q, Jankowsky E, Lambowitz AM (2007) Involvement of DEAD-box proteins in group I and group II intron splicing: biochemical characterization of Mss116p, ATP hydrolysis-dependent and -independent mechanisms, and general RNA chaperone activity. J Mol Biol 365:835–855
del Campo M, Mohr S, Jiang Y, Jia H, Jankowsky E, Lambowitz AM (2009) Unwinding by local strand separation is critical for the function of DEAD-box proteins as RNA chaperones. J Mol Biol 389:674–693
Köhler D, Schmidt-Gattung S, Binder S (2010) The DEAD-box protein PMH2 is required for efficient group II intron splicing in mitochondria of Arabidopsis thaliana. Plant Mol Biol 72:459–467. doi:10.1007/s11103-009-9584-9
Jacobs J, Glanz S, Bunse-Graβmann A, Kruse O, Kück U (2010) RNA trans-splicing: identification of components of a putative chloroplast spliceosome. Eur J Cell Biol 89:932–939
Jacobs J, Marx C, Kock V, Reifschneider O, Fränzel B, Krisp C et al (2013) Identification of a chloroplast ribonucleoprotein complex containing trans-splicing factors, intron RNA, and novel components. Mol Cell Proteomics 12:1912–1925. doi:10.1074/mcp.M112.026583
Enyeart PJ, Mohr G, Ellington AD, Lambowitz AM (2014) Biotechnological applications of mobile group II introns and their reverse transcriptases: gene targeting, RNA-seq, and non-coding RNA analysis. Mob DNA 5:2. doi:10.1186/1759-8753-5-2
Skelly PJ, Hardy CM, Clark-Walker GD (1991) A mobile group II intron of a naturally occurring rearranged mitochondrial genome in Kluyveromyces lactis. Curr Genet 20:115–120
Lazowska J, Meunier B, Macadre C (1994) Homing of a group II intron in yeast mitochondrial DNA is accompanied by unidirectional co-conversion of upstream-located markers. EMBO J 13:4963–4972
Moran JV, Zimmerly S, Eskes R, Kennell JC, Lambowitz AM, Butow RA, Perlman PS (1995) Mobile group II introns of yeast mitochondrial DNA are novel site-specific retroelements. Mol Cell Biol 15:2828–2838
Mueller MW, Allmaier M, Eskes R, Schweyen RJ (1993) Transposition of group II intron aI1 in yeast and invasion of mitochondrial genes at new locations. Nature 366:174–176
Cousineau B, Lawrence S, Smith D, Belfort M (2000) Retrotransposition of a bacterial group II intron. Nature 404:1018–1021 (Erratum: Nature 2001;414:84)
Muñoz E, Villadas PJ, Toro N (2001) Ectopic transposition of a group II intron in natural bacterial populations. Mol Microbiol 41:645–652
Cousineau B, Smith D, Lawrence-Cavanagh S, Mueller JE, Yang J, Mills D et al (1998) Retrohoming of a bacterial group II intron: mobility via complete reverse splicing, independent of homologous DNA recombination. Cell 94:451–462
Martínez-Abarca F, Barrientos-Durán A, Fernández-López M, Toro N (2004) The RmInt1 group II intron has two different retrohoming pathways for mobility using predominantly the nascent lagging strand at DNA replication forks for priming. Nucleic Acids Res 32:2880–2888
Lambowitz AM, Mohr G, Zimmerly S (2005) Group II intron homing endonucleases: ribonucleoprotein complexes with programmable target specificity. In: Belfort M, Stoddard BL, Wood DW, Derbyshire V (eds) Homing endonucleases and inteins. Springer, Heidelberg, pp 121–145
Zimmerly S, Guo H, Perlman PS, Lambowitz AM (1995) Group II intron mobility occurs by target DNA-primed reverse transcription. Cell 82:545–554
Zimmerly S, Guo H, Eskes R, Yang J, Perlman PS, Lambowitz AM (1995) A group II intron RNA is a catalytic component of a DNA endonuclease involved in intron mobility. Cell 83:529–538
Yang J, Zimmerly S, Perlman PS, Lambowitz AM (1996) Efficient integration of an intron RNA into double-stranded DNA by reverse splicing. Nature 381:332–335
Eskes R, Yang J, Lambowitz AM, Perlman PS (1997) Mobility of yeast mitochondrial group II introns: engineering a new site specificity and retrohoming via full reverse splicing. Cell 88:865–874
Mills DA, Manias DA, McKay LL, Dunny GM (1997) Homing of a group II intron from Lactococcus lactis subsp. lactis ML3. J Bacteriol 179:6107–6111
Belfort M, Derbyshire V, Parker MM, Cousineau B, Lambowitz AM (2002) Mobile introns: pathways and proteins. In: Craig NL, Gragie R, Gellert M, Lambowitz AM (eds) Mobile DNA II. ASM Press, Washington, DC, pp 761–781
Mohr G, Smith D, Belfort M, Lambowitz AM (2000) Rules for DNA target-site recognition by a lactococcal group II intron enable retargeting of the intron to specific DNA sequences. Genes Dev 14:559–573
Singh NN, Lambowitz AM (2001) Interaction of a group II intron ribonucleoprotein endonuclease with its DNA target site investigated by DNA footprinting and modification interference. J Mol Biol 309:361–386
Singh RN, Saldanha RJ, D’Souza LM, Lambowitz AM (2002) Binding of a group II intron-encoded reverse transcriptase/maturase to its high-affinity intron RNA binding site involves sequence-specific recognition and autoregulates translation. J Mol Biol 318:287–303
Yang J, Mohr G, Perlman PS, Lambowitz AM (1998) Group II intron mobility in yeast mitochondria: target DNA-primed reverse transcription activity of aI1 and reverse splicing into DNA transposition sites in vitro. J Mol Biol 282:505–523
Guo H, Zimmerly S, Perlman PS, Lambowitz AM (1997) Group II intron endonucleases use both RNA and protein subunits for recognition of specific sequences in double-stranded DNA. EMBO J 16:6835–6848
Jiménez-Zurdo JI, García-Rodríguez FM, Barrientos-Durán A, Toro N (2003) DNA target site requirements for homing in vivo of a bacterial group II intron encoding a protein lacking the DNA endonuclease domain. J Mol Biol 326:413–423
Zhuang F, Karberg M, Perutka J, Lambowitz AM (2009) EcI5, a group IIB intron with high retrohoming frequency: DNA target site recognition and use in gene targeting. RNA 15:432–449
Mohr G, Ghanem E, Lambowitz AM (2010) Mechanisms used for genomic proliferation by thermophilic group II introns. PLoS Biol 8:e1000391. doi:10.1371/journal.pbio.1000391
Rodríguez-Martínez JM, Nordmann P, Poirel L (2012) Group IIC intron with an unusual target of integration in Enterobacter cloacae. J Bacteriol 194:150–160. doi:10.1128/JB.05786-11
Eskes R, Liu L, Ma H, Chao MY, Dickson L, Lambowitz AM, Perlman PS (2000) Multiple homing pathways used by yeast mitochondrial group II introns. Mol Cell Biol 20:8432–8446
Aizawa Y, Xiang Q, Lambowitz AM, Pyle AM (2003) The pathway of DNA recognition and RNA integration by a group II intron retrotransposon. Mol Cell 11:795–805
Guo H, Karberg M, Long M, Jones JP 3rd, Sullenger B, Lambowitz AM (2000) Group II introns designed to insert into therapeutically relevant DNA target sites in human cells. Science 289:452–457
Karberg M, Guo H, Zhong J, Coon R, Perutka J, Lambowitz AM (2001) Group II introns as controllable gene targeting vectors for genetic manipulation of bacteria. Nat Biotechnol 19:1162–1167
Muñoz-Adelantado E, San Filippo J, Martínez-Abarca F, García-Rodríguez FM, Lambowitz AM, Toro N (2003) Mobility of the Sinorhizobium meliloti group II intron RmInt1 occurs by reverse splicing into DNA, but requires an unknown reverse transcriptase priming mechanism. J Mol Biol 327:931–943
Zhong J, Lambowitz AM (2003) Group II intron mobility using nascent strands at DNA replication forks to prime reverse transcription. EMBO J 22:4555–4565
Ichiyanagi K, Beauregard A, Lawrence S, Smith D, Cousineau B, Belfort M (2002) Multiple pathways for the Ll.LtrB group II intron include reverse splicing into DNA targets. Mol Microbiol 46:1259–1271
Schäfer B, Gan L, Perlman PS (2003) Reverse transcriptase an reverse splicing activities encoded by the mobile group II intron COBI1 of fission yeast mitochondrial DNA. J Mol Biol 329:191–206
Centron D, Roy PH (2002) Presence of a group II intron in a multiresistant Serratia marcescens strain that harbors three integrons and a novel gene fusion. Antimicrob Agents Chemother 46:1402–1409
Dai L, Zimmerly S (2002) Compilation and analysis of group II intron insertions in bacterial genomes: evidence for retroelement behavior. Nucleic Acids Res 30:1091–1102
Léon G, Roy PH (2009) Group IIC intron mobility into attC sites involves a bulged DNA stem-loop motif. RNA 15:1543–1553
Dème E, Nolte A, Jacquier A (1999) Unexpected metal ion requirements specific for catalysis of the branching reaction in a group II intron. Biochemistry 38:3157–3167
Mohr S, Matsuura M, Perlman PS, Lambowitz AM (2006) A DEAD-box protein alone promotes group II intron splicing and reverse splicing by acting as an RNA chaperone. Proc Natl Acad Sci U S A 103:3569–3574
Gordon PM, Fong R, Piccirilli JA (2007) A second divalent metal ion in the group II intron reaction center. Chem Biol 14:607–612
Roitzsch M, Pyle AM (2009) The linear form of a group II intron catalyzes efficient autocatalytic reverse splicing, establishing a potential for mobility. RNA 15:473–482
Zhuang F, Mastroianni M, White TB, Lambowitz AM (2009) Linear group II intron RNAs can retrohome in eukaryotes and may use nonhomologous end-joining for cDNA ligation. Proc Natl Acad Sci U S A 106:18189–18194
White TB, Lambowitz AM (2012) The retrohoming of linear group II intron RNAs in Drosophila melanogaster occurs by both DNA ligase 4-dependent and -independent mechanisms. PLoS Genet 8:e1002534. doi:10.1371/journal.pgen.1002534
Bowater R, Doherty AJ (2006) Making ends meet: repairing breaks in bacterial DNA by non-homologous end-joining. PLoS Genet 2:e8. doi:10.1371/journal.pgen.0020008
Martínez-Abarca F, Toro N (2000) RecA-independent ectopic transposition in vivo of a bacterial group II intron. Nucleic Acids Res 28:4397–4402
Dickson L, Huang HR, Liu L, Matsuura M, Lambowitz AM, Perlman PS (2001) Retrotransposition of a yeast group II intron occurs by reverse splicing directly into ectopic DNA sites. Proc Natl Acad Sci U S A 98:13207–13212
Ichiyanagi K, Beauregard A, Belfort M (2003) A bacterial group II intron favors retrotransposition into plasmid targets. Proc Natl Acad Sci U S A 100:15742–15747
Coros CJ, Landthaler M, Piazza CL, Beauregard A, Esposito D, Perutka J (2005) Retrotransposition strategies of the Lactococcus lactis Ll.LtrB group II intron are dictated by host identity and cellular environment. Mol Microbiol 56:509–524
Beauregard A, Chalamcharla VR, Piazza CL, Belfort M, Coros CJ (2006) Bipolar localization of the group II intron Ll.LtrB is maintained in Escherichia coli deficient in nucleoid condensation, chromosome partitioning and DNA replication. Mol Microbiol 62:709–722
Smith D, Zhong J, Matsuura M, Lambowitz AM, Belfort M (2005) Recruitment of host functions suggests a repair pathway for late steps in group II intron retrohoming. Genes Dev 19:2477–2487
Coros CJ, Piazza CL, Chalamcharla VR, Belfort M (2008) A mutant screen reveals RNase E as a silencer of group II intron retromobility in Escherichia coli. RNA 14:2634–2644
Beauregard A, Curcio MJ, Belfort M (2008) The take and give between retrotransposable elements and their hosts. Ann Rev Genet 42:587–617
Zhao J, Niu W, Marcotte E, Lambowitz A (2008) Group II intron protein localization and insertion sites are affected by polyphosphate. PLoS Biol 6:e150. doi:10.1371/journal.pbio.0060150
Coros CJ, Piazza CL, Chalamcharla VR, Smith D, Belfort M (2009) Global regulators orchestrate group II intron retromobility. Mol Cell 34:250–256
Edgell DR, Chalamcharla VR, Belfort M (2011) Learning to live together: mutualism between self-splicing introns and host genomes. BMC Biol 9:22. doi:10.1186/1741-7007-9-22
Yao J, Truong DM, Lambowitz AM (2013) Genetic and biochemical assays reveal a key role for replication restart proteins in group II intron retrohoming. PLoS Genet 9:e1003469. doi:10.1371/journal.pgen.1003469
Perutka J, Wang W, Goerlitz D, Lambowitz AM (2004) Use of computer-designed group II introns to disrupt Escherichia coli DExH/D-box protein and DNA helicase genes. J Mol Biol 336:421–439
García-Rodríguez FM, Barrientos-Durán A, Díaz-Prado V, Fernández-López M, Toro N (2011) Use of RmInt1, a group IIB intron lacking the intron-encoded protein endonuclease domain, in gene targeting. Appl Environ Microbiol 77:854–861. doi:10.1128/AEM.02319-10
Mohr G, Hong W, Zhang J, Cui GZ, Yang Y, Cui Q et al (2013) A targetron system for gene targeting in thermophiles and its application in Clostridium thermocellum. PLoS One 8:e69032. doi:10.1371/journal.pone.0069032
García-Rodríguez FM, Hernández-Gutiérrez T, Díaz-Prado V, Toro N (2014) Use of the computer-retargeted group II intron RmInt1 of Sinorhizobium meliloti for gene targeting. RNA Biol 11:391–401
Yao J, Lambowitz AM (2007) Gene targeting in gram-negative bacteria by use of a mobile group II intron (“targetron”) expressed from a broad-host-range vector. Appl Environ Microbiol 73:2735–2743
Park JM, Jang YS, Kim TY, Lee SY (2010) Development of a gene knockout system for Ralstonia eutropha H16 based on the broad-host-range vector expressing a mobile group II intron. FEMS Microbiol Lett 309:193–200
Yao J, Zhong J, Fang Y, Geisinger E, Novick RP, Lambowitz AM (2006) Use of targetrons to disrupt essential and nonessential genes in Staphylococcus aureus reveals temperature sensitivity of Ll.LtrB group II intron splicing. RNA 12:1271–1281
Heap JT, Pennington OJ, Cartman ST, Carter GP, Minton NP (2007) The ClosTron: a universal gene knock-out system for the genus Clostridium. J Microbiol Methods 70:452–464
Rodriguez SA, Yu JJ, Davis G, Arulanandam BP, Klose KE (2008) Targeted inactivation of Francisella tularensis genes by group II introns. Appl Environ Microbiol 74:2619–2626
Zhong J, Karberg M, Lambowitz AM (2003) Targeted and random bacterial gene disruption using a group II intron (tagetron) vector containing a retrotransposition-activated selectable marker. Nucleic Acids Res 31:1656–1664
Malhotra M, Srivastava S (2008) An ipdC gene knock-out of Azospirillum brasilense strain SM and its implications on indole-3-acetic acid biosynthesis and plant growth promotion. Antonie Van Leeuwenhoek 93:425–433
Akhtar P, Hkan SA (2012) Two independent replicons can support replication of the anthrax toxin-encoding plasmid pXO1 of Bacillus anthracis. Plasmid 67:111–117
Cheng C, Nair AD, Indukuri W, Gong S, Felsheim RF, Jaworski D et al (2013) Targeted and random mutagenesis of Ehrlichia chaffeensis for the identification of genes required for in vivo infection. PLoS Pathog 9:e1003171. doi:10.1371/journal.ppat.1003171
Frazier CL, San Filippo J, Lambowitz AM, Mills DA (2003) Genetic manipulation of Lactococcus lactis by using targeted group II introns: generation of stable insertions without selection. Appl Environ Microbiol 69:1121–1128
Alonzo F 3rd, Port GC, Cao M, Freitag NE (2009) The postranslocation chaperone PrsA2 contributes to multiple facets of Listeria monocytogenes pathogenesis. Infect Immun 77:3077–3085
Zarschler K, Janesch B, Zayni S, Schäffer C, Messner P (2009) Construction of a gene knockout system for application in Paenibacillus alvei CCM 2051 T, exemplified by the S-layer glycan biosynthesis initiation enzyme WsfP. Appl Environ Microbiol 75:3077–3085
Steen JA, Steen JA, Harrison P, Seemann T, Wilkie I, Harper M, Adler B, Boyce JD (2010) Fis is essential for capsule production in Pasteurella multocida and regulates expression of other important virulence factors. PLoS Pathog 6:e1000750. doi:10.1371/journal.ppat.1000750
Pearson MM, Mobley HL (2007) The type III secretion system of Proteus mirabilis HI4320 does not contribute to virulence in the mouse model of ascending urinary tract infection. J Med Microbiol 56:1277–1283
Enyeart PJ, Chirieleison SM, Dao MN, Perutka J, Quandt EM, Yao J et al (2013) Generalized bacterial genome editing using mobile group II introns and Cre-lox. Mol Syst Biol 9:685. doi:10.1038/msb.2013.41
Smith CL, Weiss BL, Aksoy S, Runyen-Janecky LJ (2013) Characterization of the achromobactin iron acquisition operon in Sodalis glossinidius. Appl Environ Microbiol 79:2872–2881
Kumar S, Smith KP, Floyd JL, Varela MF (2011) Cloning and molecular analysis of a mannitol operon of phosphoenolpyruvate-dependent phosphotransferase (PTS) type from Vibrio cholerae O395. Arch Microbiol 193:201–208
Palonen E, Lindstrom M, Karttunen R, Somervuo P, Korkeala H (2011) Expression of signal transduction system encoding genes of Yersinia pseudotuberculosis IP32953 at 28°C and 3°C. PLoS One 6:e25063. doi:10.1371/journal.pone.0025063
Jones JP III, Kierlin MN, Coon RB, Perutka J, Lambowitz AM, Sullenger BA (2005) Retargeting mobile group II introns to repair mutant genes. Mol Ther 11:687–694
Rawsthorne H, Turner KN, Mills DA (2006) Multicopy integration of heterologous genes, using the lactococal group II intron targeted to bacterial insertion sequences. Appl Environ Microbiol 72:6088–6093
Plante I, Cousineau B (2006) Restriction for gene insertion within the Lactococcus lactis Ll.LtrB group II intron. RNA 12:1980–1992
Mastroianni M, Watanabe K, Whit TB, Zhuang F, Vernon J, Matsuura M, Wallingford J, Lambowitz AM (2008) Group II intron-based gene targeting reactions in eukaryotes. PLoS One 3:e3121. doi:10.1371/journal.pone.0003121
Grabowski PJ, Seiler SA, Sharp PA (1985) A multicomponent complex is involved in the splicing of messenger RNA precursors. Cell 42:345–353
Cech TK (1986) The generality of self-splicing RNA: relationship to nuclear mRNA splicing. Cell 44:207–210
Patel AA, Steitz JA (2003) Splicing double: insights from the second spliceosome. Nat Rev Mol Cell Biol 4:960–970
Toor N, Keating KS, Taylor SD, Pyle AM (2008) Crystal structure of a self-spliced group II intron. Science 320:77–82
Keating KS, Toor N, Perlman PS, Pyle AM (2010) A structural analysis of the group II intron active site and implications for the spliceosome. RNA 16:1–9
Eickbush TH (1994) Origins and evolutionary relationships of retroelements. In: Morse SS (ed) The evolutionary biology of viruses. Raven, New York, pp 121–157
Rogozin IB, Carmel L, Csuros M, Koonin EV (2012) Origin and evolution of spliceosomal introns. Biol Direct 7:11. doi:10.1186/1745-6150-7-11
Mohr G, Lambowitz AM (2003) Putative proteins related to group II intron reverse transcriptase/maturases are encoded by nuclear genes in higher plants. Nucleic Acids Res 31:647–652
Koonin EV (2006) The origin of introns and the role in eukaryogenesis: a compromise solution to the introns-early versus introns-late debate? Biol Direct 1:22. doi:10.1186/1745-6150-1-22
Koonin EV (2011) The origins of eukaryotes: endosymbiosis, the strange story of introns, and the ultimate importance of unique events in evolution. In: Pearson Education Inc. (ed) The logic of chance: the nature and origin of biological evolution, 1st edn. FT Press Science, Upper Saddle River, pp 171–224
Nisa-Martínez R, Laporte P, Jiménez-Zurdo JI, Frugier F, Crespi M, Toro N (2013) Localization of a bacterial group II intron-encoded protein in eukaryotic nuclear splicing-related cell compartments. PLoS One 8(12):e84056. doi:10.1371/journal.pone.0084056, eCollection 2013
Spector DL, Lamond AI (2011) Nuclear speckles. Cold Spring Harb Perspect Biol 3:a000646. doi:10.1101/cshperspect.a000646
Dai LX, Zimmerly S (2002) The dispersal of five group II introns among natural populations of Escherichia coli. RNA 8:1294–1307
Fernandez-Lopez M, Munoz-Adelantado E, Gillis M, Willems A, Toro N (2005) Dispersal and evolution of the Sinorhizobium meliloti group II RmInt1 intron in bacteria that interact with plants. Mol Biol Evol 22:1518–1528
Leclercq S, Cordaux R (2012) Selection-driven extinction dynamics for group II introns in Enterobacteriales. PLoS One 7:e52268
Simon DM, Clarke NA, McNeil BA, Johnson I, Pantuso D, Dai L et al (2008) Group II introns in eubacteria and archaea: ORF-less introns and new varieties. RNA 14:1704–1713
Chillón I, Martínez-Abarca F, Toro N (2010) Splicing of the Sinorhizobium meliloti RmInt1 group II intron provides evidence of retroelement behavior. Nucleic Acids Res 39:1095–1104
Toro N, Martínez-Rodríguez L, Martínez-Abarca F (2014) Insights into the history of a bacterial group II intron remnant from the genomes of the nitrogen-fixing symbionts Sinorhizobium meliloti and Sinorhizobium medicae. Heredity. doi:10.1038/hdy.2014.32
Acknowledgments
This work was supported by research grants BIO2011-24401 and BIO2014-51953-P from the Spanish Ministerio de Economía y Competitividad, and CSD 2009-0006 from Consolider-Ingenio Programme, which includes ERDF (European Regional Development Funds).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer Science+Business Media New York
About this chapter
Cite this chapter
Molina-Sánchez, M.D., Nisa-Martínez, R., García-Rodríguez, F.M., Martínez-Abarca, F., Toro, N. (2015). Intron Biology, Focusing on Group II Introns, the Ancestors of Spliceosomal Introns. In: Felekkis, K., Voskarides, K. (eds) Genomic Elements in Health, Disease and Evolution. Springer, New York, NY. https://doi.org/10.1007/978-1-4939-3070-8_8
Download citation
DOI: https://doi.org/10.1007/978-1-4939-3070-8_8
Publisher Name: Springer, New York, NY
Print ISBN: 978-1-4939-3069-2
Online ISBN: 978-1-4939-3070-8
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)