Abstract
Classifying electrocardiogram (ECG) signals into different heart disease classes requires a series of computationally complex signal processing models. According to the standardization of ECG equipment, pre-processing and filtering steps are redundant, as combined, these are built into the capturing device itself to design a highly accurate ECG classification system. Feature extraction, selection, classification, and post-processing operations must be combined to be executed with utmost efficiency. In this research, a novel ensemble-based classification model that combines the efficiency of convolutional neural networks (CNN) with linear & bio-inspired classifiers like random forests (RF), support vector classifier (SVC), and k-nearest neighbor (kNN) classifier is proposed. This classifier classified arrhythmia datasets taken from Massachusetts Institute of Technology-Beth Israel Hospital (MIT-BIH) with an accuracy of 99.98%, that is currently highest amongst all the decent algorithms. The system also provides a high value of precision of 99.48%, recall of 99.73%, and 99.6% of f-Measure. Judicial selection of features became possible by combining several layers CNN network, with linear & bio-inspired classifiers. The proposed model outperformed with the decent models and affirms its real-time applicability for clinical usage over multiple patient types.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
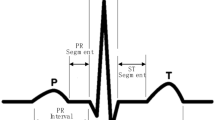
ECG classification is a multi-domain task that requires efficient signal processing blocks, although optimization of ECG signal acquisition and filtering processes have been achieved with the advancement in signal acquisition technology, and integration of on board digital signal processing (DSP) elements. Typical ECG classification system wherein the QRS feature is used to classify ECG signals into normal and arrhythmia signals. Each ECG wave is necessarily made up of following signals, P-Wave, the initial wave of the ECG signal [1]. It is a low amplitude, and short time period wave that has a typical interval of 0.12–0.22 s for regular heart beat patterns. QRS wave, also known as the heart beat pulse, is a spike-like wave with a typical duration of less than 0.12 s [1]. Difference between ‘R’ intervals for consecutive ECG signals is termed as ‘RR’ interval, and is used for identification of arrhythmic heart signals. ST segment used for measuring depression in exercise stress testing [2]. The Q duration is also known as the duration of a beat QT that represents the condition of heart. If there is a variations in these measures from the normal value they indicate the irregular rhythm called Arrhythmia [3].
All these waveforms are used for feature extraction for ECG signal. Once the features are extracted, they are fed to a feature selection unit. Some features are passed for classification purpose while others are removed based on their feature variance. These selected features with maximum variance are given to a classification block, like neural networks, SVM, RF, CNN, etc. To differentiate between signals belonging to different categories. The results of this classification are given to a post-processing block, that helps to analyse the changes in patterns of those signals, and evaluate future health risks for patients. A survey of the most recently proposed algorithms for ECG signal classification has been mentioned in next section i.e. overview of the literature. This section will help the researchers to evaluate nuances, advantages, and drawbacks of these systems. The section is followed by design of the proposed ensemble learning classifier that uses a combination of CNN with bio-inspired & linear classification algorithms like random forest, kNN & SVC.
Summary of proposed paper is given as,
-
1.
Design of a customized CNN model which is inspired by VGGNet-16 (visual geometry group network) classifier method.
-
2.
Design of the ensemble classification model that combines multiple classifiers to form effective classifier system.
The suggested algorithm as well as a comparison of that algorithm to a number of high-efficiency models that have been studied before for ECG classification. Additionally, it suggests carrying out additional studies in this area so as to further enhance the functioning of the system as a whole.
2 Overview of related literature
Review of several works based on deep learning and machine learning
Ref. No | Method used/Feature extraction technique | Description |
|---|---|---|
[4] | Morphological and dynamic features | An SVM classifier was used to classify distinct types of heartbeats using morphological and dynamic data |
[5] | Optimal orthogonal wavelet filters | It was shown that wavelet decomposition with an orthogonal filter bank decreased stop-band energy and fuzzy entropy characteristics |
[6] | Neural network (Predictive Coefficients and Probabilistic) | ECG wavelet analysis revealed an R peak. RNN and SVM classifiers use QRS linear predictive coefficients |
[7] | Dual tree complex wavelet based features | Was done using the discrete wavelet transform and dual-tree complex wavelet transform |
[8] | Feature methods with long short-term memory (LSTM) network model | An LSTM model was used to divide the MIT-BIH arrhythmia datasets into five types of arrhythmia beats based on these characteristics |
[9] | DNN (Deep Neural Network) with Unsupervised Feature Extraction Technique | Pre-training and fine-tuning were done using deep auto-encoders and deep neural networks, respectively |
[10] | DNN and engineered features | On the 2017 PhysioNet/Computing in Cardiology challenge database, the ensemble technique does a better job of identifying arrhythmia than individual classifiers |
[11] | Auto-encoders (stacked sparse) and softmax regression | An ECG arrhythmia classification algorithm based on softmax regression was reported.In-depth features were extracted from the MIT-BIH arrhythmia data set using stacked sparse auto-encoders |
[12] | Deep CNN with long duration ECG signals | There are 17 kinds of arrhythmia beats in the 1-D CNN model. The MIT-BIH arrhythmia database was used to look at 10-s long-duration ECG signal fragments from one lead of 45 patients |
[13] | CNN, LSTM | The model handled variable-length data with 98.10 percent accuracy using ten-fold cross-validation |
[14] | LSTM-Based Auto-Encoder | The auto-encoder decoder model received the reconstructed ECG signals with high-level characteristics using the LSTM model |
[15] | convolutional encoded features with bidirectional LSTM memory | A convolutional encoder initially encoded a bidirectional LSTM model to identify arrhythmias from ECG data |
[16] | Multi layer NN (Neural Network) and metaheuristic algorithm approach | Automatic ECG arrhythmia classification using multilayer perceptron neural networks (MLP) and enhanced metaheuristics was presented. The MLP classifier was trained and tested using particle swarm optimization. |
[17] | Combination of CNN, LSTM | The authors suggested a hybrid DL model using CNN and LSTM to detect six types of arrhythmia beats. To increase the hybrid model's performance, a varied number of people were used to train and test the dataset |
Researchers have proposed several powerful algorithms over the past several years to optimize the performance of ECG signal classification. Most of these algorithms use neural networks and other optimization techniques in order to optimize classification accuracy, precision, recall and f-Measure values. In order to optimize the performance of ECG signal classification, number of decent algorithms approaches have been proposed by researchers over the past several years.
For classification some researcher used machine learning [18,19,20,21,22,23,24] and deep learning [25,26,27,28,29,30] algorithms. Machine learning classifier like, Naive Bayes [31, 32], Decision Tree [33], KNN [34, 35], SVM [36, 37], RF [38], Logistic Regression, Optimization technique [39] and others are used to categories the dataset. The CNNs [38, 39, 34] are often used by many researcher in different areas. RNN, multi-layered feed-forward neural network (MLFFNN) [35] is a type of artificial neural network (ANN) [34, 40] It improves fixed-size input and output networks. After that, the suggested model's statistical evaluation and comparison with other CNN-based models are demonstrated. There are a few observations and research areas that academics might explore to enhance and adjust the recommended model.
2.1 Challenges
Selecting the appropriate feature is also essential for optimal classifier performance. The standard method for classifying diseases has a significant problem when it comes to picking the most essential attribute. Standard deep learning suffers from high processing complexity and lengthy training times because of the large dimensionality of the data used for training. In the medical literature, there are several illness-decision-help systems with varying degrees of precision. However, the vast majority of investigations have not looked into missing data and feature selection in its entirety. The lack of data used for training leads to over-fitting, which in turn leads to inaccurate predictions.
3 Materials and method
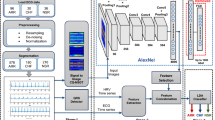
In order to enhance accuracy levels of ECG classification models, an ensemble CNN network model with bio-inspired and linear classifiers are designed in this section. This design combines VGGNet-16 inspired CNN, Discrete wavelet transform (DWT), bio-inspired RF model, linear SVM, and kNN classifier together to reduce classification errors. Architecture diagram for the proposed model can be observed from Fig. 1, wherein these classifiers are combined via weighted operations in order to achieve high accuracy, precision, recall, and F-measure.
3.1 Database used
The MIT-BIH arrhythmia database is a publicly available dataset which includes standard investigation material for the identification of cardiac arrhythmia. Since 1980, it has been utilised for the purpose of fundamental research and medical device development on heart rhythm and associated illnesses.For validating the proposed method, we have used ECG signals from MIT-BIH dataset [41] (see Tables 1, 2) (see Fig. 2).
The CNN is trained using sparse categorical cross-entropy (SCCE) loss function, and is optimized for accuracy. The CNN model has high accuracy, but is not able to differentiate between some Cardiovascular disease (CVD) classes via feature convolutions. The hyperparameters used for these convolutions are depicted in Table 3 as follows,
Thereby, the same dataset is given to a discrete wavelet transformation engine to evaluate wavelet features. These features are evaluated using Eqs. 1 and 2 as follows,
‘x’ is the ECG waveform trials, ‘\(\partial \left(k\right)\)’ is the wavelet function, ‘a’ is wavelet constant, ‘N’ is the number of samples in the ECG waveform and the output wavelet features. Using these features, kNN, SVM and Random forest classifiers are trained. After this training, all the test set values are given to each of these classifiers, and the following Eq. 3 is used to find the final class,
where, ‘w’ are the weight factors, and ‘C’ are the class outputs given by the classifier. Weight factors are evaluated from historical accuracy values obtained by these classifiers in the literature, and it is observed that the following values of weight are most optimum for the proposed model, \({w}_{cnn}=0.6, {w}_{knn}=0.2,\) & \({w}_{svm}=0.1\,and\,{w}_{rf}=0.1\) etc., Thereby, the final class for the given ECG waveform is evaluated and stored for further performance comparison In order to evaluate performance of the proposed model, the next section uses entire MIT-BIH dataset, and divides it into equal parts for evaluation of accuracy, precision, recall and f-Measure values. These values are compared using state-of-the-art methodologies.
3.2 Performance parameter
Precision measures the quality of a model's positive prediction and is used to evaluate model performance. Precision is the ratio of true positives to valid forecasts. Precision measures how many of the positively anticipated samples are meaningful.
Where, TP, is True positive (TP), and false positive (FP). False negative (FN), True negative (TN) etc.
The recall rate is the percentage of accurate positive samples to total positive samples. measures the model's ability to identify positive samples. Higher recall means more positive samples. recall, or sensitivity. It measures how many positive samples are expected to be positive.
The accuracy of a machine learning model determines which model is better at discovering correlations and patterns in a dataset based on training data. Accuracy is the ratio of correct to total classifications.
The F-measure uses the harmonic mean of accuracy and recall, weighing each variable equally. It allows a model's performance to be discussed and compared using a single score that combines accuracy and recall. The F-Measure combines accuracy and recall.
4 Results and comparative analysis
A total of 109,446 sample records were collected for this research from MIT-BIH. Each of these records is categorized into 5 different CVD classes: Premature ventricular contraction, Fusion of ventricular and normal beat, Ventricular escape beat, Paced beat, and Normal. The performance parameters like accuracy, precision, recall, and f-Measure values compared for CVD classification the proposed model and models defined in [7, 42,43,44,45,46]. Table 4 indicates the accuracy performance of the proposed model w.r.t. number of ECG entries used for testing. The dataset was divided into 70:30, wherein ~ 77 k records were used to train the model, while the remaining ~ 33 k records were used to test the model and evaluate its performance. Thus, overfitting has been taken care of by dividing the set into training & testing samples, and then evaluating testing set performance parameters. The following Table 4 showcases the test accuracy (TA) performance of all the models.
From Fig. 3a, it is observed that, Average accuracy is around better when compared to existing models wherein these accuracy values are plotted, while its ROC performance can be observed from Fig. 3b, wherein its performance can be observed. Based on this performance, the confusion matrix is evaluated, and can be observed as follows,
Confusion matrix | |||||
[[18025 | 93 | 0 | 0 | 0 | 0] |
[0 | 556 | 0 | 0 | 0 | 0] |
[0 | 0 | 144 | 4 | 0 | 0] |
[0 | 0 | 0 | 162 | 0 | 0] |
[0 | 0 | 0 | 0 | 1605 | 3] |
[0 | 0 | 0 | 0 | 0 | 0]] |
From this matrix, it can be observed that almost all the entries are properly classified, while some entries from class 1, and class 4 are not categorized into the required ECG class.
Table 5 showcases the test precision (TP) performance of all the models. From the test precision values, it can be observed that the proposed model is highly efficient and as better performance when compared to decent classification models. Average precision is around 99.47% when compared to existing models.
The following Table 6 showcases the test recall (TR) performance of all the models. From the test recall, it can be observed that the proposed model is highly efficient, and as better performance when compared to decent classification models.
Average recall is around better when compared to decent models. The following Table 7 showcases all the models' test f-Measure (TF) performance. From the test f-Measure, it can be observed that the proposed model is highly efficient, and as better performance when compared to other decent classification models. Average f-Measure is around 99.60% when compared to existing models. It can be observed that the proposed algorithm is superior in terms of all the performance parameters when compared on different datasets with different existing model. The accuracy of this model is saturated, and thus can only be improved infinitesimally by using superior deep learning models for the same datasets.
The execution of our proposed model which gives an accuracy of 99.98%. This accuracy was validated via the following process,
-
1.
The model was initially evaluated with a training & testing ratio of 70:30, wherein standard deviation of samples was considered to divide the input datasets.
-
2.
This process was repeated for \(N=10\) iterations, with different sets of training & testing samples.
-
3.
Average accuracy from these iterations was used to obtain the final performance metrics.
These metrics assisted in evaluation of the final accuracy on given dataset. To further validate this process, the following Table 8 indicate training progress in every epoch.
Based on these runs (10 runs with different training & testing sets), standard deviation of error for the model was evaluated and can be observed from Table 9 as follows,
Upon observing the standard deviation, it can be concluded that the model has low error, and thus can be used for a wide variety of real-time clinical applications.
5 Conclusion and future scope
In this research, a novel model are use to classify five heartbeat classes, namely N, S, V, F, and Q, with MIT-BIH Heartbeat Database. The proposed model network model are learn and generalise fast set-up, collective, easy execution, and better accuracy. All performance parameter values are better when compared with the recently decent algorithms. The performance of this model is confirmed on MIT-BIH standard dataset, which makes the system applicable for real-time use cases. The proposed ensemble classification model of CNN with random forest, SVM, and kNN out perform as compare to other existing models for MIT BIH heartbeat Database. The use of wavelet transforms for feature extraction, reduces the feature-length by maximizing variance between feature sets of different classes and improving overall performance. The proposed model has accuracy of 99.98%, precision of 99.48%, recall of 99.73%, which generates a high f-Measure value of 99.6%. It is advised that the system must be tested for other ECG datasets, including but not limited to Physionet, PTB Datasets, Mendeley datasets etc. The evaluation of real-time energy consumption and optimization for clinical use might be future scope of proposed research work.
References
Shan J et al (2020) Accurate classification of ECG arrhythmia using MOWPT enhanced fast compression deep learning networks. J Ambient Intell Humaniz Comput. https://doi.org/10.1007/s12652-020-02110-y
Sharma K, Kohli P, Gulati M (2012) An update on exercise stress testing. Curr Probl Cardiol 37(5 no. Cdc):177–202. https://doi.org/10.1016/j.cpcardiol.2011.11.004
Zhang Y, Zhang Y, Lo B, Xu W (2020) Wearable ECG signal processing for automated cardiac arrhythmia classification using CFASE‐based feature selection. Expert Syst 37(1):e12432. https://doi.org/10.1111/exsy.12432
Ye C, Vijaya Kumar BVK, Coimbra MT (2021) Heartbeat classification using morphological and dynamic features of ECG signals. IEEE Trans Biomed Eng 59(10):2930–2941. https://doi.org/10.1109/TBME.2012.2213253
Sharma M, Tan RS, Acharya UR (2019) Automated heartbeat classification and detection of arrhythmia using optimal orthogonal wavelet filters. Inf Med Unlock 16(May):100221. https://doi.org/10.1016/j.imu.2019.100221
Khoshnoud S, Ebrahimnezhad H (2013) Classification of arrhythmias using linear coefficients and probabilistic neural network. Appl Med Inf 33(3): 55–62. https://ami.info.umfcluj.ro/index.php/AMI/article/view/434
Thomas M, Das MK, Ari S (2015) Automatic ECG arrhythmia classification using dual tree complex wavelet based features. AEU - Int J Electron Commun 69(4):715–721. https://doi.org/10.1016/j.aeue.2014.12.013
Pandey SK, Janghel RR (2020) Automatic arrhythmia recognition from electrocardiogram signals using different feature methods with long short-term memory network model. Signal Image Video Process 14(6):1255–1263. https://doi.org/10.1007/s11760-020-01666-8
Nurmaini S et al (2019) An automated ECG beat classification system using deep neural networks with an unsupervised feature extraction technique. Appl Sci. https://doi.org/10.3390/app9142921
Hong S et al (2019) Combining deep neural networks and engineered features for cardiac arrhythmia detection from ECG recordings. Physiol Meas 40(5):10–12. https://doi.org/10.1088/1361-6579/ab15a2
Yang J, Bai Y, Lin F, Liu M, Hou Z, Liu X (2018) A novel electrocardiogram arrhythmia classification method based on stacked sparse auto-encoders and softmax regression. Int J Mach Learn Cybern 9(10):1733–1740. https://doi.org/10.1007/s13042-017-0677-5
Yıldırım Ö, Pławiak P, Tan RS, Acharya UR (2018) Arrhythmia detection using deep convolutional neural network with long duration ECG signals. Comput Biol Med 102(August):411–420. https://doi.org/10.1016/j.compbiomed.2018.09.009
Oh SL, Ng EYK, Tan RS, Acharya UR (2018) Automated diagnosis of arrhythmia using combination of CNN and LSTM techniques with variable length heart beats. Comput Biol Med 102(April):278–287. https://doi.org/10.1016/j.compbiomed.2018.06.002
Hou B, Yang J, Wang P, Yan R (2020) LSTM-based auto-encoder model for ECG arrhythmias classification. IEEE Trans Instrum Meas 69(4):1232–1240. https://doi.org/10.1109/TIM.2019.2910342
Pandey SK, Janghel RR (2021) Automated detection of arrhythmia from electrocardiogram signal based on new convolutional encoded features with bidirectional long short-term memory network classifier. Phys Eng Sci Med 44(1):173–182. https://doi.org/10.1007/s13246-020-00965-1
Bouaziz F, Oulhadj H, Boutana D, Siarry P (2019) Automatic ECG arrhythmias classification scheme based on the conjoint use of the multi-layer perceptron neural network and a new improved metaheuristic approach. IET Signal Process 13(8):726–735. https://doi.org/10.1049/iet-spr.2018.5465
Chen C, Hua Z, Zhang R, Liu G, Wen W (2020) Automated arrhythmia classification based on a combination network of CNN and LSTM. Biomed Signal Process Control 57:101819. https://doi.org/10.1016/j.bspc.2019.101819
Weimann K, Conrad TOF (2021) Transfer learning for ECG classification. Sci Rep. https://doi.org/10.1038/s41598-021-84374-8
Muthulakshmi RTSI (2021) An optimal artificial neural network based big data application for heart disease diagnosis and classification model. J Ambient Intell Humaniz Comput 12(6):6129–6139. https://doi.org/10.1007/s12652-020-02181-x
Safa M, Pandian A (2021) Intelligent big data analytics model for efficient cardiac disease prediction with IoT devices in WSN using fuzzy rules. Wirel Pers Commun. https://doi.org/10.1007/s11277-021-08788-3
Shidnal S, Latte MV, Kapoor A (2021) Crop yield prediction: two-tiered machine learning model approach. Int J Inf Technol 13(5):1983–1991. https://doi.org/10.1007/s41870-019-00375-x
Niranjan D, Kavya M, Neethi KT, Prarthan KM, Manjuprasad B (2021) Machine learning based analysis of pulse rate using Panchamahabhutas and Ayurveda. Int J Inf Technol 13(4):1667–1670. https://doi.org/10.1007/s41870-021-00690-2
Nayakwadi N, Fatima R (2021) Automatic handover execution technique using machine learning algorithm for heterogeneous wireless networks. Int J Inf Technol 13(4):1431–1439. https://doi.org/10.1007/s41870-021-00627-9
Mangrulkar A, Rane SB, Sunnapwar V (2021) Automated skull damage detection from assembled skull model using computer vision and machine learning. Int J Inf Technol 13(5):1785–1790. https://doi.org/10.1007/s41870-021-00752-5
Tyagi A, Mehra R (2021) Intellectual heartbeats classification model for diagnosis of heart disease from ECG signal using hybrid convolutional neural network with GOA. SN Appl Sci 3(2):1–14. https://doi.org/10.1007/s42452-021-04185-4
Mahajan J, Banal K, Mahajan S (2021) Estimation of crop production using machine learning techniques: a case study of J&K. Int J Inf Technol 13(4):1441–1448. https://doi.org/10.1007/s41870-021-00653-7
Bojamma AM, Shastry C (2021) A study on the machine learning techniques for automated plant species identification: current trends and challenges. Int J Inf Technol 13(3):989–995. https://doi.org/10.1007/s41870-019-00379-7
Divate MS (2021) Sentiment analysis of Marathi news using LSTM. Int J Inf Technol 13(5):2069–2074. https://doi.org/10.1007/s41870-021-00702-1
Kulkarni TR, Dushyanth ND (2021) Performance evaluation of deep learning models in detection of different types of arrhythmia using photo plethysmography signals. Int J Inf Technol 13(6):2209–2214. https://doi.org/10.1007/s41870-021-00795-8
Pandey NN, Muppalaneni NB (2021) A novel algorithmic approach of open eye analysis for drowsiness detection. Int J Inf Technol 13(6):2199–2208. https://doi.org/10.1007/s41870-021-00811-x
Pattekari A, Parveen SA (2012) Prediction system for heart disease using Naïve Bayes. Int J Adv Comput Math Sci 3(3):290–294
Dulhare UN (2018) Prediction system for heart disease using Naive Bayes and particle swarm optimization. Biomed Res 29(12):2646–2649. https://doi.org/10.4066/biomedicalresearch.29-18-620
Bhavekar GS, Das Goswami A (2022) A hybrid model for heart disease prediction using recurrent neural network and long short term memory. Int J Inf Technol. https://doi.org/10.1007/s41870-022-00896-y
Mane DT, Tapdiya R, Shinde SV (2021) Handwritten Marathi numeral recognition using stacked ensemble neural network. Int J Inf Technol 13(5):1993–1999. https://doi.org/10.1007/s41870-021-00723-w
Kumar R, Srivastava S, Dass A, Srivastava S (2019) A novel approach to predict stock market price using radial basis function network. Int J Inf Technol 13(6):2277–2285. https://doi.org/10.1007/s41870-019-00382-y
Patil AR, Subbaraman S (2021) Performance analysis of static hand gesture recognition approaches using artificial neural network, support vector machine and two stream based transfer learning approach. Int J Inf Technol. https://doi.org/10.1007/s41870-021-00831-7
Chandra MA, Bedi SS (2021) Survey on SVM and their application in image classification. Int J Inf Technol 13(5):1867–1877. https://doi.org/10.1007/s41870-017-0080-1
Sharma LD, Sunkaria RK (2019) Detection and delineation of the enigmatic U-wave in an electrocardiogram. Int J Inf Technol 13(6):2525–2532. https://doi.org/10.1007/s41870-019-00287-w
Usha Kirana SP, D’Mello DA (2021) Energy-efficient enhanced Particle Swarm Optimization for virtual machine consolidation in cloud environment. Int J Inf Technol 13(6):2153–2161. https://doi.org/10.1007/s41870-021-00745-4
Sharma LD, Chhabra H, Chauhan U, Saraswat RK, Sunkaria RK (2021) Mental arithmetic task load recognition using EEG signal and Bayesian optimized K-nearest neighbor. Int J Inf Technol 13(6):2363–2369. https://doi.org/10.1007/s41870-021-00807-7
Moody GB, Mark RG (2001) The impact of the MIT-BIH arrhythmia database. IEEE Eng Med Biol Mag 20(3):45–50. https://doi.org/10.1109/51.932724
Celin S, Vasanth K (2018) ECG signal classification using various machine learning techniques. J Med Syst 42(12):1–11. https://doi.org/10.1007/s10916-018-1083-6
Marinho LB, de Nascimento NMM, Souza JWM, Gurgel MV, Rebouças Filho PP, de Albuquerque VHC (2019) A novel electrocardiogram feature extraction approach for cardiac arrhythmia classification. Future Gener Comput Syst 97:564–577. https://doi.org/10.1016/j.future.2019.03.025
Kiranyaz S, Ince T, Gabbouj M (2015) Real-time patient-specific ECG classification by 1-D convolutional neural networks. IEEE Trans Biomed Eng 63(3):664-675. https://doi.org/10.1109/TBME.2015.2468589
Fradi M, Khriji L, Machhout M, Hossen A (2021) Automatic heart disease class detection using convolutional neural network architecture-based various optimizers-networks. IET Smart Cities 3(1):3–15. https://doi.org/10.1049/smc2.12003
Abdalla FYO et al (2020) Deep convolutional neural network application to classify the ECG arrhythmia. Signal Image Video Process. https://doi.org/10.1007/s11760-020-01688-2
Kaouter K, Mohamed T, Sofiene D, Abbas D, Fouad M (2019) Full training convolutional neural network for ECG signals classification. In AIP conference proceedings, vol. 2190, no. 1. AIP Publishing LLC, p 020055. https://doi.org/10.1063/1.5138541
Wang T, Lu C, Sun Y, Yang M, Liu C, Ou C (2021) Automatic ECG classification using continuous wavelet transform and convolutional neural network. Entropy 23(1):1–13. https://doi.org/10.3390/e23010119
Acknowledgements
This research received no governmental, commercial, or non-profit grants. The authors thank the editor and anonymous reviewers for improving this work.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflicting interests.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Goswami, A.D., Bhavekar, G.S. & Chafle, P.V. Electrocardiogram signal classification using VGGNet: a neural network based classification model. Int. j. inf. tecnol. 15, 119–128 (2023). https://doi.org/10.1007/s41870-022-01071-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s41870-022-01071-z