Abstract
Nowadays, skin disease is a major problem among peoples worldwide. Different machine learning techniques are applied to predict the various classes of skin disease. In this research paper, we have applied six different machine learning algorithm to categorize different classes of skin disease using three ensemble techniques and then a feature selection method to compare the results obtained from different machine learning techniques. In the proposed study, we present a new method, which applies six different data mining classification techniques and then developed an ensemble approach using bagging, AdaBoost, and gradient boosting classifiers techniques to predict the different classes of skin disease. Further, the feature importance method is used to select important 15 features which play a major role in prediction. A subset of the original dataset is obtained after selecting only 15 features to compare the results of used six machine learning techniques and ensemble approach as on the whole dataset. The ensemble method used on skin disease dataset is compared with the new subset of the original dataset obtained from feature selection method. The outcome shows that the dermatological prediction accuracy of the test dataset is increased compared with an individual classifier and a better accuracy is obtained as compared with subset obtained from feature selection method. The ensemble method and feature selection used on dermatology datasets give better performance as compared with individual classifier algorithms. Ensemble method gives more accurate and effective skin disease prediction.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The application of machine learning algorithms is widely used in the area of the medical field. Various disease diagnosis classification algorithms are developed to find the high accuracy for predicting the disease. Many machine learning algorithms are developed for predicting the various type of disease at early level after examining the various attributes of the disease. These algorithms are widely applicable in breast cancer, kidney diseases, thyroid disease, diabetes, cancer diseases, erythemato-squamous diseases, and many more. In this research paper, we choose erythemato-squamous disease for analysis. Various classification algorithms are applied and then ensemble methods are applied in this study. Another approach using feature selection is applied with these classification algorithms to obtain the accuracy of the prediction for the application to make an expert system [1, 2]. Various studies have been done in this field and some are discussed below.
Polat and Güneş [3] achieved 96.71% classification accurate rate on the diagnosis of erythemato-squamous diseases using a novel hybrid intelligence method based on C4.5 decision tree classifier and one-against-all approach for multi-class classification problem.
Immagulate and Vijaya [4] focus on non-melanoma skin cancer and classify types, using support vector machines (SVM) to accurately predict disease types. The chrominance and texture features are extracted pre-processed training datasets.
Chang and Chen [5] discussed the decision tree combined with neural network classification methods to construct the best predictive model of dermatology. The learning predicted and analyzed six common skin conditions. All classification techniques can predict disease fairly accurately, and the neural network model has the highest accuracy of 92.62%.
Ramya and Rajeshkumar [6] discussed the Gray-Level Co-Occurrence Matrix (GLCM) technique for finding features from segmented disease and classifying skin disease based on fuzzy classification, which is more accurate than existing ones.
Übeyli and Doğdu [7] find the results of a study where they deployed a k-means clustering approach to classifying erythemato-squamous diseases dataset. The results of the study indicate approximately 94% overall classification accuracy rate when using 5 out of the 6 decision classes (excluding Pityriasis rubra pilaris—20 instances).
Güvenir et al. [8] discussed the VFI5 classification algorithm which represents a concept description by a set of feature value intervals. The classification of a new instance is based on voting among the classifications made by the value of each feature separately.
Ahmed et al. [9] discussed clusters of pre-processed data, using k-means clustering algorithms to separate related and unrelated data into skin disease. Frequent patterns were evaluated using the MAFIA algorithm. Decision tree and AprioriTid algorithms are used to extract frequent patterns from clustered datasets.
Fernando et al. [10] discussed a disease prediction method, DOCAID, to predict malaria, typhoid fever, jaundice, tuberculosis, and gastroenteritis based on patient symptoms and complaints using the naïve Bayesian classifier algorithm. The authors reported an accuracy rate of 91% for predicting disease.
Jaleel et al. [11] extracted these features using a 2D wavelet transform method and then classified them using a back propagation neural network (BPNN). They classify the dataset as cancer or non-cancer.
Theodoraki et al. [12] developed a predictive model to predict the final outcome of a seriously injured patient after an accident. The investigation includes a comparison of data mining techniques using classification, clustering, and association algorithms. Using this analysis, they obtained results in terms of sensitivity, specificity, positive predictive value, and negative predictive value, and compared results between different predictive models.
Sharma and Hota [13] used SVM and ANN data mining techniques, to classify various types of erythema-squamous diseases. They used a confidential weighted voting scheme to combine the two technologies to achieve the highest accuracy of 99.25% in the training and 98.99% in the testing phases.
Rambhajani et al. [14] used a Bayesian classification to classify the erythemato-squamous disease dataset. The author used the best first search feature selection technology technique, and they removed 20 features from the dermatology dataset collection collected by the University of California Irving repository and then used the Bayesian technology to achieve 99.31% accuracy.
Bakpo and Kabari [15] used ANN for diagnosis of different skin diseases and they achieved 90% accuracy. There are a few unique features for skin cancer regions.
Manjusha et al. [16] predict different skin diseases using the naïve Bayesian algorithm. Automatic identification of circulatory disease dermatological features was extracted from the local binary pattern from affected skin images and used for classification.
Yadav and Pal [17] discussed about women thyroid prediction using data mining techniques. They used two ensemble techniques. The first ensemble technique generated by decision tree and the second was generated by bagging and boosting techniques. They observed dataset for thyroid symptom and find better accuracy results.
Tuba et al. [18] applied the automatic erythemato-squamous diseases detection method by optimized support vector machine. Parameters of the support vector machine were tuned by the recent swarm optimization algorithm, elephant herding optimization. They tested on a standard dataset that contains data for 366 patients with one of six different erythemato-squamous diseases. The accuracy achieved by them is 99.07%.
Zhang et al. [19] applied a hybrid approach that uses granular computing (GrC) and supports vector machines (SVM). The authors reviewed and evaluated most of the past artificial intelligence systems used for the diagnosis of erythemato-squamous disease and tabulated the classification results of all these algorithms. The results achieved averages of sensitivity and specificity as 98.43% and 99.71%, respectively.
In this study, an attempt is done to use machine learning methods to ensemble six different classifiers, which are the following:
Passive Aggressive Classifier
Passive aggressive algorithms are a family of online learning algorithms (for both classification and regression) proposed by Crammer at al. The passive aggressive classifier (PAC) algorithm is perfect for classifying massive streams of data. It is easy to implement and very fast but does not provide global guarantees like the support vector machine (SVM). Passive: if correct classification, keep the model; Aggressive: if incorrect classification, update to adjust to this misclassified example.
Linear Discriminant Analysis
Linear discriminant analysis (LDA) is closely related to the analysis of variance (ANOVA) and regression analysis, which also attempt to express one dependent variable as a linear combination of other features or measurements. LDA is also closely related to principal component analysis (PCA) and factor analysis in that they both look for linear combinations of variables which best explain the data.
Radius Neighbors Classifier
Radius neighbors classifier (RNC) is very similar to a k-neighbors classifier with the exception of two parameters. First, in radius neighbors classifier, we need to specify the radius of the fixed area used to determine if an observation is a neighbor using radius. Unless there is some substantive reason for setting radius to some value, it is best to treat it like any other hyper-parameter and tune it during model selection. The second useful parameter is outlier-label, which indicates what label to give an observation that has no observations within the radius—which itself can often be a useful tool for identifying outliers.
Bernoulli Naïve Bayesian
Bernoulli naïve Bayesian (BNB) implements the naïve Bayes training and classification algorithms for data that is distributed according to multivariate Bernoulli distributions; i.e., there may be multiple features but each one is assumed to be a binary-valued (Bernoulli, Boolean) variable. Therefore, this class requires samples to be represented as binary-valued feature vectors; if handed any other kind of data, the decision rule for Bernoulli naïve Bayes is based on:
which differs from the multinomial NB’s rule in that it explicitly penalizes the non-occurrence of a feature i that is an indicator for class y, where the multinomial variant would simply ignore a non-occurring feature.
Gaussian Naïve Bayesian
Gaussian naïve Bayesian (NB) implements the Gaussian naïve Bayes algorithm for classification. The likelihood of the features is assumed to be Gaussian:
The parameters σyand μy are estimated using maximum likelihood.
Extra Tree Classifier
An “extra trees” classifier (ETC), otherwise known as an “Extremely randomized trees” classifier, is a variant of a random forest. Unlike a random forest, at each step, the entire sample is used and decision boundaries are picked at random, rather than the best one. In real-world cases, performance is comparable with an ordinary random forest, sometimes a bit better.
Three different ensemble techniques, bagging classifier, AdaBoost classifier, and gradient boosting classifier, are applied to predict skin disease.
Methods
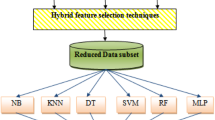
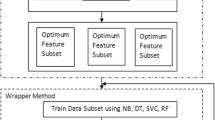
Figure 1 demonstrates the whole structure of the methodology used in this research paper. The figure demonstrates the different data mining methods (I) PAC, (II) LDA, (III) RNC, (IV) BNB, (V) NB, and (VI) ETC used in this study. The approach used in this paper is completely data driven. In this paper, we have applied the six classification algorithms to measure the accuracy and sensitivity of the predicted values of skin disease classes. The obtained values are then improved using ensemble technique by using the bagging classifier, AdaBoost classifier, and gradient boosting classifier. The same techniques are again applied on the skin disease dataset by using feature selection—in this selection, we obtained 15 important attributes using the feature importance method. Now, we reduce the dataset and take only 15 attributes and 366 instances of the dataset and again evaluate the accuracy of the prediction of skin disease dataset. A comparative study is then performed to evaluate the best prediction.
Dataset Analysis
The database used in this study is taken from the UCI machine learning repository (http://archive.ics.uci.edu/ml). Briefly, this dataset was formed to examine skin disease and classify the type of erythemato-squamous diseases. This dataset contains 34 variables; in this dataset, 33 variables are linear and 1 variable is nominal. These six classes of skin disease include the following: C1, psoriasis; C2, seborrheic dermatitis; C3, lichen planus; C4, pityriasis rosea; C5, chronic dermatitis; C6, pityriasis rubra. Biopsy is one of the basic treatments in diagnosing these diseases. A disease may also contain the properties of another class of disease in the initial stage, which is another difficulty faced by dermatologists when performing the different class of diagnosis of these diseases. Initially, patients were first examined with 12 clinical features, after which the assessment of 22 histopathological attributes was performed using skin disease samples. Histological features were identified by analyzing the samples under a microscope. If any of the diseases is found in the family, the family history attribute in the dataset constructed for the domain has a value of 1 (one), and if not found, the value is 0 (zero). The age of the patient is used to indicate age characteristics. All other attributes (clinical and histopathological both) were assigned a value in the range from 0 to 3 (0 = absence of features; 1, 2 = comparative intermediate values; 3 = highest value). There are six classes of erythemato-squamous disease, with 366 instances and 34 attributes in the domain. Table 1 summarizes the contents of the attributes.
Feature Selection
Feature selection is a process to automatically select only those features from the dataset which contribute most to the prediction variable or output. Irrelevant attributes in the dataset can decrease the accuracy of many models, especially linear algorithms like linear and logistic regression. Three benefits of performing feature selection before modelling dataset are the following:
- 1.
Reduces over fitting
- 2.
Improves accuracy
- 3.
Reduces training time
There are many feature selection techniques (univariate selection, recursive feature elimination, principal component analysis, and feature importance) available, but in this research paper, we have applied the feature importance method to choose 15 most important attributes from the skin disease dataset. The importance of attributes is shown in Fig. 2. Figure 2 shows an important score for each attribute where the larger the score, the more important the attribute.
We can get the feature importance of each feature of the dataset by using the feature importance property of the model. Feature importance gives a score for each feature of the dataset, the higher the score, the more important or relevant is the feature towards the output variable. The feature importance is calculated based on fallowing algorithm:
- 1.
Train an FR suing D _ train (all k features)
- 2.
Compute average RMSE of a model for cross-validation data (C-V)
- 3.
Rank performance by \( {VI}_k=\sum \limits_{\theta}^{\varphi}\frac{\left(\in {\theta}_k-{\epsilon \theta}_{\pi k}\right)}{\varphi \delta} \)
- 4.
For each subset of k ki = k − 1, k − 2, k − 3, ……. . , 1 do
- 5.
Train a new forest from ki features, highest VI
- 6.
Calculate the average RMSE of the model on C-V set
- 7.
Re-rank the features (k)
- 8.
Find ji with smallest RMSE
Ensembles Method
In this research paper, the ensemble method is used as a method to find the accuracy of the skin disease dataset to improve the performance of algorithms. We will apply an ensemble method to combine six different machine learning algorithms using bagging classifier, AdaBoost classifier and gradient boosting classifier.
- 1.
Bagging. Bootstrap aggregating, also known as bagging, is a machine learning model aggregation technique designed to improve the stability and accuracy and to reduce variance to avoid over fitting of machine learning algorithms applied in regression and classification methods.
- A.
Takes original dataset D with N training examples
- B.
Creates M copies \( {\left\{\overset{\sim }{D_m}\right\}}_{m-1}^M \)
- a.
Each \( \overset{\sim }{D_m} \) is generated from D by sampling with replacement.
- b.
Each dataset \( \overset{\sim }{D_m} \) has the same number of examples as in dataset D.
- c.
These datasets are reasonably different from each other.
- a.
- C.
Trains models h1, … …, hM using \( \overset{\sim }{D_1},\dots \dots .\overset{\sim }{D_M} \), respectively
- D.
Uses an averaged model \( h=\frac{1}{M}{\sum}_{m-1}^M{h}_m \) as the final model
- A.
- 2.
AdaBoost, also known as adaptive boosting, is a machine learning meta-algorithm which uses the concept of combining independent individual hypothesis in a sequential order to improve the accuracy. Basically, boosting algorithms convert the weak learners into strong learners. It is well designed to address the bias problems.
- A.
Given training data (x1, y1), … … …… … ,(xN, yN) with yn ∈ {−1, +1}, ∀ n
- B.
Initialize weight for each example \( \left({x}_n,{y}_n\right):{D}_1(n)=\frac{1}{N},\forall n \)
- C.
For round t = 1:T
- a.
Learn a weak ht(x) → {−1, +1} using training data weighted as per Dt.
- b.
Compute the weighted fraction of errors of ht on this training data \( {\in}_t=\sum \limits_{n-1}^N{D}_t(n)\coprod \left[{h}_t\left({x}_n\right)\ne {y}_n\right] \).
- c.
Set “importance” of \( {h}_t:{\alpha}_t=\frac{1}{2}\mathit{\log}\left(\frac{1-{\epsilon}_t}{\epsilon_t}\right) \).
- d.
Update the weight of each example. \( {D}_{t+1}(n)\kern0.75em \propto {\displaystyle \begin{array}{c}\left\{\begin{array}{c}{D}_t(n)\times \exp \left({\alpha}_t\right)\kern1.25em if\ {h}_t\left({x}_n\right)={y}_n\\ {}{D}_t(n)\times \exp \left({\alpha}_t\right)\kern1em if\ {h}_t\left({x}_n\right)\ne {y}_n\end{array}\right.\\ {}={D}_t(n)\times \exp \left({\alpha}_t{y}_n{h}_t\Big({x}_n\right)\Big)\kern5.25em \end{array}} \)
- e.
Normalize Dt + 1 so that it sums to 1:\( {D}_{t+1}(n)-\frac{D_{t+1}(n)}{\sum \limits_{m-1}^N{D}_{t+1}(m)} \)
- a.
- D.
Output the boosted final hypothesis \( H(x)=\operatorname{sign}\left({\sum}_{t-1}^T{\alpha}_t{h}_t(x)\right) \)
- A.
- 3.
Gradient boosting. It is an example of a generalized boosting algorithm. Gradient boosting machine learning technique works for regression and classification problems, which produces a prediction model in the form of an ensemble of weak prediction models, typically decision trees.
- A.
Given training data (x1, y1),……………, (xN, yN).
- B.
Initialize \( {\hat{f}}_0 \)with a constant.
- C.
For t = 1 to M do the following:
- a.
Compute the negative gradient gt(x).
- b.
Fit a new base-learner function h(x, θt).
- c.
Find the best gradient descent step size ρt: \( {\rho}_t=\arg \min \rho \sum \limits_{i=1}^N\Psi \left[{y}_i,{\hat{f}}_{t-1}\left({x}_i\right)+\rho h\left({x}_i,{\theta}_t\right)\right] \).
- d.
Update the function estimate \( {\hat{f}}_t\leftarrow {\hat{f}}_{t-1}+{\rho}_th\left(x,{\theta}_t\right) \).
- a.
- A.
Results
We have conducted two experiments to obtain the prediction results for skin disease datasets. In the first experiment, we have used the skin disease dataset obtained from the UCI machine repository which consists 34 variables and one target variable “Class.” Then we have applied an ensemble method to obtain the predicted values. While in the second experiment, we have used feature selection method with the same classifier and ensemble methods to check the results obtained are better or not. Same six classifiers are again used in the feature selection dataset to obtain the predictions.
Experiment 1
Before analyzing the dataset, we first visualize the distribution of values as shown in Figs. 3 and 4. Figure 3 depicts the distribution of values of skin disease used in our study containing 366 instances and 35 attributes. Each feature shows the distribution of frequency among the 4 values (0 to 3) except the feature f34 (which is age) and feature f35 (which is targeting variable class).
The density map is a smooth continuous version of the smoothed graph estimated from the data. The most common form of estimation is called the kernel density estimation. In this method, a continuous curve (core) is drawn at each individual data point, and then all of these curves are added together for a single smoothed density estimate. The most commonly used kernel is Gaussian (which produces a Gaussian bell curve at each data point). The density map of the attributes is shown in Fig. 4.
We have used the Python code to calculate the prediction on the skin disease dataset to calculate the mean value, standard deviations, and accuracy of the six different classification techniques. The values obtained by the six classifiers are shown in Table 2.
The accuracy of any classification method is calculated using the confusion matrix and is defined as:
In another term, it can be represented as:
where TP is true positives; TN, true negatives; FP, false positives; and FN, false negatives.
From Table 2, it is clear that the highest accuracy obtained is 98.64% in the case of Bernoulli naïve Bayesian classification algorithm and the least accuracy is 58.67% for radius neighbors classification algorithm with a radius of 10 (r = 10). The box and whisker plot of six classifier methods are shown in Fig. 5.
Now, we have applied three ensemble methods boosting, AdaBoost, and gradient boosting to calculate the root mean square error values of six classification methods. This is the square root of the mean of the squared errors. RMSE indicates how close the predicted values are to the actual values; hence, a lower RMSE value signifies that the model performance is better. RMSE value is calculated by the formula:
The RMSE values for each classification algorithms using ensemble techniques are shown in Table 3.
The accuracy of three ensemble methods using six classification machine learning algorithms is shown in Table 4. It is clear that the accuracy found in Table 4 is higher than the accuracy found in Table 2. The accuracy of RNC in gradient boosting ensemble method has a very high increase as compared with individual RNC accuracy of 58.67%.
Now, we ensemble all the six classification algorithms using the bagging classifier, AdaBoost classifier, and gradient boosting classifier. The results obtained after applying these three ensemble methods are shown in Table 5.
Experiment 2
In the new obtained dataset, after feature selection using the feature importance method, we obtained 10 clinical features (f1, f3, f2, f7, f6, f10, f4, f5, f8, and f9) and 5 histopathological features (f14, f16, f20, f12, and f15). This shows that clinical features are more important than histopathological features in the accuracy of disease prediction. We first visualize the distribution of values as shown in Figs. 6 and 7. Figure 6 depicts the distribution of values of skin disease used in our study containing 366 instances and 16 attributes. Each feature shows the distribution of frequency among the 4 values (0 to 3) except the feature f35 (which is targeting variable class). Figure 7 depicts the density map of the features of the dataset.
We have again used the Python code to calculate the prediction on the skin disease dataset obtained from the feature selection method to calculate the mean value, standard deviations, and accuracy of the six different classification techniques. The value obtained from the six classifiers is shown in Table 6.
From Table 6, it is clear that the highest accuracy obtained is 97.29% in the case of Bernoulli naïve Bayesian classification algorithm and the least accuracy is 81.08% in the radius neighbors classification algorithm with a radius of 4 (r = 4). The box and whisker plot of six classifier methods are shown in Fig. 8.
The root mean square error (RMSE) values obtained by a dataset from feature selection method are shown in Table 7.
The accuracy of three ensemble methods using six classification machine learning algorithms on a reduced dataset obtained after features selection is shown in Table 8.
Here, we observe that the accuracy of ensembles methods is increased in comparison to without ensemble data mining techniques. It is also clear that the accuracy obtained from experiment 2 (with feature selection on the reduced dataset) is higher than the accuracy obtained from experiment 1 (using three ensemble techniques on six classifiers on whole skin disease dataset). Now, we ensemble all the six techniques using the bagging classifier, AdaBoost classifier, and gradient boosting classifier. The results obtained after applying these three ensemble methods are shown in Table 9.
The accuracy obtained in the feature selection method has improved over the first experiment which considered the full spectrum of data. The possible causes are due to the inclusion of irrelevant features, which does not play an important role in the prediction of skin diseases.
ROC curve represents a plot of the true positive rate (sensitivity) in the function of the false positive rate (100-specificity) for different cut-off points of a parameter. Each point on the ROC curve represents a sensitivity/specificity pair corresponding to a particular decision threshold. The area under the ROC curve (AUC) is a measure of how well a parameter can distinguish between two diagnostic groups (diseased/normal). The ROC curve of the three ensemble techniques bagging, AdaBoost, and gradient boosting is shown in Fig. 9.
Discussion
In this paper, we have conducted two experiments differently one using three ensemble methods bagging, AdaBoost, and gradient boosting on six different classification algorithms PAC, LDA, RNC, BNB, NB, and ETC, and second using the feature selection method. This experiment is done on the UCI skin disease dataset which contains 366 instances and 34 attributes. The skin dataset consists of six classes of skin disease: C1, psoriasis; C2, seborrheic dermatitis; C3, lichen planus; C4, pityriasis rosea; C5, chronic dermatitis; C6, pityriasis rubra. We have analyzed the mean, standard deviation, root mean square error, and accuracy of six different machine learning algorithms to obtain the highest accuracy of 98.64% in the case of Bernoulli naïve Bayesian classification algorithm. The performance demonstrated by the ensemble data mining techniques for skin disease prediction lies in input variable choice and classification method selection. A root mean square error is calculated for each ensemble methods. After applying ensembles method, we get 99.46% accuracy in the case of the gradient boosting classifier method.
In the second experiment, we first choose the important attributes using the feature importance method to obtain the most crucial fifteen features and then reduce the dataset as a subset of the original dataset. The reduced dataset contains 15 attributes and 366 patient records. The same ensemble methods and machine learning algorithms are applied and we get different results; the highest accuracy achieved in this experiment is 99.68% in the case of the gradient boosting classifier method.
A comparison of experiment one and two are shown in Table 10 (FS stands for feature selection).
In this comparison, we execute six classifiers using three different ensemble algorithms without feature selection (all 34 features used) and we calculated the corresponding accuracy of classifiers. The accuracies obtained with all features used and after feature selection in experiment 2 are shown in Table 10. The highest accuracy of each classifier is also presented in the same row in order to make comparison better.
The comparison from Table 10 shows the efficiency of the feature selection method on the reduced dataset is better than without the feature selection in each case, and we get higher accuracy.
To illustrate the success of our approach, the results obtained in this study were compared with other results given in the literature. In order to compare the efficiency of the proposed dermatological classification, we used a large number of technical studies using the same information but using different classification techniques and then developing a multi-model ensemble method. According to these studies, the same partitions of the above test datasets were followed. To illustrate this, the classification efficiency is compared with previous studies. This is shown in Table 11.
Although the predicted values after applying feature selection in experiment 2 gives better results than in experiment 1, it does not guarantee that it will perform in all cases, if we use more instances. The new feature selection process is used on only these 366 instances, so whether the selection of features is accurate or not will depend upon more instances, which can be used in a future study.
Conclusion
Machine learning techniques play an important role in the diagnosis of diseases in the biotechnology field. Knowledge obtained using machine learning techniques can be used to develop expert systems, which provide help in predicting various types of disease. This paper describes different data mining techniques for skin disease prediction. Six machine learning classification techniques PAC, LDA, RNC, BNB, NB, and ETC are used to classify the prediction of skin disease and three ensemble techniques bagging, AdaBoost, and gradient boosting classifiers are applied to improve the accuracy obtained by machine learning algorithms. A feature selection method is also applied on skin disease dataset and obtains more accurate accuracy of 99.68% in the case of gradient boosting ensemble method applied on RNC. We got the highest accuracy in the literature available on the skin disease dataset.
This study achieved higher accuracy as compared with previous research done in this field. This study can be useful to correctly predict the skin disease. Hence, we conclude that the proposed feature selection model can be of efficient use in the detection of erythemato-squamous diseases with improvement in speed and accuracy.
Abbreviations
- PAC:
-
passive aggressive classifier
- LDA:
-
linear discriminant analysis
- RNC:
-
radius neighbors classifier
- BNB:
-
Bernoulli naïve Bayesian
- NB:
-
Gaussian naïve Bayesian
- ETC:
-
extra tree classifier
- FS:
-
feature selection
References
Chaurasia, V., Pal, S., & Tiwari, B. B. (2018). Chronic kidney disease: a predictive model using decision tree. International Journal of Engineering Research and Technology, 11(11), 1781–1794.
Chaurasia, V., Pal, S., & Tiwari, B. B. (2018). Prediction of benign and malignant breast cancer using data mining techniques. Journal of Algorithms & Computational Technology, 12(2), 119–126.
Polat, K., & Güneş, S. (2009). A novel hybrid intelligent method based on C4. 5 decision tree classifier and one-against-all approach for multi-class classification problems. Expert Systems with Applications, 36(2), 1587–1592.
Immagulate, I., & Vijaya, M. S. (2015). Categorization of non-melanoma skin lesion diseases using support vector machine and its variants. International Journal of Medical Imaging, 3(2), 34–40.
Chang, C. L., & Chen, C. H. (2009). Applying decision tree and neural network to increase quality of dermatologic diagnosis. Expert Systems with Applications, 36(2), 4035–4041.
Ramya, G., & Rajeshkumar, J. (2015). A novel method for segmentation of skin lesions from digital images. International Research Journal of Engineering and Technology, 2(8), 1544–1547.
Übeyli, E. D., & Doğdu, E. (2010). Automatic detection of erythemato-squamous diseases using k-means clustering. Journal of Medical Systems, 34(2), 179–184.
Güvenir, H. A., Demiröz, G., & Ilter, N. (1998). Learning differential diagnosis of erythemato-squamous diseases using voting feature intervals. Artificial Intelligence in Medicine, 13(3), 147–165.
Ahmed, K., Jesmin, T., & Rahman, M. Z. (2013). Early prevention and detection of skin cancer risk using data mining. International Journal of Computer Applications, 62(4), 1–6.
Fernando, Z. T., Trivedi, P., & Patni, A. (2013). DOCAID: predictive healthcare analytics using naive Bayes classification. In Second Student Research Symposium (SRS), International Conference on Advances in Computing, Communications and Informatics (ICACCI’13), 1–5.
Jaleel, J. A., Salim, S., & Aswin, R. B. (2012). Artificial neural network based detection of skin cancer. International Journal of Advanced Research in Electrical, Electronics and Instrumentation Engineering, 1(3), 200–205.
Theodoraki, E. M., Katsaragakis, S., Koukouvinos, C., & Parpoula, C. (2010). Innovative data mining approaches for outcome prediction of trauma patients. Journal of Biomedical Science and Engineering, 3(08), 791–798.
Sharma, D. K., & Hota, H. S. (2013). Data mining techniques for prediction of different categories of dermatology diseases. Journal of Management Information and Decision Sciences, 16(2), 103.
Rambhajani, M., Deepanker, W., & Pathak, N. (2015). Classification of dermatology diseases through Bayes net and best first search. International Journal of Advanced Research in Computer and Communication Engineering, 4(5), 275–86.
Bakpo, F. S., & Kabari, L. G. (2011). Diagnosing skin diseases using an artificial neural network. In Artificial Neural Networks-Methodological Advances and Biomedical Applications, Suzuki K (ed.), intech. Available from: http://www.intechopen.com/articles/show/title/diagnosing-skin-diseases-using-an-artificial-neural-network.
Manjusha, K. K., Sankaranarayanan, K., & Seena, P. (2014). Prediction of different dermatological conditions using naive Bayesian classification. International Journal of Advanced Research in Computer Science and Software Engineering, 4(1), 864–868.
Yadav, D. C., & Pal, S. (2019). To generate an ensemble model for women thyroid prediction using data mining techniques. Asian Pacific Journal of Cancer Prevention, 20(4), 1275–1281.
Tuba, E., Ribic, I., Capor-Hrosik, R., & Tuba, M. (2017). Support vector machine optimized by elephant herding algorithm for erythemato-squamous diseases detection. Procedia Computer Science, 122, 916–923.
Zhang, X., Wang, S., Liu, J., & Tao, C. (2018). Towards improving diagnosis of skin diseases by combining deep neural network and human knowledge. BMC Medical Informatics and Decision Making, 18(2), 59.
Übeyli, E. D. (2009). Combined neural networks for diagnosis of erythemato-squamous diseases. Expert Systems with Applications, 36(3), 5107–5112.
Lekkas, S., & Mikhailov, L. (2010). Evolving fuzzy medical diagnosis of Pima Indians diabetes and of dermatological diseases. Artificial Intelligence in Medicine, 50(2), 117–126.
Xie, J., & Wang, C. (2011). Using support vector machines with a novel hybrid feature selection method for diagnosis of erythemato-squamous diseases. Expert Systems with Applications, 38(5), 5809–5815.
Cataloluk, H., & Kesler, M. (2012). A diagnostic software tool for skin diseases with basic and weighted K-NN in International Symposium on Innovations in Intelligent Systems and Applications, IEEE (2012), 1-4.
Olatunji, S. O., & Arif, H. (2013). Identification of erythemato-squamous skin diseases using extreme learning machine and artificial neural network. ICTACT Journal of Softw Computing, 4(1), 627–632.
Ravichandran, K. S., Narayanamurthy, B., Ganapathy, G., Ravalli, S., & Sindhura, J. (2014). An efficient approach to an automatic detection of erythemato-squamous diseases. Neural Computing and Applications, 25(1), 105–114.
Amarathunga, A. A. L. C., Ellawala, E. P. W. C., Abeysekara, G. N., & Amalraj, C. R. J. (2015). Expert system for diagnosis of skin diseases. International Journal of Scientific & Technology Research, 4(01), 174–178.
Parikh, K. S., Shah, T. P., Kota, R., & Vora, R. (2015). Diagnosing common skin diseases using soft computing techniques. International Journal of Bio-Science and Bio-Technology, 7(6), 275–286.
Maghooli, K., Langarizadeh, M., Shahmoradi, L., Habibi-koolaee, M., Jebraeily, M., & Bouraghi, H. (2016). Differential diagnosis of erythemato-squamous diseases using classification and regression tree. Acta Informatica Medica, 24(5), 338.
Pravin, S. R., & Jafar, O. A. M. (2017). Prediction of skin disease using data mining techniques. IJARCCE, 6(7), 313–318.
Zhou, H., Xie, F., Jiang, Z., Liu, J., Wang, S., & Zhu, C. (2017). Multi-classification of skin diseases for dermoscopy images using deep learning. In 2017 IEEE International Conference on Imaging Systems and Techniques (IST) (pp. 1-5). IEEE.
Idoko, J. B., Arslan, M., & Abiyev, R. (2018). Fuzzy neural system application to differential diagnosis of Erythemato-squamous diseases. Cyprus Journal of Medical Sciences, 3(2), 90–97.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Research Involving Human Participants and/or Animals
This paper does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Verma, A.K., Pal, S. & Kumar, S. Prediction of Skin Disease Using Ensemble Data Mining Techniques and Feature Selection Method—a Comparative Study. Appl Biochem Biotechnol 190, 341–359 (2020). https://doi.org/10.1007/s12010-019-03093-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-019-03093-z