Abstract
Now-a-days Skin disease is very common worldwide problem. We have preset this study for the prediction of skin disease. Based on data from UCI data set, there are 34 attributes which plays a vital role in the skin disease diagnosis but all are not important. In this paper we have analyzed only those important attributes which give best accuracy in prediction of skin disease. To select important attributes, we have applied a new hybrid approach using three feature extraction techniques Chi Square, Information Gain and Principle Component Analysis (PCA) and then combining them to select the best possible data subset of skin disease data set. Six base learners Gaussian Naïve Bayesian (NB), K-Nearest Neighbour (KNN), Decision Tree (DT), Support Vector Machine (SVM), Random Forest (RF) and Multilayer Perceptron (MLP) are used to evaluate the prediction performance of base learners. Boosting, Bagging and Stacking ensemble techniques are applied on base learners to enhance the results of the proposed model. In this paper, a new proposed method of hybrid feature selection technique is used for evaluating the performance of base learners and we find that reduced data subset performed is higher as compared to whole data set. The metrics are necessary to evaluate the model and calculated to illustrate the performance of prediction. Hybrid feature selection technique along with base learners are then applied on Bagging, Boosting and Staking ensemble techniques to enhance the results. These results are compared with individual base learners. The result obtained in this research paper is higher than previous studies.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Erythemato-squamous disease is also called the skin disease. Skin disease can be classified in six classes C1: psoriasis, C2: seborrheic dermatitis, C3: lichen planus, C4: pityriasis rosea, C5: chronic dermatitis, C6: pityriasis rubra [1]. Skin disease diagnosis is difficult, because six classes possess identical clinical properties with very small changes. Basically Biopsy is used for treatment of these skin diseases.
In past decade many expert systems were developed for providing fruitful decisions in various fields, as data related to medical field is widely available freely on internet. Therefore, a great improvement in the clinical area for predicting the various diseases using machine learning algorithms and deep learning on data sets from the previous patient’s history. The development of expert systems using data mining in the field of medical decision support system is beneficial for patients which were do not bear high cost medical tests. Expert systems now provide assistance to doctors for helping in diagnose better [2].
Many research paper published on skin diseases with the help of decision tree [3,4,5,6,7,8]. They conducted different experiments focusing on six major skin diseases classes as its research subjects. They uses decision tree, Random Forest, Chi-square Automatic Interaction Detector (CHAID) and Extra tree of data mining classification methods to construct the best predictive model in dermatology. They used data set from UCI machine learning repository for calculating the accuracy of prediction model.
Some papers have studied the skin image and predict the accuracy of skin disease [9,10,11]. The application of deep convolution neural network (CNN) changes the quality of computer-aided supporting systems. A computer-aided diagnosis system is a young field. There are many Challenges for the researcher to achieve best Result. Current researches indicate that expert systems developed with the help of machine learning algorithms and data mining techniques required to collectively satisfactory clinical expertise to attain a best result. Convolution neural network (CNN) algorithms are used to classify four common skin disease images and obtained improved results. Besides CNN different studies are done using Adaptive boosting Boost, BayesNet, J48, Multi Layer Perceptron and NaiveBayes.
There are various studies which used ANN, KNN and SVM algorithms to solve skin disease prediction and developed model to achieve best accuracy [8, 12,13,14,15,16,17]. They developed models using popular soft computing techniques namely Artificial Neural Network, Support Vector Machine and deep leaning or combination of these techniques. These approaches are applied on the multi class skin disease data set and some comparative inferences are generated various metric like RMSE, Kappa Statistics, Accuracy, Sensitivity, F-scores and etc. The ELM and SVM are compared in the identification of erythema-squamous skin diseases was studied. The comparative studies have been done through various experiments. These experiments result show that ELM is better than SVM. In this study, the outcome of changing in the training and testing data depends on performance of classifier.
Some studies also done on skin disease data set using frequent pattern analysis [18] in this study MAFIA algorithm was used to find significant frequent patterns and discovered using to predict Skin Cancer risk level. Association, classification, clustering, Naïve Bayesian and fuzzy network are also used to predict skin disease [2, 19,20,21,22,23,24].
Ensemble methods are a way to combine various machine learning classifiers to improve the results obtained by single machine learning classifiers. There are various types of ensemble methods. Bagging, Adaptive Boosting, Gradient Tree Boosting, Stacking and Bucket of Models are popular ensemble methods. Using ensemble methods allows producing better predictions compared to a single model. There are a number of articles based on these ensemble methods to improve the accuracy as compared to single classifier [25,26,27]. Most of these articles discussed the use of ensemble methods to improve the results of various base classifiers.
Feature selection techniques are used to reduce the unnecessary attributes of data set. Generally all the attributes of a disease does not play important role in prediction results. In recent years a great importance was shown on feature selection. The recent publications [28,29,30,31,32,33], based on feature selection techniques indicate that the research scope of feature selection technique is still not finished. Now research are focused on developing new hybrid feature selection technique not only based on general feature selection methods defined in machine learning [34,35,36].
Base learners and their abbreviations which are used in paper are listed in Table 1.
2 Previous study
In this section, we have presented important previous studies on skin disease data set using various classifier and feature selection techniques. The studies are summarized in Table 2.
3 Methods
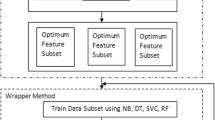
We have used three statistical methods: 1. Chi-square 2. Information Gain and 3. Principal Component Analysis to make a new hybrid feature selection technique. This hybrid feature selection technique is applied on skin disease data set which consists of 34 attributes is taken from UCI machine learning repository. Now we apply hybrid feature selection technique to choose ten important attributes to obtain new reduced data subset of skin disease data set. The six base learner classifiers 1. Gaussian Naïve Bayesian classifier, 2. K Nearest Neighbor 3. Decision Tree Classifier 4. Support Vector Machine 5. Random Forest Classifier and 6. Multilayer Perceptron classifier are applied to find the prediction. To improve the results of base learner’s three ensemble methods Bagging, Boosting and Stacking are applied.
The results obtained using ensemble methods are compared with each other and we have selected the best ensemble techniques on the basis of prediction accuracy. Figure 1 illustrates the proposed methodology applied in this research paper.
3.1 Data set analysis
The data set for skin disease is collected from [1]. The data set have 12 clinical features which are listed in Table 3a and 22 Histopathological features which are shown in Table 3b. The range of features are defined as
The clinical attribute age is nominal attribute.
3.2 Feature selection
The simples rule is garbage in then we find garbage out. This means if we provide unnecessary attribute to a classifier, we get undesired results. Important features are those features which provide efficient predictions. Feature selection is one of the most important and time consuming step in the development of a model. This is the process by which we choose the variables that will be used for predict outcomes. By ranking features according to importance can be chosen for selecting the important features. These methods are used for following reasons:
-
To reduce training time of classifiers.
-
To reduces the complexity of developed model.
-
To enhance the results of prediction through reducing unnecessary attributes from the data set.
-
To reduce over-fitting.
We have developed a new hybrid feature selection techniques to choose ten most significant attributes selected by three statistical methods chi-square, information gain and principal component analysis.
3.2.1 Chi-square feature selection
One of the main feature selection methods that is used to find the relation between attributes to target attribute is Chi-Square feature selection. To test the independence of two events χ2 test is used in statistics. Further specially in feature selection we use χ2 to test whether the occurrence of a particular attribute and the occurrence of a particular class are independent. Chi-square values are calculated using formula (1):
where, c degrees of freedom, O observed value, E expected value
3.2.2 Information gain
The feature importance is basically the mean of the individual trees’ improvement in the splitting criterion produced by each variable. It is how much the score (so-called “impurity” on the decision tree notation) was improved when splitting the tree using that specific attributes.
The most common impurities are the Gini Impurity and Entropy. An improvement on the Gini impurity is known as “Gini importance”, while an improvement on the Entropy is the Information Gain. Gini Imp (2) and Entropy (3) can be calculated with equations:
3.2.3 Principal component analysis (PCA)
PCA is a method to convert dimensionality reduction by choosing the important features that contain all most information of the data set. Important features are chosen on the basis of variance that they produce in the target variable. The feature which has highest variance is called as the first principal component. The feature which gets second highest variance is called as the second principal component, and so on. The principal components do not have any correlation with each other.
3.3 Hybrid feature selection technique
A new hybrid method is presented in this section for choosing the important features. We have chosen three feature selection methods Chi-square, Information gain and principal component analysis for selecting the important features, but no one is complete, because they perform differently on different type of data sets. Therefore, we have combined these techniques to get a new hybrid feature selection technique which performs well in all conditions. The algorithm of combing these feature selection techniques is described below:
Step 1: Normalize the values of chi-square test by finding the highest value of Chi-square and dividing the remaining values by it.
Step 2: All the three feature selection techniques gives the values in the range 0–1. Arrange the values in ascending order of three feature selection techniques.
Step 3: Merge theses values in one list using merge sorting method.
Step 4: Choose desired number of features n from the merged values.
3.4 Machine learning classifiers
In this paper, we have used six different classifiers for predicting the skin disease. The six different classifiers are selected as a combination of homogeneous and heterogeneous classifiers, because we use various type of ensemble method.
3.4.1 Gaussian Naïve Bayesian classifier
Gaussian Naive Bayes evaluate continuous values of each attribute and represent their distribution according to a Gaussian distribution which is also known as Normal Distribution. When we draw a Gaussian distribution it will give a bell shaped curve which is symmetric about the mean of the feature values and these values are calculated with Eq. (4):
The \({\sigma }_{y}\) and \({\mu }_{y}\) is evaluated using maximum likelihood.
3.4.2 K nearest neighbor
KNN base learner is used for classification and regression problems but KNN is generally used in classification problems. KNN is a lazy learning and non-parametric algorithm. If there is no assumption for underlying data distribution then it is called Non-parametric it means the model structure determined from the data set. KNN will be helpful in prediction, where the data sets do not follow mathematical hypothetical assumptions. KNN does not need any training for data for development of model; therefore, it is called Lazy learning algorithm. All training data are used in directly testing phase.
3.4.3 Decision tree classifier
Decision tree classifier is the most dominant and accepted tool for prediction and classification. A Decision tree is a tree structure, where each interior node represents a test on an feature, each branch denotes a result of the test, and each terminal node holds a class label. Decision tree can be used, because they are able to generate understandable rules. Decision tree is valuable, famous and useful because of its flowchart. Flow chart is shown in Fig. 2.
3.4.4 Support vector machine
Support Vector Machines is used for both classification and regression analysis. SVM finds the hyperplane which maximizes the margin within two classes. The vectors which define the hyperplane are called support vectors. Under favorable conditions, SVM research should construct a hyperplane that completely separates the vector into two non-overlapping classes. However, in many cases, this is not possible, so SVM will find hyperplanes that maximize margins and minimize classification errors.
3.4.5 Random forest classifier
RF is a supervised learning algorithm that can be used for classification and regression. But it is mainly used for classification problems. A forest is a collection of trees, and more trees means a strong forest. Similar to decision trees, the random forest algorithm also creates decision trees on the data, then gets predictions from each tree, and finally chooses the best solution by voting. It's called an ensemble approach, and it's better than a single decision tree, because it reduces over-fitting by averaging the results.
3.4.6 Multilayer perceptron
A Multilayer perception (MLP) is a logistic regression classifier. In this classifier input data is changed with the help of a learnt non-linear conversion. This change input data into a layer, where input data becomes linearly divisible. This layer which changes data from input is called a hidden layer. Only single hidden layer is used in multilayer perceptron otherwise it will works as ANN. Although the use of multiple hidden layer are very beneficial.
3.5 Ensemble techniques
Ensemble methods are used for combining several base learners to predict a problem enhancing the perdition of single classifiers. Ensemble methods can be broadly categorized in two types—joining multiple classifiers of similar types and joining multiple classifiers of different types. Here both types are used for evaluations.
3.5.1 Bagging
Use Bagging ensemble technique when the goal is to reduce the variance of another base learner. The goal here is to create some subset of the data from a randomly selected and replaced training set. Each subset of the data set is used to train its base learner. As a result, we have a collection of different models. Using the average of all predictions from different base learners, it is more reliable than one base learner.
3.5.2 Boosting
Boosting ensemble technique is used to create a set of predictors. In this technique, learners learnt sequentially, while near the beginning learners fit simple models to the data and then analyze error in the data. Continuous trees (random samples) are suitable, and in each step, the goal is to improve the accuracy of the previous tree. When an input is misclassified by an assumption, its weight increases, so the next assumption is more likely to be correctly classified. This process turns base learners with weak learning capabilities into better performing models.
3.5.3 Staking
Stacking ensemble method is used for combining multiple base learners of different type with the help of a meta-classifier. Six base learners NB, KNN, DT, SVM, RF and MLP are trained with whole training data set; and then, meta-classifier is applied on the outputs “meta-features” of the each base Lerner. Meta-classifier can be trained on the probabilities from the ensemble techniques or predicted class labels.
4 Results
We used hybrid features selection technique for finding the best features to predict skin disease. This hybrid feature selection techniques is based on three different feature selection techniques (Chi-square, Information Gain, Principal Component Analysis). First we apply the hybrid feature selection technique to choose the important features using the skin disease data set. 10 most important features are obtained using hybrid feature selection technique. The significant attributes selected by hybrid feature selection technique used in the paper along with the selection criteria are listed in Table 4.
In this paper, we have developed Python code for implementing the base learners and hybrid feature selection technique, ensemble methods and to calculate different metrics. To calculate the different metrics and accuracy of base learners we used the following formulas:
In another term is can be represented as
where, TN class is negative and observation is negative, FN class is negative but observation is positive, FP class is positive but observation is negative, TP class is positive and observation is positive.
Mean value, standard deviations and accuracy (5, 6) of each base learners are illustrated in Table 5.
It is observed that the highest accuracy is 97.29% in found support vector machine base learner and the lowest accuracy is 89.18% in Gaussian Naïve Bayesian base learner.
Hybrid feature selection technique gives the highest accuracy 97.29% in case of SVM base learner. So, we can say that the selection of hybrid feature selection technique is good.
A graphical representation of accuracy achieved by six base learners using hybrid feature selection technique are shown in Fig. 3.
The performance and accuracy of base learners, model and ensemble methods are closely related with metrics. In this paper, we have evaluated RMSE, KSE and AUC. These terms are defined as:
Root mean square error: RMSE is defined as the values predicted by a base learner and the values actually observed. If we have developed the good base learner then the RMSE values for both training and testing data are similar, otherwise the base learner developed is not good if the RMSE values are much higher in training and testing of data. To evaluate the RMSE value we use the following formula (7):
Kappa statistic error: KSE metric is used to compare evaluated accuracy and expected accuracy. The value of KSE always in between −1 to 1. If the calculated value of KSE is nearly 1 then the performance of classifier is more accurate rather than by observation. KSE is evaluated for a single base learner, as well as for ensemble methods. KSE is evaluated with the help of following formula (8):
Area under receiver operating characteristics (AUC): With the help of TP, FP, FN, TN we can calculate True Positive Rate (TPR) and True Negative Rate (TNR). The average of True Positive Rate and True Negative Rate (TNR) is called the area under receiver operating characteristics. These terms are calculated using following formulas:
Values of RMSE (7), KSE (8) and AUC (11) are calculated with the help of formulas for base learners applied on reduced data subset obtained by hybrid feature selection techniques and are represented in Table 6.
We have used three ensemble techniques Bagging, Boosting and Stacking to enhance the results obtained by base learners and different predication accuracy are found which are shown in Table 7.
Six base learners are combined with the help of bagging, boosting and stacking ensemble methods. The skin disease data subset obtained using hybrid feature selection technique is evaluated with three ensemble methods are discussed in Table 8. Confusion matrix precision recall and accuracy calculated by different ensemble techniques are discussed and their formulas are given by Eqs. (12), (13) and (14):
5 Discussion
In this paper, we have presented a new approach to enhance the prediction accuracy with the help of a new hybrid feature selection techniques. Much work has already done using feature selection techniques, because feature selection is an important step to improve the accuracy of prediction. Feature selection removes the attributes which does not play important role in data sets. As in the case of Skin disease data set there are 34 attributes but all of them do not play important role in skin disease perdition; therefore, it is necessary to remove those attributes by finding them using feature selection techniques. We choose only ten most important attributes for prediction of skin disease with the help of chi-square, information gain and principal component analysis. These three statistical method evaluated their score and the we combine theses scores and select ten attributes out of which two were clinical and eight histopathological attributes were selected with hybrid feature selection technique, the selected attributed are shown in Table 4. From table it is obvious that the selected attributes are combination of all three statistical methods four attributes are selected from principal component analysis, three attributes are selected with information gain and three attributes are selected with chi-square test.
At second step after selecting the skin disease reduced data subset consisting of only ten attributes, we use six base learners NB, KNN, DT, SVM, RF and MLP to evaluate the performance of these base learners. Performance of base learners are evaluated using Mean value, slandered deviation and accuracy, RMSE, KSE and AUC. These values provide information about the base learners, where the predicted values are good or not. The evaluated values of these metrics prove that the base learners taken in this study well suits for the prediction of skin disease. Tables 5 and 6 show the values of metrics. The highest accuracy obtained from base learners is 97.29% using support vector machine and second highest accuracy obtained is 95.94% in multi layer perceptron. This accuracy proves that the base learner’s performance is very good.
At third Step, we applied three ensemble methods Bagging, Boosting and Stacking on six base learners to enhance the prediction results. Ensemble methods are used to combine the results obtained by base learners to give better results. Accuracy, confusion matrix, precision, recall, f1-score and support values for three ensemble techniques are shown in Table 8. The accuracy obtained by Bagging, Boosting and staking are 95.94%, 97.70% and 99.67%, respectively, which shows that the accuracy of ensemble methods are improved as compared to base learners.
There are a number of articles based on feature selection techniques to improve the accuracy of skin disease prediction model. To compare our study using hybrid feature selection techniques we have illustrated the previous studies in Table 2. The authors classifier used in their studies and feature selection methods are shown in Table 2. The highest accuracy achieved by their studies is compared with this study.
6 Conclusion
Today, machine learning algorithms are used for knowledge discovery with the help of previous stored information from healthcare to build up expert systems for taking decisions. In this paper a hybrid new feature selection technique is developed to enhance the results of the perdition. The new hybrid feature selection technique is developed using the combination of chi-square, information gain and principal component analysis statistical techniques to select the 10 most important features. Six base learners NB, KNN, DT, SVM, RF and MLP are used to check the performance of skin disease data subset. Support vector machine provides the best result of 97.29%. Various metrics are evaluated to measure the performance of base classifiers. Mean Value, Standard Deviation, Root Mean Square Error (RMSE), Kappa Statistic Error (KSE) and Area under Receiver Operating Characteristics (AUC) are calculated to verify the results of the base classifiers. At last three ensemble methods bagging, boosting and stacking are used to combine the six base learners. The accuracy obtained by Bagging, Boosting and staking are 95.94%, 97.70% and 99.67%, respectively, which shows that the accuracy of ensemble methods are improved as compared to base learners. The highest accuracy obtained is 99.67% in staking ensemble method on skin disease data subset using hybrid feature selection technique. Therefore, we recommend that the hybrid feature selection technique is the best feature selection techniques in case of skin disease.
References
Güvenir, H.A., Demiröz, G., Ilter, N.: Learning differential diagnosis of erythemato-squamous diseases using voting feature intervals. Artif. Intell. Med. 13(3), 147–165 (1998)
Barati, E., Saraee, M., Mohammadi, A., Adibi, N., Ahamadzadeh, M.R.: A survey on utilization of data mining approaches for dermatological (skin) diseases prediction. J. Sel. Areas Heal. Inf. 2(3), 1–11 (2011)
Chang, C.L., Chen, C.H.: Applying decision tree and neural network to increase quality of dermatologic diagnosis. Expert Syst. Appl. 36(2), 4035–4041 (2009)
Yadav, D.C., Pal, S.: Decision tree ensemble techniques to predict thyroid disease. Int. J. Recent Technol. Eng. 8(3), 8242–8246 (2019)
Elsayad, A.M., Dhaifallah, M.A., Nassef, A.M.: Analysis and diagnosis of erythemato-squamous diseases using CHAID decision trees. In: 2018 15th Int. Multi-Conference Syst. Signals Devices, SSD 2018, pp. 252–262 (2018)
Oladele, T.O., Olarinoye, D.R., Adebisi, S.S.: Artificial neural network(ANN). Anal. Ser. Inf. 16, 189–193 (2018)
Maghooli, K., Langarizadeh, M., Shahmoradi, L., Habibi-Koolaee, M., Jebraeily, M., Bouraghi, H.: Differential diagnosis of erythmato-squamous diseases using classification and regression tree. Acta Inform. Med. 24(5), 338–342 (2016)
Kabari, L.G., Bakpo, F.S.: Diagnosing skin diseases using an artificial neural network. ICAST 2009—2nd Int. Conf. Adapt. Sci. Technol., pp. 187–191 (2009)
Amarathunga, A.A.L.C., Ellawala, E.P.W.C., Abeysekara, G.N., Amalraj, C.R.J.: Expert system for diagnosis of skin diseases. Int. J. Sci. Technol. Res. 4(1), 174–178 (2015)
Kanaan, K.Q.: Classification of human skin diseases using data mining. Int. J. Adv. Eng. Res. Sci. 4(1), 159–163 (2017)
Zhang, X., Wang, S., Liu, J., Tao, C.: Towards improving diagnosis of skin diseases by combining deep neural network and human knowledge. BMC Med. Inform. Decis. Mak. 18(S2), 59 (2018)
Çataloluk, H., Kesler, M.: A diagnostic software tool for skin diseases with basic and weighted K-NN. INISTA 2012 Int. Symp. Innov. Intell. Syst. Appl., pp. 0–3 (2012)
Kolkur, M.S., Kalbande, D.R., Kharkar, V.: Machine learning approaches to multi-class human skin disease detection. Int. J. Comput. Intell. Res. 14(1), 29–39 (2018)
Olatunji, S.O., Arif, H.: Identification of erythemato-squamous skin diseases using extreme learning machine and artificial neural network. ICTACT J. Soft Comput. 4(1), 627–632 (2013)
Olatunji, S., Arif, H.: Identification of erythemato-squamous skin diseases using support vector machines and extreme learning machines: a comparative study towards effective diagnosis. Trans. Mach. Learn. Artif. Intell. 2, 6 (2014)
Parikh, K.S., Shah, T.P., Kota, R.K., Vora, R.: Diagnosing common skin diseases using soft computing techniques. Int. J. Bio-Sci. Bio-Technol. 7(6), 275–286 (2015)
Putatunda, S.: A hybrid deep learning approach for diagnosis of the erythemato-squamous disease. arxiv:1909.07587v1, pp. 1–13 (2019)
Ahmed, K.: Early prevention and detection of skin cancer risk using data mining. Int. J. Comput. Appl. 62(4), 1–6 (2013)
Giveki, D.: Detection of erythemato-squamous diseases using AR-CatfishBPSO-KSVM. Signal Image Process. An Int. J. 2(4), 57–72 (2012)
Güvenir, H.A., Emeksiz, N.: Expert system for the differential diagnosis of erythemato-squamous diseases. Expert Syst. Appl. 18(1), 43–49 (2000)
Sudha, M., Poorva, B.: Predictive tool for dermatology disease diagnosis using machine learning techniques. Int. J. Inno. Technol. Explor. Eng. 8(9), 355–360 (2019)
Idoko, J. B., Arslan, M., Abiyev, R.: Fuzzy neural system application to differential diagnosis of erythemato-squamous diseases. Cyprus J. Med. Sci., pp. 90–97 (2018)
Ravichandran, K.S., Narayanamurthy, B., Ganapathy, G., Ravalli, S., Sindhura, J.: An efficient approach to an automatic detection of erythemato-squamous diseases. Neural Comput. Appl. 25(1), 105–114 (2014)
Übeyli, E.D., Doǧdu, E.: Automatic detection of erythemato-squamous diseases using κ-means clustering. J. Med. Syst. 34(2), 179–184 (2010)
Sharma, D., Hota, H.: Data mining techniques for prediction of different categories of dermatology diseases. J. Manag. Inf. Decis. Sci. 16(2), 103 (2013)
Verma, A.K., Pal, S.: Prediction of skin disease with three different feature selection techniques using stacking ensemble method. Appl. Biochem. Biotechnol. (2019). https://doi.org/10.1007/s12010-019-03222-8
Verma, A.K., Pal, S., Kumar, S.: Classification of skin disease using ensemble data mining techniques. Asian Pac. J. Cancer Prev. 20(6), 1887–1894 (2019)
Aruna, S., Nandakishore, L.V., Rajagopalan, S.P.: A hybrid feature selection method based on IGSBFS and naive bayes for the diagnosis of erythemato-squamous diseases. Int. J. Comput. Appl. 41(7), 13–18 (2012)
Kamala, F.R., Thangaiah, P.R.J., Info, A.: An improved hybrid feature selection method for huge dimensional datasets. IAES Int. J. Artif. Intell. 8(1), 77–86 (2019)
Ozcift, A., Gulten, A.: A robust multi-class feature selection strategy based on rotation forest ensemble algorithm for diagnosis of erythemato-squamous diseases. J. Med. Syst. 36(2), 941–949 (2012)
Pattanshetti, T., Attar, V.: Performance evaluation and analysis of feature selection algorithms. Adv. Intell. Syst. Comput. 808, 47–60 (2019)
Xie, J., Lei, J., Xie, W., Shi, Y., Liu, X.: Two-stage hybrid feature selection algorithms for diagnosing erythemato-squamous diseases. Heal. Inf. Sci. Syst. 1(1), 1–14 (2013)
Verma, A.K., Pal, S., Kumar, S.: Prediction of skin disease using ensemble data mining techniques and feature selection method—a comparative study. Appl. Biochem. Biotechnol. (2019). https://doi.org/10.1007/s12010-019-03093-z
Badrinath, N., Gopinath, G., Ravichandran, K.S., Soundhar, R.G.: Estimation of automatic detection of erythemato-squamous diseases through AdaBoost and its hybrid classifiers. Artif. Intell. Rev. 45(4), 471–488 (2016)
Chaurasia, V., Pal, S.: Skin diseases prediction: binary classification machine learning and multi model ensemble techniques. Res. J. Pharm. Technol. 12(8), 3829–3832 (2019)
Maryam, N.A., Setiawan, Wahyunggoro, O.: A hybrid feature selection method using multiclass SVM for diagnosis of erythemato-squamous disease. AIP Conf. Proc. 1867, (2017)
Xie, J., Wang, C.: Using support vector machines with a novel hybrid feature selection method for diagnosis of erythemato-squamous diseases[1] J. Xie and C. Wang, “Using support vector machines with a novel hybrid feature selection method for diagnosis of erythemato-squamou. Expert Syst. Appl. 38(5), 5809–5815 (2011)
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Verma, A.K., Pal, S. & Tiwari, B.B. Skin disease prediction using ensemble methods and a new hybrid feature selection technique. Iran J Comput Sci 3, 207–216 (2020). https://doi.org/10.1007/s42044-020-00058-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42044-020-00058-y