Abstract
Proteins play a vital role by booming out a number of activities within an organism to withstand its life. The field of Natural Language Processing has successfully adapted deep learning to get a better insight into the semantic nature of languages. In this paper, we propose semantic approaches based on deep learning to work with protein amino acid sequences and compare the performances of these approaches with traditional classifiers to predict their respective families. The Bidirectional Encoder Representations from Transformers (BERT) approach was tested over 103 protein families from UniProt consortium database. The results show the average prediction accuracy to 99.02%, testing accuracy to 97.70%, validation accuracy to 97.69%, Normalized Mutual Information (NMI) score on overall data to 98.45, on test data to 96.99, on validation data 96.93 with high weighted average F1 scores of 99.02 on overall data, 97.72 on test data and 97.70 on validation data, and high macro average F1 scores of 99.00 on overall data, 98.00 on test data and 98.00 on validation data. From the results, it is justified that our proposed approach is outperforming well when compared to the existing approaches.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Amino acids are the structural blocks of protein. Although there are innumerable proteins, the basic building blocks are limited to 20 amino acids (as described in Table 1). These amino acids are arranged in different orders and compositions to form a specific protein sequence. The task of assigning proteins to their respective families makes use of a range of sources, like protein family databases, sequence analysis tools, scientific literature, and sequence similarity search tools. The protein sequences are known to implicitly encode structural information of the proteins, which in turn encodes functional properties and proteins with similar functional properties tend to belong to the same protein family [1,2,3,4,5,6].
Thus, the sequence analysis tools make use of this property to classify proteins into their families [7]. They mainly compare the two sequences token by token, using methods like Edit distance, Hamming distance, and a combination of these. Algorithms like Needleman–Wunsch [8] is used for global alignment, Smith–Waterman for local alignment of protein sequences. In terms of Natural Language Processing (NLP), these algorithms perform character by character matching for two sentences. The global alignment performs alignment based on comparing the entire sequences (end to end alignment) whereas, local alignment finds local regions with a maximum level of similarity between the two sequences. For example, we have two sentences—‘I am a man’, and ‘I am not a man’, then these tools will match over the characters to get the similarity score. Even the most dynamic algorithms in sequence similarity would give irrational results in many such cases. They fail to encompass the meaning behind the way the amino acid sequences are arranged. Like here, the two sentences express two opposite ideas, but the similarity scores after alignment say a different story. Similarly, when we take another pair of sentences—‘My name is Dave’ and ‘Dave is my name’. Here, the two sentences express the same meaning but have a different structure of representation. Here, we observe that even though the two sentences express similar ideas, the similarity score suggests differently.
However, most of the published methods tend to be computationally expensive sequence alignment methodologies [9]. One of the popular approach is based on the Hidden Markov Model (HMM) [10]. This technique provides a strong statistical base for building classification models that use multiple sequence alignment. This act as extremely discriminative models of biological sequences that have a formal probabilistic basis. It does not depend on the detections of intracellular loops and is publicly available through a web-server. This method return an 89.7% accuracy. This is very expensive in terms of computational time because each protein sequence has to be aligned with all the other protein sequences in the training set before making the predictions.
Although alignment-independent techniques have been developed for the classification of proteins, most of these methods are based on feature extraction and the derived features are encoded into feature vectors to apply suitable machine learning models for training a model for classification [1, 2, 4, 5, 11]. Although this method is effective, it still needs to extract the features of the proteins from their sequences first and then encoding them into vector representations to apply machine learning models to them. The features obtained are based on the knowledge we have about the working of the biological and chemical phenomena of the protein sequences, instead in this paper, the method we propose is free of this knowledge base. Instead of using any pre-existing knowledge modeling procedure, the methods proposed here gain insight into the protein sequences based on their morphological features, i.e., the arrangement of the amino acids and their contexts, the models itself form its own knowledge base from the distribution in the dataset and then use this knowledge to make further predictions [12].
One such method we use here is the document embedding technique where fixed size vectors are created from the protein sequences based on the amino acid content and arrangement of each sequence. We can then apply the classification algorithms such as Logistic Regression, Support Vector Classifier (SVC), K-Nearest Neighbors (KNN), Extra Tree Classifier, Random Forest Classifier, Gaussian Naïve Bayes on the obtained protein vectors for classification. Another approach that we applied to the protein sequences for family prediction is Bidirectional Encoder Representations from Transformers (BERT) [13]. It is a transformer-based bidirectional training approach for language modeling. Unlike the previous algorithms which looked at texts from left to right or right to left while training, BERT combines both of these methodologies to perform bidirectional training on the text. Research shows that the bidirectionally trained language models have a deeper understanding of the flow of language and context. The quality of predictions made by these algorithms was then further compared with each other.
Honglei Liu et al. [14] applied the deep learning techniques and Natural Language Processing (NLP) methods to find out the liver cancer. They have used the deep learning methods and NLP methods to extract the radiological features. They have developed a computer-aided method for diagnosing the liver cancer. The authors have used the BERT model to identify the phrases like hypointense in the portal phase and hyperintense enhancement in the arterial phase. Shivaji Alaparthi et al. [15] investigated the effectiveness of the BERT in supervised deep learning frameworks. The authors gave a clear study on sentiment analysis which will help for the people in analytics company and researchers who are working in text analysis. The authors have concluded that BERT model is performing well for the sentiment classification since it has high computational capabilities. Alaa Joukhadar et al. [16] proposed model to detect Arabic language dialogue acts and the experiment was done on the Arabic datasets. Existing researches show that the identification of Arabic dialogue acts is little bit complicated. The authors [16] presented the impact of BERT model to identify the Arabic language dialogue acts using different models like AraBERT base, AraBERT large, and AraBERT original. The authors found out the Arabic corpus and it contains more than 21 K tweets. The authors also exploited the LevInt Arabic corpus that contains eight speech acts. The dysregulation of glutarylation leads to many human diseases and identification of the dysregulation of glutarylation is becoming an important task. Chuan-Ming Liu et al. [17] presented a deep-learning word-embedding-based framework to identify the dysregulation of glutarylation. The authors have done the experiments to improve the protein sequence data representation and they identified that deep neural network works well to handle complicated problems in protein identification. The method [17] is used to find the new sites of glutarialisations and shows the relationship between glutarial and protein acetylation. Wazib Ansar et al. [18] presented a new transformer encoder framework which has less complexity and computational overhead when compared to the BERT. Parinnary Chaudhry [19] has utilized the sentiment analysis capability of BERT to make a quantitative relation between the news and reporting of an industry. Also, the authors [19] used BERT to predict the stock market price and analyze the human psychology. Jairus Mingua et al. [20] used the BERT model to classify the Filipino tweets and the results show some significant difference in the accuracy.
Methodology
Protein Sequences as Language
In this paper, we treat the protein sequences as a part of a language that our body communicates in. If we consider language processing, the primary task is the identification of the morphemes of the language. Figure 1 shows the outline of the proposed work. The morphemes of the protein sequence language are the amino acids, the structural blocks of protein. The single-letter symbol representations (as shown in Table 1) will act as the alphabets of this language. So, now we have a new language with 20 alphabets and a vast vocabulary of a lot more than a million texts, which now would be regarded as the corpora of this language. So, we treat the amino acid symbols (as shown in Table 1) A, V, T, etc., as the alphabets of the protein language and the protein sequence is formed as a sentence. Now, with a new language at hand, a wide range of possibilities opens up. The very first major issue would be with tokenization. In this work, we have tried to solve sentence tokenization by assuming every protein sequence as a sentence. The problem with word tokenization is that there is no specific delimiter that acts as a tokenizer (Like in English, space—‘’ acts as a delimiter between two words, thus facilitating the simple tokenization). To solve this problem of word tokenization, we analyzed tokenization techniques in existing non-tokenized Asian language like Chinese [21].
We presumed that all the single characters, i.e., amino acids are also words themselves. Then we applied a tokenization algorithm based on the Viterbi decode and a lexicon built of frequent k-mers from all the sequences from the Swiss-Prot data file [22]. K-mer is a subsequence of length ‘k’. So, in the sequence AVTLAD, the 2-mers are (AV, VT, TL, LA, AD). K-mers from k = 2 to k = 20 is computed and then to reduce the size of the set of all k-mers, drop the k-mers with frequency probability less than e−15, thus removing the less frequent k-mers and making the process memory efficient. These k-mers, along with the characters (amino acid symbols), are now treated as words.
Document Embedding with Protein Sequences
Now, with protein sequences established as a language, we can move ahead to apply similar algorithms to it to gain insight. Document embedding is a deep learning algorithm that is used to create fixed-length vectors from variable length text documents (in our case, protein sequences). We created a custom dataset of all the protein sequences tokenized and tagged with their respective families. Here we have used the doc2vec algorithm [23] to create document embeddings using the gensim [24] library in python. Doc2vec performs robustly on a large scale of data, and as we know, the biological data are colossal and increasing every day. Doc2vec was proposed in two primary forms—a Distributed Bag of Words representation (DBOW), and a Distributed Memory (DM) version. Le and Mikolov [25] stated that DM alone usually is efficient for most tasks, but others have found a combination of DM and DBOW to perform better [23, 25]. In this specific case, the combination of DM and DBOW algorithms was found to perform with better accuracy, than the standalone DM or DBOW algorithm-based model. The architecture diagram is shown below in Fig. 2. The obtained word vectors using the Doc2Vec algorithm can be further used for training a prediction model. We used these sequence vector data to model a K-Nearest Neighbor (KNN), Logistic Regression (LR), Extra Tree Classifier (ETC), Random Forest Classifier (RFC), Gaussian Naïve Bayes (GNB), Support Vector Machine (SVM) models with linear and radial basis function (RBF) kernels.
The architecture diagram of the complete model, from the raw input of protein fasta sequence to the prediction of a protein family, is illustrated in Fig. 2. The process flow of the model can also be observed in Fig. 2. The system first takes in a protein amino acid sequence input and then passes it to the DBOW Doc2Vec model. The sequence vectors obtained from here are then passed into a classification model for family prediction using the sequence vectors. The classification model here represents the different classification models of Logistic Regression, Support Vector Classifier (SVC), K-Nearest Neighbor, Extra Tree, Random Forest, and Gaussian Naïve Bayes Classifier.
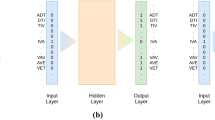
Figure 3 shows architecture of BERT-based model. For tokenization, BERT uses a WordPiece tokenizer. In WordPiece tokenization, a word can be broken down into more than one subword [26]. This type of tokenization proves to be useful when dealing with protein sequence vectors where we have to work with a lot of out of vocabulary words. The tokenized data are then encoded into token ids with the help of the pre-trained vocabulary dictionary mapping tokens to respective ids. In this work, we use the bert-base-uncased model created by Hugging Face. The model has 12 layers, 768 hidden features, 110 million parameters, and is pre-trained using Masked Language Modeling (MLM) on English language [27]. Yes, you read correctly a language model trained on plethora of English data available. We used this pre-trained model on English data because amino acid uses a subset of alphabets of English language. Each protein sequence fed into the model has a special token (CLS) added at the beginning of the sequence and the end of a sequence is marked by the special token (SEP), thus separating two inputs from each other. The model takes a fixed size input of 512 tokens. The sequences shorter than this requirement are padded up to a length of 512 and the longer sequences are truncated to a size of 512. Apart from the input, there exists an attention mask which tells the model which tokens should be taken and which should be ignored while training.
Results and Discussion
The family data of the protein sequences were downloaded separately from the UniProt website [22, 28], which contained about 5,63,000 proteins, along with their sequences and annotations (i.e., additional information as specified in Table 2). Recently, it has been used by Chang Woo Ko [29]. Swiss-Prot was used because the content here is reviewed and maintained by experts, and has minimal redundancy. The family to which the protein sequence belonged was present in a separate file downloaded from the UniProt website. In this work, we first dealt with 36 protein families with a document embedding model, the rest of the families with number of protein sequences less than 1000 were dropped. Later, we deal with 103 protein families with a BERT model, the rest of the families with number of protein sequences less than 700 were dropped.
The protein sequences were tokenized as explained in the methodology and stored alongside their respective protein family in training, validation, and testing sets. The size of these training, validation, and testing sets was in the following ratios. The sequence data were initially used to train Doc2Vec models, both DM and DBOW. After designing sequence vectorizing models, the protein sequence vector obtained from both DM and DBOW models was stored alongside their protein family in training, validation, and testing data files. To test the document embedding method, 36 protein families were selected from the Swiss-Prot database, such that number of protein sequences in each family was greater than 1,000. Redundant sequences were eliminated before model construction and testing. There were 72,208 protein sequences in 36 families. The data were shuffled randomly and split into training, validation, and testing datasets. To test the BERT method, 103 protein families were selected from the Swiss-Prot database, such that number of protein sequences in each family was greater than 700. There were 112,401 protein sequences in 103 families.
Document Embedding Model Performance
The obtained accuracies indicate in Table 3 that the DBOW algorithm of Doc2Vec modeling is most suitable to understand and work with biological sequence data. The best average classification accuracy of 0.8827 is achieved by applying Support Vector Classifier with radial basis function kernel over the protein vector data obtained from Doc2Vec (DBOW) model. Also, the highest testing accuracy of 0.8346 is achieved using this specific combination. Although the accuracy is high, accuracy alone cannot be used as a metric for validating a model. This specific problem can be considered as a clustering problem, wherein we have to create clusters out of the given protein sequences into 36 protein families’ clusters. So, the models can be evaluated using clustering metrics like NMI score. It is a good measure for determining the quality of clustering. It is an external measure because we need the class labels of the instances to determine the NMI. Support Vector Classifier with RBF kernel combined with DBOW vectors performs well on NMI metric too. The NMI score achieved with this combination is 0.7092 over the testing data.
The document embedding models are also evaluated using precision–recall curves and Receiver Operating Characteristics (ROC) curves on one vs. all classification forms of these models. The ROC curves can be seen below in Figs. 4, 5, and 6. Considering the highest testing accuracy and highest NMI of the SVC (RBF kernel) model on DBOW vectors, the ROC curve of this model must be performing better than all the other models on DBOW vectors, but from the graph displayed in Fig. 4, one can infer that the Gaussian Naïve Bayes model performed better than any other model in case of DBOW vectors. From this, we could infer that even though the accuracy of Support Vector Classifier is greater than Gaussian Naïve Bayes Classifier, the reliability of Gaussian Naïve Bayes Classifier is greater than other models, but the dataset in consideration here is imbalanced, and in case of imbalanced dataset, precision–recall curve is considered as a much better testing metric than ROC curve.
One can see the precision–recall curves of different classifiers combined with sequence vectors obtained using DBOW, DM and DBOW + DM algorithms of Doc2Vec algorithms in the following Figs. 7, 8, 9, 10, 11, 12, and 13. In case of precision–recall curve, the perfect test will have a curve that passes through the upper right corner corresponding to 1.0 precision and 1.0 recall. Generally, it is stated that closer the precision–recall curve is to the upper right corner, the better the model is. Thus, on observing the graphs in Figs. 7, 8, 9, 10, 11, 12, and 13, one can infer that the most reliable models are Logistic Regression Classifier combined with DBOW Doc2Vec model and SVC with RBF kernel combined with DBOW Doc2Vec model.
So, considering the metrics such as accuracy score, NMI score, precision and recall curve, it can be concluded that SVC with RBF kernel applied on the sequence vectors obtained using a DBOW form of Doc2Vec algorithm is so far the best performing model in this paper for protein family prediction using protein sequences alone. With more sophisticated data, this model can be re-scaled based on new inputs. The elegance of this model is the way it works with new data input, which was never used before for training either the document embedding or the later classification methods like K-Nearest Neighbor, Logistic Regression, Extra Trees Classifier, Random Forest Classifier, Gaussian Naïve Bayes, Support Vector Classifier models. The architecture of this model can be applied to any variations of biological sequences (with sufficient data) for different purposes using the concept of transfer learning.
Table 4 represents the overall accuracy of the proposed work. The BERT model performs exceptionally well over the protein sequence data with decent accuracy, F1 scores, and NMI scores. Here, we use a very imbalanced dataset, and still achieve a decent weighted average F1 score of 0.9772 and macro-averaged F1 score of 0.9800 on test data. The high NMI metric value of 0.9699 signifies good clustering. The BERT model used was pre-trained on English language corpora but still was able to work with protein data on training the model on the available tagged protein sequence data. Moreover, Table 5 is used to identify the significance of the proposed method through average precision (AP), F1 score (F1), area under receiver operating characteristic curve (AUROC), area under precision–recall curve (AUPR) and Matthews correlation coefficient (MCC). To verify our approach, we compared our model with the state-of-art method BLAST and FUTUSA. We have selected three major activities namely oxidoreductase, the acetyltransferase, and demethylase activity. BERT model depicted higher values of accuracy than existing model. Although, the BERT model presented on 103 protein families, with the BERT’s property of transfer learning, it can be re-scaled to work on any number of protein families as long as the sequences per family are significant enough for training. Apart from this, it can also be inferred that BERT model performs much better than document embedding model, and the metrics tested on validation data also emphasize the fact that the model is not overfitting on the available data.
Conclusion
A BERT model could be used to effectively encompass the information in a protein sequence in a context-based manner rather than the traditional sequential manner to predict its family. From the results, it is identified that the BERT model yields the results with an average accuracy of 0.9902, test accuracy of 0.9770, average F1 score of 0.9702, and testing F1 score of 0.9772 (as shown in Table 6). In this work, we also inferred that a standalone DBOW Doc2Vec model performed better over biological sequence data, irrespective of the popular opinion of Doc2Vec modeling. The DM model works better than any other model and DM supersedes the performance of other Doc2Vec models on text data. This new strategy has a wide scope of future extensions, because these methods tend to assign more meaning to protein sequences. Unlike the traditional method of dealing with sentences, now we can use these techniques to gain insight into these sequences. In this paper, we have applied BERT technique for protein family prediction, but this method can be transcended to determine the similarity between sequences and give better results than any traditional pairwise sequence alignment techniques. This new strategy has a wide scope of future extensions, because the sequences are converted to vectors which tend to assign more meaning to them. In future, with the help of more non-redundant data, suitable hyper-parameter tuning and scaling, this BERT architecture, a robust model, for prediction of protein family for all protein sequences of all known families can be built.
References
Liu B, Gao X, Zhang H. BioSeq-Analysis2.0: an updated platform for analyzing DNA, RNA and protein sequences at sequence level and residue level based on machine learning approaches. Nucl Acids Res. 2019;47(20):e127.
Hong H, Hong Q, Perkins R, Shi L, Fang H, Su Z, Tong W. The accurate prediction of protein family from amino acid sequence by measuring features of sequence fragments. J Comput Biol. 2009;16(12):1671–88.
McClure MA, Vasi TK, Fitch WM. Comparative analysis of multiple protein-sequence alignment methods. Mol Biol Evol. 1994;11(4):571–92.
Seo S, Oh M, Park Y, Kim S. DeepFam: deep learning based alignment-free method for protein family modelling and prediction. Bioinformatics. 2018;34(13):254–62.
Beigi MM, Behjati M, Mohabatkar H. Prediction of metalloproteinase family based on the concept of Chou’s pseudo amino acid composition using a machine learning approach. J Struct Funct Genomics. 2011;12(4):191–7.
Caragea C, Silvescu A, Mitra P. Protein sequence classification using feature hashing. Proteome Sci. 2012;10:1–14.
Wang Y, Zhang H, Zhong H, Xue Z. Protein domain identification methods and online resources. Comput Struct Biotechnol J. 2021;19(2):1145–53.
Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970;48(3):443–53.
Yan Y, Chaturvedi N, Appuswamy R. Accel-Align: a fast sequence mapper and aligner based on the seed-embed-extend method. BMC Bioinform. 2021;22(1):257–68.
Sgourakis NG, Bagos P, Papasaikas PK, Hamodrakas SJ. A method for the prediction of GPCRs coupling specificity to G-proteins using refined profile Hidden Markov Models. BMC Bioinform. 2005;6(1):104.
Mallika V, Sivakumar KC, Jaichand S, Soniya EV. Kernel based machine learning algorithm for the efficient prediction of type III polyketide synthase family of proteins. J Integr Bioinform. 2010;7(1):47–54.
Ofer D, Brandes N, Linial M. The language of proteins: NLP, machine learning & protein sequences. Comput Struct Biotechnol J. 2021;19:1750–8.
Kades K, Sellner J, Koehler G, Full PM, Lai T, Kleesiek J, Maier-Hein KH. Adapting bidirectional encoder representations from transformers (BERT) to assess clinical semantic textual similarity: algorithm development and validation study. JMIR Med Inform. 2021;9(2):22795.
Liu H, Zhang Z, Xu Y, Wang N, Huang Y, Yang Z, Jiang R, Chen H. Use of BERT (bidirectional encoder representations from transformers)-based deep learning method for extracting evidences in Chinese radiology reports: development of a computer-aided liver cancer diagnosis framework. J Med Internet Res. 2021;23(1):118–26.
Alaparthi S, Mishra S. Bidirectional encoder representations from transformers (BERT): a sentiment analysis odyssey. J Market Anal. 2021;9(8):118–26.
Joukhadar A, Ghneim N, Rebdawi G. Impact of using bidirectional encoder representations from transformers (BERT) models for Arabic dialogue acts identification. Int Inf Eng Technol Assoc. 2021;26(5):469–75.
Liu C-M, Ta V-D, Le NQK, Tadesse DA, Shi C. Deep neural network framework based on word embedding for protein glutarylation sites prediction. Life. 2022;12(8):1213.
Ansar W, Goswami S, Chakrabarti A, Chakraborty B. A novel selective learning based transformer encoder architecture with enhanced word representation. Appl Intell. 2022. https://doi.org/10.1007/s10489-022-03865-x.
Parinnay C. Bidirectional encoder representations from transformers for modelling stock prices. J Res Appl Sci Eng Technol. 2022;10(2):896–901.
Mingua J, Padilla D, Celino EJ. Classification of fire related tweets on twitter using bidirectional encoder representations from transformers (BERT). In: 2021 IEEE 13th international conference on humanoid, nanotechnology, information technology, communication and control, environment, and management (HNICEM), Manila, Philippines. 2021. p. 1–6
Li H, Bai S, Lin Z (2005) Chinese sentence tokenization using Viterbi decoder. In: International symposium on Chinese spoken language processing. Singapore, December 7–9, 1998.
UniProt. Swiss-Prot protein knowledgebase. SIB Swiss Institute of Bioinformatics. https://www.uniprot.org/docs/similar.txt (2021). Accessed 01 Oct 2021.
Lau JH, Baldwin T. An empirical evaluation of doc2vec with practical insights into document embedding generation. In: Proceedings of the 1st workshop on representation learning for NLP, Berlin, Germany. 2016. p. 78–86.
Řehůřek R, Sojka P. Gensim—statistical semantics in python. genism.org (2011). Accessed 01 Oct 2021.
Le Q, Mikolov T. Distributed representations of sentences and documents. In: Proceedings of the 31st international conference on international conference on machine learning China. 2014. p. 1188–1196.
Devlin J, Chang MW, Lee K, Toutanova K. BERT: Pre-training of deep bidirectional transformers for language understanding. arXiv:1810.04805 [Preprint]. 2018.
Wolf T, Debut L, Sanh V, Chaumond J, Delangue C, Moi Rush AM. HuggingFace’s transformers: state-of-the-art natural language processing. arXiv:1910.03771 [Preprint]. 2019.
Bairoch A, Apweiler R. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucl Acids Res. 2000;28(1):45–8.
Ko CW, Huh J, Park J-W. Deep learning program to predict protein functions based on sequence information. MethodsX. 2022;9(1):1016–22.
Funding
N/A.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
We declare that there is no conflict of interest.
Ethical Approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Balamurugan, R., Mohite, S. & Raja, S.P. Protein Sequence Classification Using Bidirectional Encoder Representations from Transformers (BERT) Approach. SN COMPUT. SCI. 4, 481 (2023). https://doi.org/10.1007/s42979-023-01980-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s42979-023-01980-1