Abstract
Diabetes is a chronic progressive spectrum of diseases with debilitating comorbidities that is one of the top ten leading causes of deaths globally. The development and application of drug–disease models have made significant contributions to all stages of discovery and development for the treatment of diabetes. There are various reliable biomarkers that describe the physiological processes involved in the glucose homeostasis and pathophysiological state of inadequate glucose control. These biomarkers can be classified as fast and slow biomarkers that are highly predictive of the long-term clinical outcomes. Drug–disease models have been developed that describe the dynamics of these biomarkers with varying complexities ranging from systems pharmacology to disease progression and cardiovascular outcome measures. In this chapter, the readers are introduced to the disease of diabetes, well understood predictive biomarkers associated with diabetes, available target-specific therapeutic interventions, drug–disease models and their applications, including a detailed case example in systems pharmacology model. Several key drug–disease models are described with details to facilitate implementation for common therapeutic targets. The availability of predictive biomarkers and quantifiable clinical endpoints has made the development and application of drug–disease modeling highly efficient and valuable in drug discovery and development of important therapies for the treatment of diabetes and associated comorbidities.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Diabetes drug–disease model
- Systems pharmacology
- Indirect-response model
- Glucose homeostasis
- Disease progression
5.1 Introduction
Diabetes is a chronic, progressive disease that is estimated to be one of the top ten leading causes of death globally (WHO fact sheet number 310). Type 2 diabetes is the most common type of diabetes, affecting 90–95 % of the US diabetes population. According to the latest International Diabetes Federation (IDF) report, about 8.3 % of the global adult population, or 382 million people, have diabetes. The number of newly diagnosed cases worldwide continues to grow every year and the global figure is expected to rise to 595 million by 2035. The majority of the 382 million people with diabetes are aged between 40 and 59, and 80 % of them live in low- and middle-income countries. The economic burden of diabetes is approximately 548 billion US $ in health spending (11 % of the total spent worldwide) in 2013 (IDF report 2013). Therefore, improving the treatment and management of diabetes, its comorbidities, and associated complications continue to be an important focus of pharmaceutical research and development (R&D).
Diabetes is a metabolic disease that is rich with quantitative biomarkers and well understood regulatory and counter-regulatory processes. For these reasons, diabetes is one of the therapeutic research areas rapidly gaining R&D efficiencies with the use of predictive drug and disease models. The mathematical modeling of glucose–insulin homeostasis has provided significant insights into the underlying mechanisms of the disease and is becoming a critical component of the pharmaceutical R&D program (Ajmera et al. 2013). Driven by the increasing cost of drug development and the high rate of late-stage failures (over 90 % of clinical candidates never make it to market), the scientific leaders and regulators, advocated for the incorporation of model-based approaches into drug development processes (Woodcock and Woosley 2008) to improve the efficiency and the quality of decision-making: to select targets, predictive biomarkers, drug candidates, clinical trial designs, dosing range or regimens, and development programs with high probabilities of success in all the stages of drug discovery and development. Fueled by the recent technical advances in computational power, aided by the arsenal of predictive drug and disease models and study design efficiency-enhancing tools, pharmacometricians have been able to contribute more effectively in the early terminations of the “bad” drugs and the optimization of the development program to expedite the delivery of the “good” drugs to the patient.
All drug-disease models developed should be fit for purpose, specifically, provide answers to the questions of interest. Models should be developed with well-characterized basic physiology and biochemical regulatory aspects of the disease in mind in order to have predictive fidelity. It is consequently important that the pharmacometricians have a basic understanding of the disease, including that the term “diabetes” does not characterize a single, homogenous disease but rather encompasses a group of metabolic disorders, which are all characterized by hyperglycemia that result from defects in insulin secretion, insulin action, or both (American Diabetes Association 2008). It is a condition in which a person has higher than normal blood glucose levels either because the body does not produce enough insulin in response to meal intake (impaired beta cell functions), or because the body does not properly respond to the insulin that is produced (insulin resistance). Insulin is a hormone produced by the beta cells in the Islets of Langerhans located in the pancreas which promotes the uptake of glucose by tissues such as muscles and adipose thereby mediating the clearance of glucose. If there is a diminished uptake of glucose by tissues due to resistance to insulin, the beta cell will compensate by secreting just enough insulin to normalize glucose level (euglycemia). Over time, the acute insulin secretory response to glucose is exhausted and the homeostatic feedback control diminishes progressively, leading to hyperglycemia and ultimately diabetes mellitus. A persistent state of hyperglycemia is associated with complications, including an increased susceptibility to infections, ketoacidosis, microvascular diseases, such as nephropathy or retinopathy and may lead to early macrovascular complications, such as heart attack and stroke (Morghissi et al. 2007). In recent years, there has been a significant emergence of type 2 diabetes driven by lifestyle factors leading to increased body weight and obesity .
Type 2 diabetes can be controlled with various treatment modalities. Regimented treatment of diabetes is important and, generally, a holistic approach that includes blood glucose and blood pressure control, and lifestyle changes, such as maintaining a healthy body weight is recommended. Therefore, the choice of treatments depends on the disease status and often includes more than one antihyperglycemic medication.
5.2 Therapeutic Interventions
An important component in developing drug–disease models of diabetes is that soon after diagnosis, patients with type 2 diabetes mellitus may be able to manage their glucose levels on diet and exercise alone for a few years. However, the disease usually progresses over time requiring multiple drugs to be prescribed concomitantly. Numerous drugs of different mechanisms of action are available by oral or subcutaneous routes of administration. The therapeutic combination for the treatment of type 2 diabetes may include insulin, to provide better glycemic control in combination with the more convenient oral agents. The antihyperglycemic medicines that are available in the market by pharmacologic class include:
-
Biguanides for inhibition of hepatic gluconeogenesis (metformin)
-
Insulin secretagogues (sulfonylureas)
-
Insulin sensitizers (thiazolidinedione)
-
Alpha-glucosidase inhibitors for glucose or starch absorption (acarbose)
-
Incretin mimetics for glucose-dependent insulin secretion (glucagon-like-peptide-1, or GLP-1 anlogues)
-
Dipeptidyl peptidase-4 (DPP-4) inhibitors
-
Sodium-dependent glucose co-transporter 2 (SGLT2) inhibitors
-
Insulins (including long-acting basal insulins)
In addition, there are many investigational agents in various stages of drug discovery and development, which are targeting different pathways for glucose control (Verspohl 2012), such as glucagon receptor antagonist, glucokinase activator, incretin hormones , sodium-dependent glucose co-transporters, G-protein-coupled receptor agonists, etc. Many newer agents are designed to have pleiotropic effects and have beneficial attributes in addition to glucose lowering to provide the additional benefit in the management of multiple facets of this complex metabolic disorder.
5.3 Biomarkers and Clinical Surrogates
Diabetes disease models are developed using a plethora of quantitatively predictive and clinically relevant biomarkers . The standard biomarker panel is not limited to fasting blood glucose (FBG) and postprandial glucose (PPG). There are numerous biomarkers and pharmacodynamic measurements available to assess glycemic status and pancreatic beta cell health as well as to evaluate the effects of pharmacologic interventions. The pancreas releases insulin which is produced in the pancreatic beta cells, and glucagon is produced in the alpha cells. Glucagon is an antagonist to insulin causing hepatic glucose output to increase either by gluconeogenesis or glycogen breakdown—its effect detectable following prolonged hypoglycemia. In addition, hormones (somatostatin, growth hormone, cortisol, gastrointestinal hormones, etc.), amino and fatty acids also play roles in this complex metabolic system.
The choice of a pharmacodynamic biomarker in model development is dependent on the drug’s mechanism of action, the duration of the trial or the stage of development and the objective of the assessment. In addition, during the translational phase of development, the choice of animal models of disease is important and is often dependent on the mechanism of action of the pharmacologic agent and known inter-species differences in the target expression and biomarker response (Shafrir 2007, 2010).
Acute biomarkers (measured in minutes, hours, or days) are measured in preclinical and early clinical phases of drug development in trials of short duration (e.g., in phase 1). The most common biomarkers assessed in early trials are FBG, PPG, C-peptide, or insulin, in response to meals or glucose challenges. Biomarkers of the target engagement may include glucagon, dipeptidyl peptidase-4 [DPPIV] enzyme inhibition, glucagon-like peptide-1 (GLP-1), gastric inhibitory polypeptide (GIP), and other hormones . In addition, complex systems pharmacology models of diabetes may also incorporate information based on biomarkers of pleiotropic effects, such as, cholesterols, free fatty acids and hemodynamic measures (e.g., blood pressure and heart rate) as well as standard laboratory assessment of cardio-renal functions .
Glycosylated hemoglobin (HbA1c) is formed through a nonenzymatic and irreversible reaction between glucose and hemoglobin. HbA1c is a clinical surrogate for long-term disease progression and treatment effects and is, thus, well accepted as an efficacy endpoint in long-term trials (e.g., months to years). Fasting insulin and C-peptide levels, on the other hand, are measures of endogenous insulin production and are used to assess insulin resistance and beta cell function as well as markers of disease progression. Long-term outcomes of diabetes and diabetes complications, such as strokes, coronary heart disease , neuropathy or nephropathy, might take several years to present themselves. Outcomes are often assessed using empirical or Bayesian probability models, rather than drug–disease models.
Each of these biomarkers, whether fast or slow turnover markers, carry importance at different stages of the drug discovery and development process. It is important to have a good understanding of the translatability and reproducibility of these biomarkers such that the use of these biomarkers is reliable and has predictive fidelity.
To evaluate the drug effects, pharmacokinetics-pharmacodynamics relationships are developed in the forms of models linking the concentration of drug to biomarkers of interest or outcomes, as shown in Fig. 5.1.
Diabetes is well known as a risk factor for cardiovascular disease. Despite glucose control, the risk of cardiovascular mortality and morbidity remains high in patients with diabetes. Drug and disease modeling approaches may incorporate cardiovascular biomarkers in combination with glycemic parameters to assess the impact of a new therapeutic intervention on both diabetes and cardiovascular outcomes (Vlasakakis and Pasqua 2013).
Due to increasing awareness in the holistic treatment of the comorbidities of diabetes, namely, obesity and cardiovascular diseases, novel therapies not only treat hyperglycemia but also aim to manage the symptoms of these comorbidities, often referred to as “diabetes plus”. Therefore, the biomarker panels have been expanded to include measures of cardiometabolic health (e.g., body mass index, waist circumference, body fat composition, lipids, triglycerides), assessment of hemodynamics (e.g., blood pressure, heart rate), and risk of arrhythmias (ECG changes). This chapter focuses on the modeling of glycemic parameters; however, similar modeling concepts may be applied to cardiovascular biomarkers.
5.4 Drug–Disease Models of Diabetes
Leveraging the range of available biomarkers, computational models play an increasingly important role in understanding the dynamic behaviors and the mechanisms underlying diverse and complex biological systems, leading to better drug candidate selection, study design, dose and dosing regimen decision, and ultimately, better control and treatment for diabetes .
Computational models of diabetes, either mathematical or statistical, in the published literature can broadly be classified into clinical and nonclinical categories, based on complexity, depth of biological description, and the type of data (individual or population level; Landersdorfer and Jusko 2008; Ajmera et al. 2013). Analysis models of clinical data are mostly empirical in nature and emulate clinical data by considering only essential biological descriptors. Due to the purpose of their uses, these models are useful in understanding effects of dose (concentration) of new treatments, time course in changes of response, understanding disease progression, and predicting risks for complications. Nonclinical physiologically based models are more complex in nature and account for the mechanistic description of the biological systems, eventually, through translational sciences, aimed at being used for simulating clinical scenarios. Recently, semi-mechanistic and mechanistic systems pharmacology models have also been used in clinical setting as described later in this chapter.
As emphasized earlier, the choice of the model really depends on the questions that need to be answered and the nature of available data. For example, simple models without mechanistic understanding that are developed to describe observed clinical data can be used for retrospective hypothesis testing and clinical trial optimization using simulations. On the other hand, a mechanistic model may be necessary to generate prospective hypothesis to be evaluated where changes in biomarkers can be assessed by altering specific biochemical pathways .
In the following section, we summarize key types of models that have to be utilized with nonclinical and clinical data to describe the dynamics of biomarkers of interest. Most of these models can be adequately modified to characterize and predict different biomarkers data and their interactions.
5.4.1 Systems Pharmacology Models
Conceptually, the systems approach is a mathematical representation of the pertinent physiology that comprise of the key pathways or targets of interest. Systems pharmacology models usually employ a “bottom up” modeling approach to represent the physiology and disease states. The approach requires physiological or systems-level information as well as the biological pathways and mechanisms. These physiologically based models aimed to quantitatively integrate relevant biology across the systems, with targets or pathways, are expressed as state variables and parameters. The parameters in these complex models typically include those reported in the literature and those calibrated to match subsystem and/or system-level behaviors. Each unique set of model parameterization represents one “virtual patient,” and each virtual patient response is qualified by comparing simulated responses to experimentally observed or published data. This approach focuses on finding biologically feasible parameterizations that reproduce critical behaviors, rather than on exact characterization of numerous difficult-to-measure parameters (Kansal 2004; Klinke 2008; Shoda et al. 2010; Schaller et al. 2013). The PK and PD properties of the drug(s) of interest are subsequently evaluated on specific pathways or targets in a systematic fashion. Together, these integrated drug–disease models are used to simulate the expected physiological and pharmacological responses to a novel therapy or combination of therapies or clinical trial simulations in virtual patient populations (Waters et al. 2009). A detailed case study is presented at the end of this chapter to describe the development and application of systems pharmacology models using a glycogen phosphorylase inhibitor (GPi) for the treatment of type 2 diabetes.
5.4.2 Models for Glucose–Insulin Interaction
An advanced model of glucose–insulin regulation was developed using data from both healthy and type 2 diabetic subjects in glucose provocation experiments (Jauslin et al. 2007, 2011; Silber et al. 2007). Briefly, the glucose model is described using a two-compartment model with a glucose absorption component. As shown in Fig. 5.2, the glucose model also includes two effect compartments accounting for effects on glucose production and insulin secretion. The model for insulin incorporates both secretion and distribution. Baseline glucose and insulin values are represented as population values with inter-subject variability terms.
One application of this model is for evaluation of combination of treatments with different mechanisms of action. An example of this application is the prediction of glucose response to investigational insulins or incretin mimetics, when added to metformin (a drug that affects hepatic glucose production) in combination with sulfonylurea (a drug that increases insulin secretion). The pharmacokinetic component of drug treatment can be introduced into the model by linking to the site of action. This model characterizes the fast biomarkers, thus the application of this model to predicting long-term steady-state biomarker response is limited. For such an application, the placebo response with respect to inter-occasion glycemic variability and links to HbA1c response will need to be considered. As glucose input is the driver for the biomarker dynamics, this model requires reliable details about the glucose (as OGTT, IVGTT, MGTT or meals) intake.
5.4.3 Models for Glucose–Insulin–Glucagon Interaction
The first models exploring the glucagon counter-regulation (GCR) mechanism were proposed by Farhy and McCall (2009) based on rodent studies. As all the components of these models were clinically measurable, these models identify the role of delayed feedback illustrating the relationship between basal glucagon level and different aspects of GCR responses to insulin-induced hypoglycemia in T1DM conditions. Hetherington et al. (2011) and Sumner et al. (2011) developed a composite model for glucagon/insulin driven liver glucose homeostasis by linking a series of sub-system models corresponding to different aspects of physiology. This model has been used further to explore the behavior of glucose homeostasis systems by modulating the liver insulin sensitivity and diet glucose level. Kim et al. (2007) developed a multi-scale model illustrating hormonal control of whole-body glucose homeostasis during exercise and can be envisioned as a roadmap towards achieving a holistic mechanistic view of the glucose homeostasis system from sub-cellular to a “whole-body” level.
Schneck et al. (2013) extended the insulin–glucose interaction model described in previous section to incorporate the key counter-regulatory hormone glucagon; this model was utilized to investigate the effect of a novel glucokinase activator on glycemic control. Baseline glucagon secretion, the inhibitory influence of glucose and insulin, and the stimulatory influence of ingested exogenous protein on glucagon secretion were combined in a differential equation to describe glucagon dynamics. An effect compartment was utilized to represent a delayed effect of glucagon within the system. This model describes the dynamics of fast biomarkers (e.g., glucose, insulin, and glucagon), thus limits the ability to utilize this model for predicting long-term HbA1c effects. As glucose input is the driver for the biomarker dynamics, this model requires reliable details about the glucose intake.
5.4.4 Time Course Models—Fasting Blood Glucose or HbA1c
In clinical trials, glycemic parameters (fasting blood glucose or HbA1c) are measured at intervals during the course of the trial. These time courses of the glycemic parameters are used to evaluate the effects of an intervention relative to placebo or an active comparator. A typical profile of FBG as a function of time is shown in Fig. 5.3. The time course profile of HbA1c or any other biomarker can also be generated and modeled in similar fashion.
At randomization, prior antidiabetic treatments may be washed out to allow better evaluation of the active treatments. Wash-out of prior medication may cause a baseline excursion in fasting glucose. Specifically, trials to evaluate a new agent for a monotherapy indication may include a lead-in phase, during which patients discontinue and washout their previous antidiabetic agents. In these trials, during the lead-in phase, FBG levels will rise as shown by the placebo response curve in Fig. 5.3. Upon treatment, FBG decreases from baseline to reach a maximum possible effect (E max) for the assigned dose. The following model can be used to describe the rise in FBG in the placebo group and the fall in FBG levels with drug treatment.
There are various ways of describing the placebo and drug effects. One of the ways we can describe this relationship is by the following equation:
where \({{P}_{max}}\) is the maximum change in FBG for placebo; \({{E}_{max}}\) is the maximal drug effect, \(E{{C}_{50}}\) is the drug exposure that produces half maximal effect. Exposure is the drug exposure (which may be the area-under-curve or average concentration during a doing interval, or dose); \(FB{{G}_{Base}}\) is the baseline FBG; \({{k}_{eff}}\) is a rate constant of glucose turnover in determining the time required to achieve the maximum treatment or placebo effect; γ is a concentration–response steepness parameter (Hill coefficient); OAD stands for oral antidiabetic drug which is the excursion between initiation of washout of antihyperglycemic medications to baseline FBG.
In some cases, when the time-course of biomarker response may not be critical, for example, when the dose (exposure)–response relationship to be evaluated is at steady state, a model describing the changes at a predefined endpoint (for example, at 12 or 26 weeks) may be sufficient. The equation described above may be modified to:
where all the terms are as described earlier. It should be noted that the term that characterized the time course (with \({{k}_{eff}}\) and time) has been removed in this equation.
In longer clinical trials, where HbA1c values are measured to evaluate the effects of a therapeutic intervention, the time course model described in this section for FBG can be easily applied to HbA1c.
In trials of add-on therapy during which patients continue taking their antihyperglycemic medications, without washout, FBG levels are stabilized at baseline.
5.4.5 Indirect Response Models—Insulin, Glucose, and HbA1c
As previously described, the biomarkers that are frequently measured in clinical trials for antidiabetic medications are fasting serum insulin (FSI), FBG, and HbA1c. A generalized approach can be taken to link FSI, FBG, and HbA1c to describe the time courses of these biomarkers . Figure 5.4 shows a representation of the key components and the relationships that can be used in the model.
A generalized model structure linking fasting serum insulin, fasting blood glucose, and glycosylated hemoglobin (HbA1c). Open boxes show the stimulation and the solid box represents suppression of biomarker production rates. Shaded boxes represent the rates which are impacted by drugs working via different mechanisms
Møller et al. (2013) used a similar approach of linking glucose to HbA1c where the mean plasma glucose (from 24-h glucose measurements) instead of FBG was used to develop an indirect response model and predict long-term HbA1c changes based on short-term mean plasma glucose data.
5.4.6 Physiological Linked Fasting Glucose and HbA1c Model
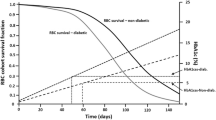
Hamrén et al. (2008) published a model that linked fasting glucose and HbA1c using a transit compartment model informed by physiology which is a mechanistically better approach versus using an indirect response relationship between these two biomarkers (as described in Sect. 5.4.5). In this model, a series of four transit compartments describe red blood cell (RBC) aging with a zero-order release of RBCs into the circulation. A first-order rate constant defines the transition of RBCs from one stage to the next until the cell dies as shown in Fig. 5.5.
Fasting glucose—glycosylated hemoglobin (HbA1c) transit model using red blood cells (RBCs) lifespan. (Adapted from Hamrén et al. 2008)
5.4.7 Models for Progression
Efforts towards the development of novel antidiabetic agents are directed at the drugs that can alter diabetes progression. In studies of greater than 1 year duration, it is important to include disease progression in the model in order to study the long-term effect of antidiabetic agent at different stages of progression. Disease progression models specific for diabetes incorporating long-term population studies with antidiabetic agents have been developed (Frey et al. 2003; de Winter et al. 2006). De Winter et al. (2006) utilized clinical data with pioglitazone, metformin, and gliclazide to assess the disease progression and drug effect using indirect response model linking FSI, FBG, and HbA1c as described in Sect. 5.4.5 (Fig. 5.6). The model described the rate of disease progression through the drugs effect on beta cell function and insulin sensitivity.
Schematic representation of the structure of the mechanism-based population disease progression model, including the homeostatic feedback between fasting serum insulin (FSI) and fasting blood glucose (FBG) and the feed-forward between FBG and glycosylated hemoglobin (HbA1c). Open boxes show the stimulation and the solid box represents suppression of biomarker production rates. Shaded boxes represent the rates which are impacted by drugs and disease progression
Topp et al. (2000) developed a model of disease progression by considering beta cell mass together with insulin and glucose concentrations. This model was described by three nonlinear ordinary differential equations, where glucose and insulin dynamics were fast relative to beta cell mass dynamics. Extending this model, Ribbing et al. (2010) proposed a semi-mechanistic pharmacokinetic/pharmacodynamic model, illustrating the dynamics of fasting plasma glucose, fasting insulin, insulin sensitivity, and beta-cell function in a heterogeneous population.
5.4.8 Models for Diagnostic Tests
In order to evaluate the diabetic and prediabetic condition in an individual, different glucose tolerance tests, such as intravenous glucose tolerance test (IVGTT), oral glucose tolerance test (OGTT), and mixed meal glucose tolerance test (MMTT) have been devised. The aims of these tests are to obtain estimates of insulin sensitivity (IS), glucose effectiveness (potency or potentiation), insulin secretion, and beta cell function. Since the liver metabolizes more than half of the secreted insulin before it is utilized by other body tissues, accurate estimation of pre-hepatic insulin secretion, hepatic insulin extraction and clearance are essential for evaluating insulin secretion and beta cell function under normal and diseased conditions. Plasma C-peptide, a part of preproinsulin peptide and therefore secreted in equimolar amounts as insulin by beta cell, acts as an indicator for insulin secretion. However, peripheral C-peptide has a longer half-life and can limit the accurate estimation of insulin secretion. Consequently, a model-based approach may be recommended for greater accuracy.
The assessment of insulin sensitivity has been conducted using either the glucose clamp technique or the minimal models (Bergman et al. 1979, 1989) which had insulin sensitivity and glucose effectiveness as the main parameters. Cobelli et al. (2009) incorporated peripheral compartment for glucose distribution which modeled the glucose and insulin data simultaneously. These models do not take into account dynamic control mechanisms and were not ideal for predictive purposes. Significant improvement over the minimal models was achieved incorporating an additional compartment for glucose kinetics and data from labeled IVGTT experiments. Although these bi-compartment models allow precise estimation of IS and potency, the additional cost and technology involved in using labeled IVGTT make it impractical for application in large clinical trials or patient care settings. Silber et al. (2007, 2010) proposed an integrated insulin–glucose model to describe IVGTT data from healthy as well as diabetic individuals, using an insulin–glucose feedback mechanism. As OGTT closely resembles the physiological condition, this model was extended further by incorporating the description for glucose absorption and incretin effects following a meal or 24-h glucose and insulin profile following multiple test meals.
5.5 Case Study: Systems Pharmacology Model
A case example of systems pharmacology model aims to assess the therapeutic potential of a glycogen phosphrylase inhibitor (GPi) for treatment of type 2 diabetes. Glycogen phosphorylase is the rate-limiting enzyme in glycogenolysis and thus is responsible for roughly 50 % of hepatic glucose output (HGO). Glycogenolysis is thought to be elevated in type 2 diabetes and several publications have described GPi as a promising therapeutic strategy for type 2 diabetes (Martin et al. 1998; Baker et al. 2005; Torres et al. 2011). The general approach to describe the pertinent physiology is to start with a “core” model of fundamental mechanisms that have been well characterized clinically. An example of such a systems or physiological model that would enable evaluation of various targets of glucose regulation is presented in Fig. 5.7.
As illustrated in Fig. 5.7, glucose enters the blood from the gastrointestinal tract following a meal (SGLT-1 transporter mediated absorption) or through hepatic glucose production (via G6Pase enzyme). The regulation of HGO has been well characterized, which can be completely suppressed by high glucose, high insulin, or low glucagon levels. Conversely, low glucose, low insulin, or high glucagon can increase HGO by about fourfold. Glucose is utilized by major tissues of the body, most notably the brain , muscle, and abdominal organs (splanchnic tissue). The brain takes up glucose at a roughly constant rate via GLUT2 transporter. Muscle glucose uptake occurs via GLUT4 and GLUT1 transporters. Splanchnic glucose uptake is thought to be glucose dependent (via GLUT2 transporter). Finally, glucose is filtered, and then reabsorbed at the kidney via SGLT-1 and SGLT-2 transporters. However, the reabsorption process begins to saturate when glucose rises above 180 mg/dl resulting in urinary glucose excretion.
This core model of glucose regulation is coupled to simple models of insulin and glucagon dynamics. Insulin secretion is driven largely by plasma glucose levels and incretin hormones (GLP-1, GIP, etc.). It should also be noted that insulin is secreted directly into the liver via the portal vein where about 50 % is cleared on first pass through the liver. Thus, hepatic insulin levels are roughly twice as high as plasma levels. Muscle insulin levels are similar to plasma, but there is a time delay required for insulin to diffuse through the tight capillary junctions. Glucagon is a counter-regulation hormone that increases three- to fourfold during the development of hypoglycemia. A schematic of simple insulin model is represented in Fig. 5.8.
Baseline parameters for this mechanistic model of glucose and insulin dynamics represent the estimated population mean value for healthy volunteers derived from a meta-analysis of public literature. Virtual patients with type 2 diabetes are created by incorporating a real-world distribution of insulin resistance (muscle and liver), and insulin secretory defects. As a specific example, if the therapy of interest targets liver, then the core model can be expanded to include target specific liver physiology; in this case the glycogenolysis, glycogen synthesis, gluconeogenesis, and glycolysis (Fig. 5.9).
Available data suggest that glycogenolysis and gluconeogenesis contribute roughly equally to total HGO. Glucose, insulin, glucagon and carbohydrate metabolites (represented in this model by G6Pase process) all contribute to the regulation of glycogenolysis and glycogen synthesis. Glycolysis is thought to be a substrate driven process while gluconeogenesis is thought to be roughly constant throughout the day. However, gluconeogenesis has been shown to increase during prolonged fast and has been linked to glycogenolysis via hepatic auto-regulation.
After representing the physiology mechanistically, the next step is to link the drug effect to a PK/PD model. For representative purpose, we can assume the new drug (GPi) exhibits a 100 % inhibition of GP for the entire 24-h interval in a typical patient. The maximally effective GPi is projected to have a dramatic effect on glycogenolysis (and thus HGO) overnight, but minimal impact during the day (as glycogenolysis is highly inhibited by postprandial glucose and insulin excursions). As a result, GPi is projected to display impressive glucose lowering overnight but little to no effect during the day (Fig. 5.10). Overall, chronic dosing with a maximally effective GPi is projected to result in noncompetitive HbA1c reductions (Fig. 5.10).
In addition to limited efficacy, chronic GPi therapy may be associated with metabolic adverse events. Following an acute dose, glycogen accumulates during the day but does not decrease overnight. Thus, following multiple doses, liver glycogen levels will likely increase to a point where they inhibit glycogen synthesis. At this point, the glucose that is normally converted to glycogen will be redirected to either lactate (risk of lactic-acidosis) or be converted to triglyceride via de novo lipogenesis (risk of hepatic steatosis). These pathways also suggest that GPi may not combine well with metformin (inhibition of gluconeogenesis may exacerbate lactate or triglyceride change) or sulfonylureas (GPi may impede the counter-regulatory response to hypoglycemia). Since these drugs are the two most commonly prescribed diabetes therapies, there is a chance that GPi therapy may be limited to a small subset of the diabetes population.
Overall, this analysis, together with expert knowledge of metabolism and physiology, illustrated using a systems pharmacology model of diabetes, suggests that GPi would be unlikely to become a viable therapy for type 2 diabetes. Thus, detailed mechanistic modeling, although tedious and resource intensive, provides a rigorous methodology for integrating our present knowledge of human pathophysiology and extrapolating to expected clinical outcomes. While the predictions using such complex models may not always have high predictive accuracy, making decisions based on a rigorous analysis of the available data, informed by expert knowledge, is likely to be more effective, and less costly in the long run, than a trial-and-error method of discovery. In addition, mechanistic models can identify key knowledge gaps for strategic expansion of our knowledge.
As described in this case study, mechanistic modeling can effectively inform both efficacy and safety aspects of therapeutic interventions. Lesko et al. (2013) described how systems approaches can be leveraged for understanding adverse drug events and ability to predict them, thus taking a step towards personalized medicine by enabling better identification of risk factors for an individual or subgroup.
5.6 Applications of Drug–Disease Models
The drug–disease models and the associated simulations that include parameter uncertainty and variability are powerful tools in decision-making in drug discovery and development. After the drug–disease models are developed based on accruing experimental data and informed by expert knowledge, simulations are performed to explore alternative or expanded scenarios of patient populations, clinical trial designs or disease outcome. The common questions or what-if scenarios that can be addressed through model-based analysis or simulations in diabetes R&D may include:
-
What is the predicted efficacy in patients based on in vitro or animal data?
-
What is the appropriate biomarker of pharmacology, based on variability, sensitivity, and time course of response?
-
What is the power or sample size of the study to detect a target response (difference from placebo at endpoint) for a specific biomarker or mechanism of action?
-
What is the minimum study duration to demonstrate the target response?
-
What is the dose to differentiate or achieve superiority to placebo?
-
What is the dose to demonstrate competitive or target response to marketed comparators?
-
What is the probability of demonstrating superiority to marketed comparator at the selected dose?
-
Are there subpopulations of responders or nonresponders based on the mechanism of action (for efficacy or safety)?
-
What is a clinically relevant drug–drug interaction or food effect?
An example application of patient response simulation was illustrated previously using a systems pharmacology model to support compound “go/no-go” decision. Example applications of trial simulations to support design optimization have been extensively published (Chien and Sinha 2010; Zhang et al. 2013). Figure 5.11 shows a conceptual example of application of modeling and simulation to support optimum dose selection.
Based on Fig. 5.11, Table 5.1 shows the probability of each dose meeting the predefined target criteria (superiority to comparator). The dose selected to advance to phase 3 confirmatory trial was selected based on a combination of high probability of competitive success and low probability of safety risk (e.g., cardiovascular or dose-limiting adverse events).
Pharmacometricians have been developing drug–disease models of diabetes to facilitate “rational target selection.” This is akin to “rational drug design” where high throughput trial-and-error methods for identifying chemicals that bind to receptors or enzymes have been replaced with methods that use knowledge of target structure to build ideal inhibitors or activators. For target selection, the current strategy is to replace trial-and-error methods of testing all reasonable targets preclinically (and many clinically) with an approach that leverages our knowledge of integrated human physiology, and strategically expand our knowledge of physiology, to identify therapeutic strategies that have a higher probability of success. A central part of this approach is mathematical models of human physiology. These ideas can be expanded to include the identification of optimal combination therapy , responder populations, and even personalized medicine (rational drug development). Philosophically, this approach represents a shift from “discovery” to “design.” Ultimately, the model-based approaches, through the use of predictive biomarkers, basic science and expert disease knowledge, aim to improve the efficiency and the quality of decision-making in selecting the right targets, drug candidates, dosing range and regimens, optimal clinical trial designs, and a more efficient, more cost-effective development programs with high probabilities of success in drug discovery and development for the treatment and management of diabetes and its comorbidities.
5.7 Key Highlights of the Chapter
-
Diabetes is a chronic progressive disease with robust quantitative biomarkers and well understood regulatory/counter-regulatory processes.
-
Drug–disease models of varying degrees of complexity can be employed to describe fast biomarkers, slow biomarkers and clinical outcomes with high predictive ability at different stages of drug development.
-
Antidiabetic therapies target specific pathways, thus enabling application of mechanistic systems pharmacology models to inform novel therapies.
-
Drug–disease models can effectively inform both efficacy and safety aspects of therapeutic interventions.
-
Development of drug–disease models should start early in the development program to answer key questions or address uncertainties from early discovery to clinical development in the evaluation of a novel therapy.
References
Ajmera I, Swat M, Laibe C, Le Novère N, Chelliah V (2013) The impact of mathematical modeling on the understanding of diabetes and related complications. CPT: Pharmacometrics Syst Pharmacol 2:e54
American Diabetes Association (2008) Diagnosis and classification of diabetes mellitus. Diabetes Care 31(1):562
Baker DJ, Timmons JA, Greenhaff PL (2005) Glycogen phosphorylase inhibition in type 2 diabetes therapy—a systematic evaluation of metabolic and functional effects in rat skeletal muscle. Diabetes 54:2453–2459
Bergman RN, Ider YZ, Bowden CR, Cobelli C (1979) Quantitative estimation of insulin sensitivity. Am J Physiol 236:E667–E677
Bergman RN, Hope ID, Yang YJ, Watanabe RM, Meador MA, Youn JH, Ader M (1989) Assessment of insulin sensitivity in vivo: a critical review. Diabetes Metab Rev 5:411–429
Chien JY, Sinha VP (2010) The application of drug-disease models in the development of anti-hyperglycemic agents. In: Kimko HHC, Peck CC (eds) Clinical trial simulations: applications and trends (AAPS advances in the pharmaceutical sciences series) Springer, USA 175–198
Cobelli C, Dalla Man C, Sparacino G, Magni L, De Nicolao G, Kovatchev BP (2009) Diabetes: models, signals, and control. IEEE Rev Biomed Eng 2:54–96
de Winter W, DeJongh J, Post T, Ploeger B, Urquhart R, Moules I, Eckland D, Danhof M (2006) A mechanism-based disease progression model for comparison of long-term effects of pioglitazone, metformin and gliclazide on disease processes underlying type 2 diabetes mellitus. J Pharmacokinet Pharmacodyn 33:313–343
Farhy LS, McCall AL (2009) Pancreatic network control of glucagon secretion and counter regulation. Meth Enzymol 467:547–581
Frey N, Laveille C, Paraire M, Francillard M, Holford NH, Jochemsen R (2003) Population PKPD modelling of the long-term hypoglycaemic effect of gliclazide given as a once-a-day modified release (MR) formulation. Br J Clin Pharmacol 55:147–157
Hamrén B, Björk E, Sunzel M, Karlsson MO (2008) Models for plasma glucose, HbA1c, and hemoglobin interrelationships in patients with type 2 diabetes following tesaglitazar treatment. Clin Pharmacol Ther 84(2):228–235
Hetherington J, Sumner T, Seymour RM, Li L, Rey MV, Yamaji S, Saffrey P, Margoninski O, Bogle ID, Finkelstein A, Warner A (2011) A composite computational model of liver glucose homeostasis. I. Building the composite model. J R Soc Interface 9(69):689–700
IDF Report (2013) IDF diabetes atlas, 6th edn. http://www.idf.org/diabetesatlas. Accessed 7 May 2014
Jauslin PM, Silber HE, Frey N, Gieschke R, Simonsson US, Jorga K, Karlsson MO (2007) An integrated glucose-insulin model to describe oral glucose tolerance test data in type 2 diabetics. J Clin Pharmacol 47:1244–1255
Jauslin PM, Frey N, Karlsson MO (2011) Modeling of 24-hour glucose and insulin profiles of patients with type 2 diabetes. J Clin Pharmacol 51:153–164
Kansal AR (2004) Modeling approaches to type 2 diabetes. Diabetes Technol Ther 6(1):39–47
Kim J, Saidel GM, Cabrera ME (2007) Multi-scale computational model of fuel homeostasis during exercise: effect of hormonal control. Ann Biomed Eng 35:69–90
Klinke DJ (2008) Integrating epidemiological data into a mechanistic model of type 2 diabetes: validating the prevalence of virtual patients. Ann Biomed Eng 36(2):321–334
Landersdorfer CB, Jusko WJ (2008) Pharmacokinetic/Pharmacodynamic modelling in diabetes mellitus. Clin Pharmacokin 47(7):417–448
Lesko LJ, Zheng S, Schmidt S (2013) Systems approaches to risk assessment. Clin Pharmacol Ther 93(5):413
Martin WH, Hoover DJ, Armento SJ, Stock IA, McPherson RK, Danley DE, Stevenson RW, Barrett EJ, Treadway JL (1998) Discovery of a human liver glycogen phosphorylase inhibitor that lowers blood glucose in vivo. Proc Natl Acad Sci U S A 95:1776–1781
Møller JB, Overgaard RB, Kjellsson MC, Kristensen NR, Klim S, Ingwersen SH, Karlsson MO (2013) Longitudinal modeling of the relationship between mean plasma glucose and HbA1c following antidiabetic treatments. CPT Pharmacometrics Syst Pharmacol 2:e82
Morghissi ES, Korytkowski MT, DiNardo M, Einhorn D, Hellman R, Hirsch IB, Inzucchi SE, Ismail-Beigi F, Kirkman MS, Umpierez GE (2007) American association of clinical endocrinologists and American diabetes association consensus statement on inpatient glycemic control. Diabetes Care 30(suppl 1):S42–S47
Ribbing J, Hamrén B, Svensson MK, Karlsson MO (2010) A model for glucose, insulin, and beta-cell dynamics in subjects with insulin resistance and patients with type 2 diabetes. J Clin Pharmacol 50:861–872
Schaller S, Willmann S, Lippert J, Schaupp L, Pieber TR, Schuppert A, Eissing T (2013) A generic integrated physiologically based whole-body model of the glucose-insulin-glucagon regulatory system. CPT Pharmacometrics Syst Pharmacol 2(e65):1–10
Schneck KB, Zhang X, Bauer R, Karlsson MO, Sinha VP (2013) Assessment of glycemic response to an oral glucokinase activator in a proof of concept study: application of a semi-mechanistic, integrated glucose-insulin-glucagon model. J Pharmacokinet Pharmacodyn 40:67–80
Shafrir E (2007) Animal models of diabetes, frontiers of research. CRC, Boca Raton, p 365
Shafrir E (2010) Contribution of animal models to the research of the causes of diabetes. World J Diabetes 1(5):137–140
Shoda L, Kreuwel H, Gadkar K, Zheng Y, Whiting C, Atkinson M, Bluestone J, Mathis D, Young D, Ramanujan S (2010) The type 1 diabetes physioLab® platform: a validated physiologically based mathematical model of pathogenesis in the non-obese diabetic mouse. Clin Exp Immunol 161(2):250–267
Silber HE, Jauslin PM, Frey N, Gieschke R, Simonsson US, Karlsson MO (2007) An integrated model for glucose and insulin regulation in healthy volunteers and type 2 diabetic patients following intravenous glucose provocations. J Clin Pharmacol 47:1159–1171
Silber HE, Frey N, Karlsson MO (2010) An integrated glucose-insulin model to describe oral glucose tolerance test data in healthy volunteers. J Clin Pharmacol 50:246–256
Sumner T, Hetherington J, Seymour RM, Li L, Varela Rey M, Yamaji S, Saffrey P, Margoninski O, Bogle ID, Finkelstein A, Warner A (2011) A composite computational model of liver glucose homeostasis. II. Exploring system behaviour. J R Soc Interface 9(69):701–706
Topp B, Promislow K, deVries G, Miura RM, Finegood DT (2000) A model of beta-cell mass, insulin, and glucose kinetics: pathways to diabetes. J Theor Biol 206:605–619
Torres TP, Sasaki N, Donahue EP, Lacy B, Printz RL, Cherrington AD, Treadway JL, Shiota M (2011) Impact of a glycogen phosphorylase inhibitor and metformin on basal and glucagon-stimulated hepatic glucose flux in conscious dogs. J Pharmacol Exp Ther 337(3):610–620
Verspohl EJ (2012) Novel pharmacological approaches to the treatment of type 2 diabetes. Pharmacol Rev 64(2):188–237
Vlasakakis G, Pasqua OD (2013) Cardiovascular disease: the other face of diabetes. CPT Pharmacometrics Syst Pharmacol 2:e81
Waters SB, Topp BG, Siler SQ, Alexander CM (2009) Treatment with sitagliptin or metformin does not increase body weight despite predicted reductions in urinary glucose excretion. J Diabetes Sci Technol 3(1):68–82
WHO fact sheet number 310. http://www.who.int/mediacentre/factsheets/fs310/en/. Accessed 7 May 2014
Woodcock J, Woosley R (2008) The FDA critical path initiative and its influence on new drug development. Ann Rev Med 59:1–12
Zhang X, Schneck K, Bue-Valleskey J, Yeo KP, Heathman M, Sinha V (2013) Dose selection using a semi-mechanistic integrated glucose-insulin-glucagon model: designing phase 2 trials for a novel oral glucokinase activator. J Pharmacokinet Pharmacodyn 40:53–65
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2014 American Association of Pharmaceutical Scientists
About this chapter
Cite this chapter
Garhyan, P., Topp, B., Chien, J., Sinha, V., Danhof, M., Schmidt, S. (2014). Drug–Disease Model-Based Development of Therapeutic Agents for Treatment of Diabetes. In: Schmidt, S., Derendorf, H. (eds) Applied Pharmacometrics. AAPS Advances in the Pharmaceutical Sciences Series, vol 14. Springer, New York, NY. https://doi.org/10.1007/978-1-4939-1304-6_5
Download citation
DOI: https://doi.org/10.1007/978-1-4939-1304-6_5
Published:
Publisher Name: Springer, New York, NY
Print ISBN: 978-1-4939-1303-9
Online ISBN: 978-1-4939-1304-6
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)