Abstract
The study of Protein-DNA binding sites is one of the fundamental problems in genome biology research. It plays an important role in understanding gene expression and transcription, biological research, and drug development. In recent years, language representation models have had remarkable results in the field of Natural Language Processing (NLP) and have received extensive attention from researchers. Bidirectional Encoder Representations for Transformers (BERT) has been shown to have state-of-the-art results in other domains, using the concept of word embedding to capture the semantics of sentences. In the case of small datasets, previous models often cannot capture the upstream and downstream global information of DNA sequences well, so it is reasonable to refer the BERT model to the training of DNA sequences. Models pre-trained with large datasets and then fine-tuned with specific datasets have excellent results on different downstream tasks. In this study, firstly, we regard DNA sequences as sentences and tokenize them using K-mer method, and later utilize BERT to matrix the fixed length of the tokenized sentences, perform feature extraction, and later perform classification operations. We compare this method with current state-of-the-art models, and the DNABERT method has better performance with average improvement 0.013537, 0.010866, 0.029813, 0.052611, 0.122131 in ACC, F1-score, MCC, Precision, Recall, respectively. Overall, one of the advantages of BERT is that the pre-training strategy speeds up the convergence in the network in migration learning and improves the learning ability of the network. DNABER model has advantageous generalization ability on other DNA datasets and can be utilized on other sequence classification tasks.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
- Protein-DNA binding sites
- Transcription factor
- Traditional machine learning
- Deep learning
- Transformers
- BERT
1 Introduction
Protein-DNA binding site refers to a fragment of a protein macromolecule that specifically [1] binds to a DNA sequence of approximately 4–30 bp [2,3,4] in length. And transcription factors, as a common type of protein macromolecule, are an important issue for Protein-DNA binding site prediction, and when transcription factors bind to these specific regions, the sites are called transcription factor binding sites (TFBS) [5, 6]. During the transcription of a gene, transcription factor binds specifically to a segment of DNA sequence as a protein macromolecule, and the region forms the transcription factor binding site. Transcription factors are of great importance in gene regulation, transcription, and biological research and drug design [7,8,9]. Therefore, accurate prediction of Protein-DNA binding sites is very important for genomic understanding, description of gene specific functions, etc. [10, 11].
In the past decades, sequencing operations were performed using traditional biological methods, especially ChIP-seq [12] sequencing technology, which greatly increased the quantity and quality of available sequences and laid the foundation for subsequent studies. With the development of sequencing technology, the number of genomic sequences has increased dramatically, and traditional biological sequencing techniques are costly and slow, therefore, machine learning [13] ideas have been applied to Protein-DNA binding site prediction, such as, Wong et al. proposed the kmerHMM [14] model based on Hidden Markov (HMMs) and belief propagations, and Li et al. [15] proposed the fusion pseudo nucleic acid composition (PseNAC) model based on SVM. However, with the gradual accumulation of sequences, traditional machine learning methods cannot meet the requirements in terms of prediction accuracy and computational speed, and deep learning has performed well in other fields such as machine vision [2, 16, 17]. so researchers have gradually applied deep learning to bioinformatics [4, 18,19,20], DeepBind has applied convolutional neural networks to Protein-DNA binding site prediction for the first time, and Zeng et al. further explored the number of convolutional layers and pooling methods to validate the value of Convolutional Neural Network (CNN) for Protein-DNA binding sites. KEGRU is a framework model that is fully based on RNN using Bidirectional Gated Recurrent Unit (Bi-GRU) and K-mer embedding. DanQ utilizes a hybrid neural network combining CNN and Recursive Neural Network (RNN) with the addition of Bi-directional Long-Short Term Memory (Bi-LSTM) layers for better long distance dependencies in sequence relations for learning.
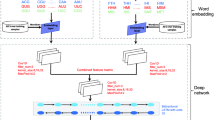
In our work, we utilized DNABERT for feature extraction of the dataset and classification by fully connected layers. First, we segment the DNA sequences using the K-mer representation, as opposed to the One-hot encoding commonly utilized in previous deep learning, we only segment it, and later utilize the processed data add the location information as the input to BERT. Then feature extraction is performed using BERT based on the Multi-headed Self-attention mechanism, with 101x768 dimensions for the input data and no change in the dimensionality of the output data. Finally, the input is fed into the fully connection and activated using the softmax function for binary classification prediction. In order to verify the generalization ability of the model, we utilized fine-tuning model to predict different cell line transcription factor datasets and verified the effectiveness of the model.
2 Materials and Methods
2.1 Benchmark Dataset
To better evaluate the performance of the model, we selected 45 public transcription factor ChIP-seq datasets of Broad cell lines from the ENCODE dataset, which were previously utilized in DeepBind, CNN-Zeng, and DeepSEA model frameworks, each with a DNA sequence sample length of 101 bp and a positive to negative sample number ratio of approximately 1:1. These data can be found in http://cnn.csail.mit.edu/motif_discovery/.
2.2 Model
Tokenization
We utilize K-mer for DNA sequences, and for each deoxyribonucleic acid base concatenate it with subsequent bases, integrating better contextual information for each deoxyribonucleic acid. Different K values correspond to different tokenization of DNA sequences, and we set the value of K to 6, i.e. {ACGTACGT} can be tagged as {ACGTAC, CGTACG, GTACGT}. In the utterance, in addition to all permutations indicated by K-mer, five other special tokens are included, the categorical CLS token inserted into the head, the SEP token inserted after each sentence, the MASK token that masks the words, the placeholder pad token, and UNK token that stands for unknown in the sequence, when K = 6, there are \({4}^{6}+5\) token.
The DNABERT Mode
Bert is a transformer-based pre-trained language representation model that is a milestone in NLP. It introduces an idea of pre-training and fine-tuning, where after pre-training with a large amount of data, an additional output layer is added for fine-tuning using small task-specific data to obtain state-of-the-art performance in other downstream tasks. The innovation of BERT is the use of a new technique of masked language model (MLM), which uses a bi-directional Transformer for language modeling, where the bi-directional model will outperform the uni-directional model in language representation. BERT models can also be used in question-and-answer systems, language analysis, document clustering, and many other tasks. We believe that BERT can be applied to Protein-DNA binding site prediction to better capture the hidden information in DNA sequences, as shown in Fig. 1.
3 Result and Discussion
3.1 Competing Methods
In order to ensure the fairness of the experiment, we used three deep learning-based models to compare performance with DNABERT model, namely DeepBind, DanQ and WSCNNLSTM. Through comparison, it is found that DNABERT model has better performance in the evaluation indexes we used. Table 1 shows the performance comparison of DNABERT in the data set of each cell line we selected. As can be seen from the Table 1, DNABERT is higher than existing models in the evaluation indexes ACC, F1-Score, MCC, Precision and Recall. ACC is 0.013537 higher than other methods on average, and F1-score increases by 0.010866. MCC increased by 0.029813, Precision and Recall increased by 0.052611 and 0.122131, respectively. Experimental results show that our method is superior to existing networks. Table 1 is the setting of hyper-parameters in the experiment.
4 Conclusion
In recent years, transformer-based series models have had state-of-the-art performance in the field of NLP. As the research gradually progressed, researchers migrated it to other fields and achieved equally desirable results. In our work, we demonstrate that the performance of DNABERT for Protein-DNA binding site prediction greatly exceeds that of other existing tools. Due to the sequence similarity between genomes, it is possible to transfer data of biological information to each other using the DNABERT pre-trained model. DNA sequences cannot be directly translated on the machine, and DNABERT gives a solution to the problem of deciphering the language of non-coding DNA, correctly capturing the hidden syntactic semantics in DNA sequences, showing excellent results. Although DNABERT has excellent performance in predicting Protein-DNA binding sites, there is room for further improvement. CLS token represents the global information of the sequence, and the rest token represents the features of each part of the sequence, we can consider separation processing to better capture the sequence features and achieve better results. However, so far, the BERT pre-training method for Protein-DNA binding site prediction has the most advanced performance at present, and the use of DNABERT introduces the perspective of high-level language modeling to genomic sequences, providing new advances and insights for the future of bioinformatics.
References
Rohs, R., Jin, X., West, S.M., Joshi, R., Honig, B., Mann, R.S.: Origins of specificity in protein-DNA recognition. Annu. Rev. Biochem. 79, 233–269 (2010). https://doi.org/10.1146/annurev-biochem-060408-091030
Jordan, M.I., LeCun, Y., Solla, S.A. (eds.): Advances in Neural Information Processing Systems: Proceedings of the First 12 Conferences. MIT Press, Cambridge (2004)
Liu, Y., et al.: RoBERTa: A Robustly Optimized BERT Pretraining Approach (2019)
Liu, Y., Zhu, Y.-H., Song, X., Song, J., Yu, D.-J.: Why can deep convolutional neural networks improve protein fold recognition? A visual explanation by interpretation. Brief Bioinform. 22, bbab001 (2021). https://doi.org/10.1093/bib/bbab001
Karin, M.: Too many transcription factors: positive and negative interactions. New Biol. 2, 126–131 (1990)
Latchman, D.S.: Transcription factors: an overview. Int. J. Biochem. Cell Biol. 29, 1305–1312 (1997). https://doi.org/10.1016/s1357-2725(97)00085-x
Jolma, A., et al.: DNA-binding specificities of human transcription factors. Cell 152, 327–339 (2013). https://doi.org/10.1016/j.cell.2012.12.009
Tuupanen, S., et al.: The common colorectal cancer predisposition SNP rs6983267 at chromosome 8q24 confers potential to enhanced Wnt signaling. Nat. Genet. 41, 885–890 (2009). https://doi.org/10.1038/ng.406
Wasserman, W.W., Sandelin, A.: Applied bioinformatics for the identification of regulatory elements. Nat. Rev. Genet. 5, 276–287 (2004). https://doi.org/10.1038/nrg1315
Lambert, S.A., et al.: The human transcription factors. Cell 172, 650–665 (2018). https://doi.org/10.1016/j.cell.2018.01.029
Basith, S., Manavalan, B., Shin, T.H., Lee, G.: iGHBP: computational identification of growth hormone binding proteins from sequences using extremely randomised tree. Comput. Struct. Biotechnol. J. 16, 412–420 (2018). https://doi.org/10.1016/j.csbj.2018.10.007
Furey, T.S.: ChIP-seq and beyond: new and improved methodologies to detect and characterize protein-DNA interactions. Nat. Rev. Genet. 13, 840–852 (2012). https://doi.org/10.1038/nrg3306
Manavalan, B., Shin, T.H., Lee, G.: DHSpred: support-vector-machine-based human DNase I hypersensitive sites prediction using the optimal features selected by random forest. Oncotarget 9, 1944–1956 (2017). https://doi.org/10.18632/oncotarget.23099
Wong, K.-C., Chan, T.-M., Peng, C., Li, Y., Zhang, Z.: DNA motif elucidation using belief propagation. Nucleic Acids Res. 41, e153 (2013). https://doi.org/10.1093/nar/gkt574
Li, L., et al.: Sequence-based identification of recombination spots using pseudo nucleic acid representation and recursive feature extraction by linear kernel SVM. BMC Bioinform. 15, 340 (2014). https://doi.org/10.1186/1471-2105-15-340
Angermueller, C., Pärnamaa, T., Parts, L., Stegle, O.: Deep learning for computational biology. Mol. Syst. Biol. 12, 878 (2016). https://doi.org/10.15252/msb.20156651
Graves, A., Mohamed, A., Hinton, G.: Speech recognition with deep recurrent neural networks. In: 2013 IEEE International Conference on Acoustics, Speech and Signal Processing, pp. 6645–6649 (2013). https://doi.org/10.1109/ICASSP.2013.6638947
Hong, J., et al.: Convolutional neural network-based annotation of bacterial type IV secretion system effectors with enhanced accuracy and reduced false discovery. Brief. Bioinform. 21, 1825–1836 (2020). https://doi.org/10.1093/bib/bbz120
Hong, J., et al.: Protein functional annotation of simultaneously improved stability, accuracy and false discovery rate achieved by a sequence-based deep learning. Brief Bioinform. 21, 1437–1447 (2020). https://doi.org/10.1093/bib/bbz081
Min, S., Kim, H., Lee, B., Yoon, S.: Protein transfer learning improves identification of heat shock protein families. PLoS ONE 16, e0251865 (2021). https://doi.org/10.1371/journal.pone.0251865
Acknowledgments
This work was supported in part by the University Innovation Team Project of Jinan (2019GXRC015), the Natural Science Foundation of Shandong Province, China (Grant No. ZR2021MF036).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Zhang, Y., Chen, Y., Chen, B., Cao, Y., Chen, J., Cong, H. (2022). Predicting Protein-DNA Binding Sites by Fine-Tuning BERT. In: Huang, DS., Jo, KH., Jing, J., Premaratne, P., Bevilacqua, V., Hussain, A. (eds) Intelligent Computing Theories and Application. ICIC 2022. Lecture Notes in Computer Science, vol 13394. Springer, Cham. https://doi.org/10.1007/978-3-031-13829-4_57
Download citation
DOI: https://doi.org/10.1007/978-3-031-13829-4_57
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-13828-7
Online ISBN: 978-3-031-13829-4
eBook Packages: Computer ScienceComputer Science (R0)