Abstract
In Arabidopsis thaliana the putative mitochondrial RNA helicases PMH1 and PMH2 are members of the large DEAD-box protein family. Our previous characterization of these proteins revealed that PMH1 and/or PMH2 are part of high molecular weight complexes. Now T-DNA insertion lines were established and characterized for each of these genes. Immunodetection analysis of cell suspension cultures established from pmh1-1 and pmh2-1 mutants revealed that indeed both DEAD-box proteins are detectable in large protein complexes with PMH2 being much more abundant than PMH1. In plants the knockout of PMH2 leads to reduced group II intron splicing efficiency. In addition the steady-state levels of several mature mitochondrial mRNAs are decreased while transcription is not influenced. This molecular phenotype suggests that PMH2 acts at the posttranscriptional level with a potential function as RNA chaperone required for formation or maintenance of complex RNA secondary structures of introns rather than a direct role in splicing. In contrast, the investigation of a pmh1-1 knockout line did not reveal any influence of this protein on processing and abundance of mitochondrial transcripts.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Mitochondria of angiosperms contain their own genomes encoding about 50–60 essential genes (Kubo and Newton 2008). For expression of this organellar genetic information a large set of proteins is required. This includes many different polypeptides for posttranscriptional processes such as splicing, RNA editing as well as 5′ and 3′ end maturation of rRNA, tRNA and mRNA (Hoffmann et al. 2001; Binder and Brennicke 2003; Marchfelder and Binder 2004; Gagliardi and Binder 2007; Bonen 2008; Takenaka et al. 2008). In recent years a number of proteins involved in these posttranscriptional processes have been identified, however, the vast majority of the factors required for mitochondrial gene expression is still unknown. For the identification of such proteins, biochemical approaches have been found difficult or even impossible, while major progress in this research field has been made by genetic approaches. Using cytoplasmic male sterility systems in different plant species, a number of genes for RESTORERS OF FERTILITY (RF) have been identified using forward genetics. These genes almost exclusively encode pentatricopeptide repeat (PPR) proteins, which take part in plant mitochondrial RNA metabolism (Bentolila et al. 2002; Brown et al. 2003; Desloire et al. 2003; Kazama and Toriyama 2003; Koizuka et al. 2003; Wang et al. 2006).
In addition reverse genetic approaches have also been successfully applied to identify and characterize several genes involved in posttranscriptional processes in plant mitochondria. For instance two mitochondrial exoribonucleases and a PPR protein required for trans-splicing of an nad1 intron in Arabidopsis thaliana were characterized by such experimental strategies (Perrin et al. 2004a, b; Falcon de Longevialle et al. 2007). A similar approach is also feasible for DEAD-box proteins, which can be readily identified in silico by several typical conserved motifs (Rocak and Linder 2004; Linder 2006). These proteins are involved in virtually all processes dealing with RNA in all kingdoms of life and are often found in association with other proteins. In A. thaliana 58 genes for DEAD-box proteins have been identified (Boudet et al. 2001; Mingam et al. 2004).
In a previous study we analyzed two DEAD-box proteins, designated PMH1 (At3g22310, also called AtRH9) and PMH2 (At3g22330, also called AtRH53). These proteins share 77% identical amino acids, are located in mitochondria and are part of high molecular weight complexes (PMH1 and/or PMH2). The PMH1 and PMH2 genes exhibit distinct spatiotemporal expression patterns. PMH2 is constitutively transcribed at high levels throughout the plant. Transcript levels of this gene are moderately increased by cold and promoter activity of PMH2 can be triggered by wounding. In contrast, PMH1 is basically transcribed at low levels, but is strongly induced by cold (Matthes et al. 2007). These distinct expression patterns suggest that despite the high similarity, the two DEAD-box proteins might have at least partially different biological roles.
Here we established T-DNA insertion lines for both of these genes. Analysis of these knockout mutants shows that PMH2 is much more abundant in the high molecular weight complexes than PMH1 even after cold induction. In the pmh2 knockout mutants, splicing efficiency of the majority of introns is reduced and several mature mRNAs accumulate to decreased steady-state levels. To the contrary, no apparent differences were observed between the mitochondrial transcripts in the pmh1-1 mutant and wild type.
Materials and methods
Plant cultivation
Arabidopsis thaliana plants ecotype Col-0 including the mutants were grown on soil composed of 80% (v/v) Fruhstorfer Erde SoMi 537 Traysubstrat (HAWITA GRUPPE GmbH, Vechta, Germany), 20% (v/v) Vermiculite grain size 2–3 mm (Isola-Mineralwolle-Werke GmbH, Sprockhövel, Germany) and 1.5 g/l Osmocote Exact Mini fertilizer (Scotts Deutschland GmbH, Nordhorn, Germany). Plants were cultivated either in a growth room at 21°C, 50% rH and a light flux of 80–160 μmol*m−2*s−1 under a 16/8 h light/dark regime or in Percival growth chambers under a 16/8 h light (160–200 μmol/m2s)/dark cycle at 21°C. For the characterization of the pmh1-1 and pmh2-1 mutants, seedlings and adult plants were grown under 16 h light (160–200 μmol/m2s)/8 h dark cycle at 21°C (normal conditions) or 16 h light at 21° (160–200 μmol/m2s)/8 h dark at 4°C regime (cold treatment). Seedlings for northern analyses were cultivated for about 14 days in the growth chamber under normal conditions and kept for 24 h at 4°C prior to extraction of RNA. Seeds for RNA isolation were imbibed on filter paper for 3 h at 21°C (160–200 μmol/m2s) followed by 3 h at 4°C.
Cell suspension cultures were established from homozygous pmh1-1 or pmh2-1 seedlings. To this end seedlings were grown in the dark for 6 days on MS medium containing Gamborg B5 vitamins, 0.5% sucrose and 5 mM MES at 21°C. Hypocotyls obtained from these etiolated seedlings were then placed on MS medium containing 3% sucrose, 4.5 nM 2,4-D, 4 μM nicotinic acid, 2.4 μM pyridoxine hydrochloride, 0.3 μM thiamine hydrochloride, 0.55 mM myoinositol and 1.33 mM BAP and incubated at 21°C in the dark for 4–5 weeks until calli occurred. About 3 g of callus were then transferred into 50 ml liquid MS medium. Cultures were grown as described previously (Forner et al. 2007). For cold treatment, cultures were incubated under normal conditions (21°C) for 3 days and 24 h at 4°C prior to isolation of mitochondria.
Nucleic acid analysis
Total DNA or total RNA was extracted from plants with the DNeasy Plant Mini Kit and the RNeasy Plant Mini Kit, respectively, according to the instructions given in the manuals (QIAGEN GmbH, Hilden, Germany). Isolation of total RNA from seeds was performed according to a protocol described previously (Birtic and Kranner 2006). Genotyping of plants and RT-PCR analysis of circularized RNA (CR-RT-PCR) were done following established procedures (Kuhn and Binder 2002; Forner et al. 2007). PCRs, RT-PCRs and northern blot hybridizations were performed according to standard protocols (Sambrook and Russel 2001). For the semi-quantitative analysis of splicing efficiency PCRs were performed with three primers.
Real-Time quantitative RT-PCR was performed using Transcriptor High Fidelity cDNA Synthesis and Light-Cycler 480 SYBR Green I Master Kits on a Light-Cycler 480 Real-Time PCR System according to instructions given by the manufacturer (Roche). Data were evaluated using the Light-Cycler 480 software (1.5). Relative transcript levels were measured in respect to the validated reference genes encoding ubiquitin conjugating enzyme 9 (UBC9, At4g27960) and protein phosphatase 2A subunit 3 (PP2A, At1g13320) with established primer pairs (Czechowski et al. 2005). Oligonucleotides for the PMH2 expression analysis were selected using the primer design tool at the NCBI home page (http://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi?LINK_LOC=BlastHomeAd). All primer sequences are given in Supplemental Table S1.
Protein analysis
Mitochondria were isolated from cell suspension culture and fractionated on discontinuous sucrose gradients as reported elsewhere (Klein et al. 1998; Matthes et al. 2007). Immunodetection assays were done using standard protocols (Sambrook and Russel 2001). The PMH1/2 antiserum also binds to the 60 kDa PMH proteins but also to an approximately 65 kDa protein, which seems to be unrelated to the PMH proteins (Matthes et al. 2007). To generate recombinant protein PMH1 and PMH2, cDNAs were amplified with primer pairs AtPMH1ÜEX1.5/AtPMH1ÜEX1.3 and AtPMH2ÜEX1.5/AtPMH2ÜEX3.3, cloned into pET32a vectors (Novagen) and expressed in E. coli following standard procedures and protocols provided by the manufacturer, respectively.
Results
Establishment of PMH1 and PMH2 knockout mutants
To study the function of the PMH1 and PMH2 proteins in Arabidopsis thaliana, we analyzed several insertional mutants from the SALK collection (Sessions et al. 2002; Alonso et al. 2003). In line 035421 (pmh1-1) from this collection, PCRs with primers annealing to PMH1 sequences (pmh1RT5′ and pmh1.2 h.3′) and with oligonucleotide Lb2a complementary to T-DNA left border sequences revealed a T-DNA insertion with left borders on both sides within exon 6 of the PMH1 gene. Sequencing of the 930 and 570 bp long products confirmed the location of the T-DNA in the PMH1 gene (data not shown). In SALK line 056387 (pmh2-1), genotyping by PCR (primer pairs SALK021834/LBXL (product length 830 bp) and pmh2hy3′/LBXL (1433 bp)) identified a T-DNA insertion with left border sequences at both extremities in exon 4 of the PMH2 gene. In SAIL line 628C06 (pmh2-2), PCRs with primer pairs PMH2FLR5/SynLB and PMH2GFP2.3/SynRB amplified products of 650 and 620 bp, respectively. Sequencing of the PCR 650 bp product identified the T-DNA 97 nucleotides upstream of the ATG of PMH2 (data not shown, Fig. 1). To test whether the T-DNA insertions disrupt expression of PMH1 and PMH2, northern blot analyses with total RNA from plants homozygous for the T-DNA alleles were performed. In wild type plants, mRNAs of the expected sizes were observed, (2100 and 2150 nucleotides for PMH1 and PMH2, respectively, Fig. 1 right panels). In the pmh1-1 and pmh2-1 mutants these transcripts were undetectable demonstrating that these T-DNA insertions cause the knockout of the PMH1 and PMH2 genes, respectively. In the pmh2-2 mutant, a weak hybridization signal indicated the presence of a minor amount of mature PMH2 transcripts. Thus the PMH2 mRNA level was determined by Real-Time quantitative RT-PCR and found to be 11.5 (±0.1)% of the wild type level. This result confirms the pmh2-2 to be a knockdown allele. A macroscopic investigation of the mutants revealed that the pmh2-1 mutant is indistinguishable from wild type. The pmh1-1 plants show a slightly retarded development and reduced root lengths, however, it remains unclear whether this phenotype is indeed linked to the knockout of the PMH1 gene (data not shown).
PMH1 and PMH2 T-DNA insertion mutants. Diagrams of the PMH genes with exons indicated as grey boxes. T-DNA insertions with left or right border sequences (triangle, LB or RB) are indicated for the pmh1-1 (SALK 035421), the pmh2-1 (SALK 056387), and the pmh2-2 allele (SAIL 628C06). Northern analyses were done with total RNA from wild type (wt), pmh1-1 (1-1), pmh2-1 (2-1) and pmh2-2 (2-2) plants with PMH-specific N-terminal probes (black bars, PMH1 or PMH2) or with an oligonucleotide complementary to 18S rRNA (18S rRNA)
PMH2 is abundant in mitochondrial high molecular weight complexes
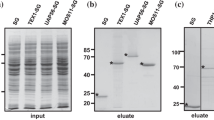
To further characterize the PMH proteins, we established cell suspension cultures from hypocotyls of etiolated seedlings homozygous for pmh1-1 and pmh2-1, respectively, as described in Materials and Methods. Cultured cells facilitate the isolation of larger amounts of highly pure mitochondria, which can then be analyzed in detail. For immunodetection analyses we used an antiserum that was raised against almost identical peptides present in PMH proteins (Matthes et al. 2007). Immunodetection assays of recombinant PMH-S-tag fusion proteins with both the PMH1/2 antiserum and the S-protein horseradish peroxidase conjugate, respectively, showed that the PMH1/2 antiserum reacts with both recombinant PMH proteins with the same efficiencies (Fig. 2a). The subsequent analysis of total mitochondrial protein extracts with the PMH1/2 antiserum revealed a signal consistent with the 60 kDa PMH proteins in wild type and both mutants (Fig. 2b, upper panel, indicated by an arrow). However, there are differences in the signal intensities. Considering the slightly unequal loading of the gel indicated by the differing amounts of Porin (Fig. 2b, lower panel), PMH2 is at least two times more abundant than PMH1 (Matthes et al. 2007).
Immunodetection analysis of the PMH proteins. a Immunodetection analysis of recombinant PMH-S-tag fusion proteins expressed in E.coli with the PMH1/2 antiserum (@PMH1/2, upper panel) and the S-protein horseradish peroxidase conjugate (S-protein HRP, lower panel). b The PMH1/2 antiserum detects the PMH proteins (indicated by an arrow) in the mitochondrial protein fractions from wild type (wt), pmh1-1 (1-1) and pmh2-1 (2-1) cell cultures (upper panel). An antiserum raised against Porin (@Porin), a protein of the outer mitochondrial membrane, was used to control the loading of the gel
We have previously found that PMH1 and/or PMH2 are part of high molecular weight complexes, but the proportion of both proteins within these complexes remained unclear. We thus analyzed high molecular weight complexes from the respective mutant and wild type cell cultures. Mitochondria isolated from these cell cultures were solubilized in the presence of 2% dodecylmaltoside and fractionated on sucrose step gradients. Equal amounts of each fraction from both mutants were analyzed in parallel with the respective protein fractions from wild type (Fig. 3). The immunodetection analysis of the pmh1-1 culture identified PMH2 in decreasing amounts in fractions 2–6, while in wild type the PMH proteins are present in equal amounts in fractions 2–6 and in minor amounts in fraction 7 (Fig. 3a). This experiment showed that PMH2 is present in the large complexes in substantial amounts. In contrast, a different pattern is revealed by the immunodetection analysis of the respective protein fractions obtained from organelles from the pmh2-1 culture. Mitochondrial proteins from sucrose gradient fractions 1–7 of the pmh2-1 culture were co-electrophoresed with fractions 1 and 2 obtained after separation of mitochondrial protein from wild type (Fig. 3b). This allowed a clear identification of the detected proteins and revealed that the PMH1 protein (lower band in the upper panel in Fig. 3b) is present only in very low amounts consistent with the result of the above mentioned immunodetection analysis (Fig. 2b). PMH1 was seen in fractions 2–5 suggesting that it is also present in large protein complexes. Thus both PMH proteins are present in different amounts in mitochondrial high molecular weight complexes. The relatively high abundance of PMH2 in the high molecular weight complexes is consistent with the generally high transcript level of this gene, while the low levels of PMH1 in these complexes correlates with low expression level.
Association of PMH1 and PMH2 with large complexes detected in pmh1-1 and pmh2-1 cell cultures. Total mitochondrial proteins from the two knockout cell lines and the wild type cell culture were size-fractionated on discontinuous sucrose gradients. Fraction 1 corresponds to the loaded sample after centrifugation of the gradient. Fraction 2 corresponds to 15%, 3–20%, 4–25%, 5–30%, 6–35%, 7–40%, 8–45% and 9–50% sucrose, respectively. Equal amounts of each fraction were analyzed by immunodetection with the PMH1/2 antiserum. a Pmh1-1 and wild type cell cultures. b Cold treated pmh2-1 and wild type cell cultures. For a clear identification of the PMH1 protein, sucrose gradient fractions 1–7 of the pmh2-1 culture were co-electrophoresed with fractions 1 and 2 obtained after separation of mitochondrial protein from wild type
Splicing efficiency of mitochondrial introns is reduced in the pmh2 mutants
The pmh1 and pmh2 knockout and knockdown mutants enabled us to search for molecular phenotypes that can indicate the function of the two DEAD-box proteins. PMH1 and PMH2 are highly expressed in early phases of germination (https://www.genevestigator.ethz.ch/gv/index.jsp) and their expression increases either up to ten-fold (PMH1) or about three-fold (PMH2) when plants are exposed to 4°C (Matthes et al. 2007). Thus we analyzed all mitochondrial mRNAs from cold treated germinating seeds since we expected a stronger phenotype in this tissue that is additionally challenged by cold.
First, we examined a potential function of PMH1 or PMH2 in 5′ and 3′ end processing of mitochondrial mRNAs. The analysis of all mitochondrial mRNAs by CR-RT-PCR as established previously (Forner et al. 2007) did not reveal any difference between transcript extremities from the mutants and from wild type (data not shown). This result suggests that both DEAD-box proteins are not required for 5′ and 3′ end formation of mitochondrial mRNAs.
Second, we investigated whether PMH1 or PMH2 play a role in splicing of mitochondrial introns. Efficiency of all 23 splicing events was examined by multiplex RT-PCRs with three primers in each reaction and with RNA obtained from cold treated seeds. One primer anneals to intron sequences, while the two other oligonucleotides are complementary to the flanking exons. When compared to wild type an increase of the PCR product originating from unspliced RNA accompanied by a decrease of the product amplified from the spliced mRNA would indicate reduced splicing efficiency in the mutant. We found that 15 of 23 group II introns had reduced splicing efficiencies in pmh2-1 plants, while six splicing events were not affected in this mutant (Fig. 4b, Supplemental Fig. S1). In addition, the results of two events remained ambiguous as differences between the pmh mutants and the wild type controls are too small or results are inconsistent between the different lines. As shown for nad2, splicing efficiency can differ within an mRNA. For introns a/b, b/c and d/e, the reductions of the spliced products were accompanied by increases of unspliced transcripts indicating that the removal of these introns is impaired in the pmh2 mutants (Fig. 4b, panels 1, 2 and 4). In contrast, no increase of the unspliced product was seen for intron c/d (Fig. 4b, panel 3). These results suggest that in the pmh2 mutants the reduction of the spliced product was caused by the reduced level of the mature nad2 mRNA as seen in the PCR covering all four splicing events (Fig. 4b, panel 5). When only a single exon was covered in the RT-PCR analysis, no reduction of nad2 transcripts was observed the mutants (Fig. 4b, panel 6). This result suggests that the amount of all nad2 transcripts including precursor molecules of the second part of the gene is not reduced.
RT-PCR analysis of nad2 transcripts from cold treated seeds. a The nad2 gene consists of 5 exons (a–e, grey boxes) encoded at two distant genomic loci. b Panels 1–4, triplex PCRs covering all nad2 introns. Calculated product sizes and the primer pairs for products originating from spliced and unspliced RNA are given in the right margin. Panel 5, PCR covering almost the complete mature nad2 mRNA. Panel 6, PCR covering exon d. Approximate localizations of the primers are given in (a). Primer sequences are listed in Supplemental Table S1. Designations of the lanes are as follows: 1-1, pmh1-1; 2-1, pmh2-1; 2-2, pmh2-2; wt1, independent Col wild type; wt2, wild type selected from the SAIL 628C06 seed probe provided by the SALK institute
To investigate mitochondrial steady-state transcripts by an independent method, northern blot analyses were preformed for various mitochondrial mRNAs (Fig. 5). In the pmh2 mutants, the steady-state levels of mature nad2 and nad4 mRNAs were reduced (Fig. 5a, b). At the same time, slight increases of larger RNAs of different sizes are observed, suggesting that precursor RNAs accumulate to slightly higher amounts in the pmh2 mutants. We also investigated the cox2 transcript. Although a reduction of the mature cox2 mRNA is not seen in the pmh2 mutants, a weak increase in levels of precursor RNA supports less efficient splicing consistent with the results of the RT-PCR analysis (Fig. 5c, upper panel and Supplemental Fig. S1). The identity of the increased cox2 precursor molecule was confirmed by hybridization with an intron-specific probe (Fig. 5c, middle panel). The detected precursor has a size of about 2600 nucleotides, which is consistent with the calculated length of 2595 nucleotides based on previously mapped mRNA termini (Forner et al. 2007). In addition, the level of the intron-less nad9 mRNAs was lowered in the pmh2 mutants (Supplemental Fig. S2). However, a general influence of the PMH2 protein on steady-state mRNA levels can be excluded since the northern analysis did not show reductions of the intron-less atp8 or nad6 transcript levels in the mutants (Supplemental Fig. S2). In contrast to pmh2-1 and pmh2-2, no substantial effects on splicing or mRNA accumulation have been observed in pmh1-1 plants.
Northern blot analysis of nad2 (a), nad4 (b) or cox2 (c) transcripts in total RNA from cold treated seedlings. Mature mRNAs corresponding to the calculated sizes (nad2: 1657, nad4: 1746 and cox2: 1250 and 1304 nucleotides) are indicated by black arrows. The cox2 precursor molecule (indicated by dashed arrows) was detected both with a probe corresponding to cDNA without intron (c, upper panel, exon) or with an intron-specific probe (c, lower panel, intron). The same membranes were probed with an oligonucleotide complementary to 18S rRNA (lower panels). Size markers are given in kilobases [kb]. Designations of the lanes are identical to those in Fig. 4. Probes used for hybridizations (solid black lanes) are indicated in diagrams given in the bottom part. The dotted line indicates the intron, which is absent from the cox2 exon probe
Finally we investigated RNA editing in the nad2 transcripts. We choose this transcript because the knockout of PMH2 has strong effects on splicing and accumulation of this RNA. Inspection of all RNA editing sites did not reveal any difference between the three pmh mutants and wild type (Supplemental Table S2 and Supplemental Fig. S3). Even partially edited sites show nearly identical efficiencies in the mutants and wild type. This demonstrates that both PMH proteins are dispensable for RNA editing of nad2 mRNA.
Discussion
PMH1 and PMH2, two mitochondrial DEAD-box proteins sharing 77% identical amino acids, are both encoded on chromosome 3 separated only by a single gene. The extreme sequence similarity in the core region of the proteins containing the conserved motifs of DEAD-box proteins implies similar or even identical functions of both proteins. But there are also arguments that suggest different biological roles of both proteins. For instance the distinct Arg/Ser/Gly-rich C-termini might allow interactions with different proteins so that complexes with different functions might be formed. Also distinct spatiotemporal expression patterns of the PMH genes might indicate different biological roles. PMH2 constitutively expressed in all tissues, is only moderately induced by cold. In contrast, PMH1 expression is strongly triggered by cold and transcription of this gene in adult plants occurs predominantly in flowers (Matthes et al. 2007). To gain information about the relevance of the PMH proteins for mitochondrial gene expression, we now investigated T-DNA knockout mutants for both genes, which allowed an individual functional examination of these genes and their gene products.
Both PMH proteins are present in mitochondrial high molecular mass complexes
We have previously found that PMH1 and/or PMH2 are part of mitochondrial high molecular mass complexes (Matthes et al. 2007). Now our analysis shows that indeed both proteins are detectable in these complexes, but the two DEAD-box proteins are clearly present in different amounts. PMH2 is found at relatively high levels, while PMH1 is present only in low quantities even after cold induction (Fig. 2b and 3). This difference in the accumulation of both proteins in these complexes is consistent with the basically higher transcription of PMH2 throughout the plant, suggesting a dominant role of PMH2 in tissue culture and in plants.
Function of PMH2
DEAD-box proteins have been shown to be involved in all RNA metabolic processes (Rocak and Linder 2004; Linder 2006) and, therefore, we hypothesized that PMH1 and PMH2 might have functions in the unique RNA processing events required for gene expression in mitochondria of Arabidopsis thaliana.
In this study we tested for alterations in all 23 mitochondrial splicing events, all major mRNA extremities, levels of several steady-state transcripts and all RNA editing events in nad2 mRNAs. The results obtained in these analyses revealed the following tendencies:
In the pmh mutants none of the analyzed processing events is completely abolished. Instead the efficiencies of many splicing events and the steady-state levels of several mature mRNAs are reduced in the pmh2 mutants. These observations indicate that at least PMH2 does not have a preference for a particular RNA substrate, but rather exhibits a relaxed specificity for many different RNAs. This broad substrate spectrum is typical for RNA chaperones, which facilitate transitions from non-functional to active conformations of structured RNA (Russell 2008). Indeed some DEAD-box proteins have been characterized as RNA chaperones. Among them the best described proteins are CYT-19 from Neurospora crassa and the related MSS116 from Saccharomyces cerevisiae (Mohr et al. 2002; Huang et al. 2005). Both proteins are required for efficient splicing of many mitochondrial group I and group II introns by converting non-native structures of these introns into catalytically active conformations. The knockout of MSS116 reduced splicing efficiency from 20 to 97% of wild type levels. Although the splicing efficiency is less severely reduced in the pmh2 mutants the phenotype is similar to what has been found for these DEAD-box proteins from fungi, strongly suggesting that PMH2 functions as RNA chaperone as it is required for efficient splicing of group II introns. The weak effect observed in the pmh2 mutants might be explained by redundant functions by other mitochondrial DEAD-box proteins such as PMH1, ISE1 and At4g09730 (Stonebloom et al. 2009) (S. Schmidt-Gattung and S. Binder, unpublished results). In addition to the contribution of CYT-19 and MSS116 to splicing, these proteins are also important for 5′ and 3′ end processing and translation. Likewise PMH2 seems to have another function that somehow influences the steady-state level of several mature mitochondrial mRNAs. In some cases this might be linked to splicing efficiency but the fact that intron-less RNAs are reduced in pmh2 mutants indicates PMH2 likely has additional functions. The reduction of the nad2 steady-state transcript level seems to be independent from transcription since an RT-PCR detecting mature as well as precursor RNAs originating from the second part of the gene do not show any reduction in total nad2 RNAs (nad2 exon d, Fig. 4 panel 6). So it appears that PMH2 in one way or another influences RNA stability.
Biological role of PMH1
In contrast to the pmh2 mutants, no molecular phenotype has so far been detected in the pmh1-1 mutant and we can presently only speculate about the function or biological role of PMH1. The location of both genes in close proximity separated only by a single gene and the extreme sequence similarity of both proteins sharing 77% identical amino acid residues suggest that the two PMH genes in A. thaliana originate from a recent duplication event. Thus one would assume that both genes have similar or even identical functions. However, identical functions for instance as RNA chaperones do not exclude that both PMH proteins are involved in different posttranscriptional processes and thus have different biological roles. Basically, three scenarios for the function and biological role of PMH1 are likely:
First, PMH1 is an RNA chaperone with a biological role very similar or even identical with PMH2. In this scenario, the knockout of PMH1 would have been fully compensated by the redundant function and biological role of PMH2. There would be full compensation even under cold stress, which is consistent with the moderate but still substantial increase of PMH2 transcription in response to cold (Matthes et al. 2007). Identical functions of both PMH proteins with nearly or even identical biological roles would be also consistent with the relatively weak molecular phenotype of the pmh2 mutants, which might be not fully compensated by PMH1, because of its generally low expression. Presumably further investigations of the RNA metabolism in a pmh1/pmh2 double knockout or knockdown mutants would allow further conclusions about redundant functions or roles of both genes. However, the knockout or knockdown of both PMH genes might lead to a strong molecular phenotype with severe consequences for plant viability. In line with that our attempts to constitutively knockdown PMH2 in the pmh1-1 mutant reduced PMH2 transcripts maximally by 60% of wild type level, while plants with stronger reductions have not been obtained. The relatively moderate decrease of the PMH2 transcript level in these knock-down plants did not even affect splicing indicating that probably also other molecular phenotypes cannot be detected in these plants (D. Köhler and S. Binder, unpublished results).
Second, PMH1 is an RNA chaperone but is relevant only under specific conditions and/or in distinct tissues. This scenario would be consistent with the generally low levels of PMH1 transcripts, which are elevated only in flowers and in response to abiotic stress like cold (Matthes et al. 2007). Thus PMH1 would have a function identical with PMH2 but would fulfill a distinct biological role. To elucidate this different role of PMH1, RNA from other tissues and/or from plants challenged by various stress conditions has to be examined.
Third, it can presently not be excluded that PMH1 has a completely different function than PMH2. PMH1 might specifically bind to a single or only very few RNA targets being required for a particular process in generation, maturation or even translation. Binding specificity of the PMH proteins might be determined by the C-termini. Although these C-terminal parts of both PMH proteins are composed of the same amino acids, i.e. mainly Ser and Gly, they differ considerably in sequence. In A. thaliana a very similar C-terminus of a nuclear DEAD-box protein was identified as an RNA binding module (Lorkovic et al. 1997) indicating that the C-termini of the PMH proteins have similar functions with potentially different substrate preferences.
Further studies of pmh mutants combined with inducible knockdowns of the otherwise intact PMH gene are required to unambiguously elucidate the exact molecular functions of PMH1 and PMH2 in the RNA metabolism in mitochondria of A. thaliana.
References
Alonso JM, Stepanova AN, Leisse TJ et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301:653–657
Bentolila S, Alfonso AA, Hanson MR (2002) A pentatricopeptide repeat-containing gene restores fertility to cytoplasmic male-sterile plants. Proc Natl Acad Sci USA 99:10887–10892
Binder S, Brennicke A (2003) Gene expression in plant mitochondria: transcriptional and posttranscriptional control. Phil Trans R Soc Lond B 358:181–199
Birtic S, Kranner I (2006) Isolation of high-quality RNA from polyphenol-, polysaccharide- and lipid-rich seeds. Phytochem Anal 17:144–148
Bonen L (2008) Cis- and trans-splicing of group II introns in plant mitochondria. Mitochondrion 8:26–34
Boudet N, Aubourg S, Toffano-Nioche C, Kreis M, Lecharny A (2001) Evolution of intron/exon structure of DEAD helicase family genes in Arabidopsis, Caenorhabditis, and Drosophila. Genome Res 11:2101–2114
Brown GG, Formanova N, Jin H et al (2003) The radish Rfo restorer gene of Ogura cytoplasmic male sterility encodes a protein with multiple pentatricopeptide repeats. Plant J 35:262–272
Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139:5–17
Desloire S, Gherbi H, Laloui W et al (2003) Identification of the fertility restoration locus, Rfo, in radish, as a member of the pentatricopeptide-repeat protein family. EMBO Rep 4:588–594
Falcon de Longevialle A, Meyer EH, Andres C, Taylor NL, Lurin C, Millar AH, Small ID (2007) The pentatricopeptide repeat gene OTP43 is required for trans-splicing of the mitochondrial nad1 intron 1 in Arabidopsis thaliana. Plant Cell 19:3256–3265
Forner J, Weber B, Thuss S, Wildum S, Binder S (2007) Mapping of mitochondrial mRNA termini in Arabidopsis thaliana: t-elements contribute to 5′ and 3′ end formation. Nucleic Acids Res 35:3676–3692
Gagliardi D, Binder S (2007) Expression of the plant mitochondrial genome. In: Logan D (ed) Plant mitochondria. Blackwell, Ames, pp 50–95
Hoffmann M, Kuhn J, Däschner K, Binder S (2001) The RNA world of plant mitochondria. Prog Nucleic Acid Res Mol Biol 70:119–154
Huang HR, Rowe CE, Mohr S, Jiang Y, Lambowitz AM, Perlman PS (2005) The splicing of yeast mitochondrial group I and group II introns requires a DEAD-box protein with RNA chaperone function. Proc Natl Acad Sci USA 102:163–168
Kazama T, Toriyama K (2003) A pentatricopeptide repeat-containing gene that promotes the processing of aberrant atp6 RNA of cytoplasmic male-sterile rice. FEBS Lett 544:99–102
Klein M, Binder S, Brennicke A (1998) Purification of mitochondria from Arabidopsis. Methods Mol Biol 82:49–53
Koizuka N, Imai R, Fujimoto H, Hayakawa T, Kimura Y, Kohno-Murase J, Sakai T, Kawasaki S, Imamura J (2003) Genetic characterization of a pentatricopeptide repeat protein gene, orf687, that restores fertility in the cytoplasmic male-sterile Kosena radish. Plant J 34:407–415
Kubo T, Newton KJ (2008) Angiosperm mitochondrial genomes and mutations. Mitochondrion 8:5–14
Kuhn J, Binder S (2002) RT-PCR analysis of 5′ to 3′-end-ligated mRNAs identifies the extremities of cox2 transcripts in pea mitochondria. Nucleic Acids Res 30:439–446
Linder P (2006) Dead-box proteins: a family affair–active and passive players in RNP-remodeling. Nucleic Acids Res 34:4168–4180
Lorkovic ZJ, Herrmann RG, Oelmuller R (1997) PRH75, a new nucleus-localized member of the DEAD-box protein family from higher plants. Mol Cell Biol 17:2257–2265
Marchfelder A, Binder S (2004) Plastid and plant mitochondrial RNA processing and RNA stability. In: Daniell H, Chase C (eds) Molecular biology and biotechnolgy of plant organelles. Springer, Dordrecht, pp 261–294
Matthes A, Schmidt-Gattung S, Köhler D, Forner J, Wildum S, Raabe M, Urlaub H, Binder S (2007) Two DEAD-box proteins may be part of RNA-dependent high-molecular-mass protein complexes in Arabidopsis mitochondria. Plant Physiol 145:1637–1646
Mingam A, Toffano-Nioche C, Brunaud V, Boudet N, Kreis M, Lecharny A (2004) DEAD-box RNA helicases in Arabidopsis thaliana: establishing a link between quantitative expression, gene structure and evolution of a family of genes. Plant Biotechnol J 2:401–415
Mohr S, Stryker JM, Lambowitz AM (2002) A DEAD-box protein functions as an ATP-dependent RNA chaperone in group I intron splicing. Cell 109:769–779
Perrin R, Lange H, Grienenberger JM, Gagliardi D (2004a) AtmtPNPase is required for multiple aspects of the 18S rRNA metabolism in Arabidopsis thaliana mitochondria. Nucleic Acids Res 32:5174–5182
Perrin R, Meyer EH, Zaepfel M, Kim YJ, Mache R, Grienenberger JM, Gualberto JM, Gagliardi D (2004b) Two exoribonucleases act sequentially to process mature 3′-ends of atp9 mRNAs in Arabidopsis mitochondria. J Biol Chem 279:25440–25446
Rocak S, Linder P (2004) DEAD-box proteins: the driving forces behind RNA metabolism. Nat Rev Mol Cell Biol 5:232–241
Russell R (2008) RNA misfolding and the action of chaperones. Front Biosci 13:1–20
Sambrook J, Russel DW (2001) Molecular cloning: a laboratory manual. Cold Spring Habror Laboratory Press, Cold Spring Harbor
Sessions A, Burke E, Presting G et al (2002) A high-throughput Arabidopsis reverse genetics system. Plant Cell 14:2985–2994
Stonebloom S, Burch-Smith T, Kim I, Meinke D, Mindrinos M, Zambryski P (2009) Loss of the plant DEAD-box protein ISE1 leads to defective mitochondria and increased cell-to-cell transport via plasmodesmata. In: Proceedings of the national academy of science USA. Epub 21 Sept 2009, doi:10.1073/pnas.0909229106
Takenaka M, Verbitskiy D, van der Merwe JA, Zehrmann A, Brennicke A (2008) The process of RNA editing in plant mitochondria. Mitochondrion 8:35–46
Wang Z, Zou Y, Li X et al (2006) Cytoplasmic male sterility of rice with boro II cytoplasm is caused by a cytotoxic peptide and is restored by two related PPR motif genes via distinct modes of mRNA silencing. Plant Cell 18:676–687
Acknowledgments
An antiserum raised against Porin from potato was kindly provided by Hans-Peter Braun (Hannover). We thank Conny Guha and Uli Tengler for excellent technical assistance. We are also very grateful to Solomon Stonebloom (Berkeley) for his very helpful comments on the manuscript. This work was supported by grants Bi 590/7-1 and 7-2 from the Deutsche Forschungsgemeinschaft.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Köhler, D., Schmidt-Gattung, S. & Binder, S. The DEAD-box protein PMH2 is required for efficient group II intron splicing in mitochondria of Arabidopsis thaliana . Plant Mol Biol 72, 459–467 (2010). https://doi.org/10.1007/s11103-009-9584-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-009-9584-9