Abstract
With the advent of integrated computational materials engineering, there is a drive to exchange statistical confidence in a design obtained from repeated experimentation with one developed through modeling and simulation. Since these models are often missing physics or include incomplete knowledge or simplifying assumptions into their mathematical construct, they may not capture the physical system or process adequately over the entire domain. This can lead to a systematic discrepancy, otherwise known as model misfit, between the model output and the system it represents over at least part of the domain. However, by accounting for this discrepancy in uncertainty analyses, reliable inference, and prediction on the model parameters and material behavior can be made despite the missing physics. The previous statement is contingent on two conditions: (1) the discrepancy is systematic, and (2) the structure of the discrepancy is well understood, which is required to minimize issues with non-identifiability among unknown model components. We illustrate these insights via a case study of inference and prediction in a phenomenological meso-scale VPSC crystal plasticity model, which does not contain physics describing the elastic regime of deformation. Inference is performed via a Bayesian approach enabled by posterior simulation. Posterior uncertainty about unknown model parameters takes into account observation error, uncertainty stemming from aleatoric sample-to-sample variability, and model form error. Posterior uncertainty in the unknown Voce hardening parameters is propagated to generate a distribution of the stress–strain response in both the elastic and plastic regimes. Additionally, posterior predictive distributions are simulated to establish uncertainty bounds for future unobserved data.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
A critical component to advancing the goals of integrated computational materials engineering (ICME) is the quantification, propagation, and mitigation of the sources of uncertainty influencing model simulations and their predictive power, formally referred to as uncertainty quantification in the materials computational community. This is of particular interest and importance with efforts to develop model linkages where the input to large scale component models is generated from the output of models at smaller scales. The underlying goal is to account for the multitude of mechanisms and phenomena occurring at scales ranging from chemical bonds at the quantum and atomistic levels and grain interactions at the mesoscale contributing to the macroscopic properties and performance of a design component. While the benefits of such an approach are clear (improved model fidelity and design reliability), the effectiveness is contingent on the adoption and routine use of rigorous and reliable techniques for uncertainty quantification (UQ), propagation (UP), and mitigation (UM). Unaccounted error or uncertainty at the lower level models will lead to macroscopic model outputs with compounded uncertainty and predictions with an unknown level of reliability.
Figure 1 summarizes the various types of uncertainty which are grouped into two primary categories: epistemic and aleatory. Both sources play a role in the model output and quality of parameter inference and, consequently, the propagation of uncertainty for predictions (or in the case of ICME, model linkages). Aleatory uncertainty is a description of the randomness in the system and could be due to natural physical randomness, which may manifest in the sample-to-sample variability among experimental tests, or due to the use of stochastic numerical methods. This form of uncertainty, while quantifiable, is not reducible. On the other hand, epistemic uncertainty is a result of omitted or missing knowledge or physics in the constitutive models, or from the use a variety of numerical techniques to simplify the computations. Since a model can always be improved by learning more about the system and incorporating the understanding into the model, this type of uncertainty is reducible.
Mature techniques for UQ, UP, and UM are being adopted with increased frequency within the materials modeling community [1,2,3,4,5,6,7,8,9,10,11,12,13,14,15]. A recent paper by Ricciardi et al. [16] proposed using a random effects Bayesian hierarchical statistical model for quantifying parameter uncertainty in material models. The authors argue that this statistical model reflects the inherent structure of an important class of problems in materials modeling and simulation since the aleatory uncertainty caused by microstructure heterogeneity in the material samples tested is naturally accounted for. A Bayesian inferential approach was used to recover information about unknown model parameters. Posterior functionals were estimated from a posterior sample generated via a Metropolis–Hastings Markov Chain Monte Carlo (MH-MCMC) simulation. Posterior uncertainty was then propagated to induce a posterior distribution in the model output and predict the distribution of possible future outcomes (prediction). This approach was demonstrated for two fundamentally different modeling problems: (1) crystal plasticity modeling at the mesoscale and (2) thermodynamic calculations of phase equilibrium.
The work presented here addresses estimation and uncertainty quantification of the random effects model in the presence of model misfit, also known as model discrepancy. Model misfit is a consequence of missing physics, modeling simplifications, or numerical methods that may lead to systematic discrepancy between the model output and observations. Improperly accounting for model misfit, or simply ignoring misfit when performing uncertainty quantification can lead to bias in calibration and under-estimation of associated uncertainty, which propagate as error to property or response predictions.
One crucial application is when model parameters have a clear connection to intrinsic material properties (such as diffusivities or mobilities, defect energies, elastic constants, etc.). In an ideal world, the model is a perfect representation of reality and, when properly calibrated, will exactly predict the material response under varying initial and boundary conditions. However, in practice, models are often based on some assumptions or approximations. As a thought experiment, the measured experimental material response can be decomposed into three components: (1) the portion that is well-represented by the model, (2) the portion of “true” material response not captured by the model due to discrepancy, and (3) noise or measurement error. Mathematically the discrepancy should be independent of the portion represented by the model. When the model has enough degrees of freedom, i.e., fitting parameters, it is often possible to find a set of parameters that allow the model to closely mimic the experimental response. When fitting is performed without accounting for the discrepancy, the values of the parameters are biased by this misfit (meaning they attain a value that is systematically different than that obtained from an ideal model). Using such biased parameter estimates for response prediction under different conditions than are used for calibration will necessarily lead to a persistent error in predictions and inaccurate error bounds, leading to overly optimistic design tolerances or safety factors.
Quantifying this source of uncertainty is rendered challenging by the requirement of balancing a flexible discrepancy model with the requirement that the complete model be identifiable given the data [17]. Discussion on model discrepancy and techniques to model this form of error are provided in "Methods" section and applied to a case study in "Case Study" section.
Methods
We begin with a brief discussion on the Bayesian paradigm, statistical model, assumptions and simulation methods applied in this work. The interested reader is referred to [16] for a full discourse on the theory and techniques extended in this work. For consistency, the same notation will be adopted. Bayesian inference is used in this study to recover unknown components of the data generation process, while taking into account various sources of uncertainty. The analysis yields a posterior probability distribution describing our understanding about the unknown model components or model parameters, \(\theta \), given the available data, y. A statistical model for the data (not to be confused with material or constitutive model of material response) is a collection of probability distributions \({\mathscr {M}} = \{f(y \mid \theta ); \; \theta \in {\varTheta } \}\), where \({\varTheta }\) is the parameter space of which \(\theta \) is an element. The first ingredient in a Bayesian analysis is a prior probability model with density \(\pi (\theta )\) over the unknown components, which is simply an epistemic model of our subjective uncertainty before any data is observed. The prior distribution is then updated by conditioning on observed data to establish the posterior distribution through Bayes’ rule,
The posterior distribution is the updated belief about \(\theta \) after data has been observed and is proportional to the product of the prior density over \(\theta \) and the likelihood, \(f\left( y \mid \theta \right) \), of observing data y given \(\theta \). This construct can be extended to make predictions about future observations, \(\tilde{y}\), by establishing the posterior predictive distribution given observed data y. Its density is a continuous mixture of the likelihood of the unobserved data \({\tilde{y}}\) weighted by the posterior of the unknown model parameters \(\theta \),
The posterior and posterior predictive distributions are all that is required for inference and prediction, respectively. Although posterior densities are often not analytically tractable, they can be estimated from a Monte Carlo sample. Markov chain Monte Carlo (MCMC) [18,19,20,21] is a class of simulation techniques for constructing a Markov chain with stationary distribution equal to the posterior distribution of interest. Monte Carlo estimates of desired posterior functionals, such as the mean, mode, and highest posterior intervals (HPIs), can then be computed from the resulting sample. Our MCMC sampling approach consists of an adaptive Metropolis–Hastings algorithm [22, 23] with conditional Gibbs updates [24] where full conditional distributions are available.
Modeling Assumptions
We consider a random effects model, which formally accounts for the variability in the response associated with the effect of the material sample tested. This statistical model is an excellent candidate for cases where the observed data do not directly represent the true underlying property or state, \(\theta \), also be called the overall effective property. Instead, the state of each material sample varies around the overall state. The sample states are called random effects and are denoted by \(\theta ^{[s]}, s = 1,\ldots ,S\), where S is the number of samples tested. Though they are not of interest in and of themselves (they are often referred to as nuisance parameters), they must be estimated in order to infer the overall effect \(\theta \).
The random effects, \(\theta ^{[s]}, s = 1,\ldots ,S\), and overall effect, \(\theta \), are related through the following probability model,
which also depends on additional parameters \({\varLambda }\) to define the distribution. Though the choice of this distribution is application-specific, a Multivariate Normal (or Gaussian) distribution is often a flexible choice to model the distribution of random effects around the overall effect since it implies that (1) The random effects are symmetrically distributed about the overall effect and (2) The random effects are more likely to lie in a region that is close to the overall effect \(\theta \). Thus, for D-dimensional parameters \(\theta ,\theta ^{[s]}, s = 1,\ldots ,S\), we assume,
where \({\varLambda }\) is a \(D\times D\) symmetric positive-definite inverse variance–covariance (precision) matrix, which controls how tightly dispersed the random effects are about the overall effect \(\theta \).
A commonly used prior distribution for the precision \({\varLambda }\) is the Wishart, a distribution on symmetric positive-definite matrices. Since this choice does not allow precise control over the prior model through the hyperparameters, we take a more flexible modeling approach. The random effects precision \({\varLambda }\) is decomposed into two components: a \(D \times D\) positive-definite and symmetric correlation matrix, R, and a \((1 \times D)\) vector of standard deviations, \(t = (t_{1},\ldots ,t_{D})^\top \), which are then modeled separately following [25, 26]. The decomposition,
allows extra flexibility in modeling known features of the distribution while allowing enough dispersion. We assign a Wishart prior to the correlation matrix, R, which preserves conjugacy and allows R to be sampled via Gibbs updates,
where \(R_{o}\) is a \(D \times D\) symmetric positive-definite scale matrix and \(r_{o}\) represents the distribution’s degrees of freedom. For computational convenience, the variances, \(t^{2}_{d}, d=1,\ldots ,D\) rather than the standard deviations are modeled as,
where \(a_t\) and \(b_t\) are fixed hyperparameters.
For each of the S samples tested, the likelihood of the N-dimensional data \(y^{[s]}\) given the corresponding random effect \(\theta ^{[s]}\) is modeled as a Multivariate Normal distribution,
centered at the physical model output \(m\left( \theta ^{[s]}\right) \) with dispersion defined through the \(N \times N\) error precision matrix \({\varPsi }\).
Model Misfit
So far, a key assumption has been that the physical model m adequately describes the mean of the data-generating process. In reality, this is either not known with certainty, or is known and the structure of this model discrepancy is understood to some degree. To capture both these scenarios, we denote by \(\zeta (x)\) the ‘ground truth’, or true state of the system, at location x. Furthermore, we make the assumption that this signal has been contaminated with additive Gaussian error \(e_n, n=1,\ldots , N\) at the observation locations \(x_1,\ldots , x_N\). This typically represents observation error or some other type of variability. The resulting data-generating model may be written as,
One possible assumption is that the physical model of the system, m, which accepts a vector (or scalar) of parameters \(\theta \) and is evaluated at location x, is representative of the true process such that \(m(x,\theta ) = \zeta (x)\). This assumption was made in [16]. The present work diverges here and will adopt a different, more realistic, data-generating model which accounts for the effect of model discrepancy. If we take into account the discrepancy between the model representation of the system, \(m(x,\theta )\), and the true mean process, \(\zeta (x)\), and denote it by \({\varDelta }(x)\), we can relate these terms as:
The model for the observations now becomes,
where \(m\left( \cdot \right) \) and \({\varDelta }\left( \cdot \right) \) are assumed to have no parameters in common and where \(\theta \) is the true but unknown vector of calibration parameters.
Among the first to formally incorporate model discrepancy into an inferential framework is the work of Kennedy and O’Hagan [27]. Model discrepancy was adopted in this framework to mitigate bias and over-confidence in inference based on reduced-order representations of expensive simulators. [17] illustrate similar issues arising from a failure to take into account systematic model discrepancy in general inference problems. Importantly, the role of the discrepancy model and its link to parameter identifiability is explored.
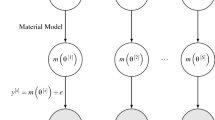
With all components of the statistical model now introduced, a schematic of the statistical model can be found in Fig. 2, in which the direction of the arrows represents the direction of conditional dependence between model components. Shaded nodes indicate that model components are observed, with everything else fixed and unknown. Under the natural assumption of conditional independence between samples \(y^{[s]} = (y^{[s]}_1, \ldots , y^{[s]}_N)^\top \) given the random effects \(\theta ^{[s]} \in {\mathbb {R}}^D\), the density of the posterior distribution is,
where \(f\left( y^{[s]} \mid \theta ^{[s]}, {\varPsi }, {\varDelta } \right) \) is the density of,
A flexible prior model for the discrepancy term \({\varDelta }\) is discussed in detail in "Gaussian Process Prior for Model Discrepancy" section. Propagating this posterior uncertainty through to prediction of the data leads to the posterior predictive density,
Gaussian Process Prior for Model Discrepancy
As an unknown model component, the discrepancy function \({\varDelta }\) must be assigned a prior model within the Bayesian hierarchy. The importance of carefully incorporating all prior knowledge about \({\varDelta }\) into this probability model was demonstrated by Brynjarsdóttir and O’Hagan [17], where bias and over-confidence in the estimates resulted from using priors that were too flexible. The problem, called lack of identifiability in the statistical literature, results from the ability of different model components (in this case the discrepancy function and model parameters) to trade off in ways that can describe the data equally well. When one of the model components is empirical (in this case the discrepancy), this lack of identifiability hinders our ability to obtain useful estimates of the physically meaningful model components. One way to overcome lack of identifiability is by defining sufficiently informative prior models, subject of course to the availability of such information. Informally, an informative prior choice serves to penalize different configurations or trade-offs and guide the posterior towards regions that we know a-priori to be more probable or physically reasonable.
A Gaussian process (GP) is a stochastic process, which is a collection of random variables such that any finite sub-collection of those variables have a jointly Gaussian distribution. For an accessible introduction to these models, the reader is referred to [28]. GPs can be used to define prior distributions over functions, such as our discrepancy function \({\varDelta }: {{\mathscr {D}}} \rightarrow {\mathbb {R}}\). They can be thought of as a functional generalization of the multivariate Gaussian distribution, which is characterized by a mean vector and positive definite covariance matrix. Likewise, a GP is fully characterized by a mean function, \(\mu : {{\mathscr {D}}} \rightarrow {\mathbb {R}}\) and a positive definite covariance function, \(c: {{\mathscr {D}}}\times {{\mathscr {D}}} \rightarrow {\mathbb {R}}\). Therefore, we can specify a GP prior model for the discrepancy function \({\varDelta }\) as,
It is often realistic to model the discrepancy as having prior mean function \(\mu (x)=0\), as this incorporates the neutral and often reasonable assumption that the magnitude of the systematic deviation from the physical model across the domain is either close to zero or not known a-priori. The choice of the covariance function is an important modeling consideration, as it controls the degree to which correlation decays with distance between inputs over the input domain. As such, the covariance controls the smoothness of realizations (called sample paths) of a GP. More information on this choice is described in "Choice of Covariance Structure" section.
A convenient feature of GPs is the joint Gaussianity of any finite sub-collection of points. While the function \({\varDelta }\) is in theory continuous, it can be probed at a finite set of variable inputs since
where \(\mathbf{0}\) is the N-dimensional zero vector and,
and the covariance between the process at any two points \(x_i\) and \(x_j\) is related to the covariance function through \(c(x_i,x_j) = \text {Cov}({\varDelta }(x_i), {\varDelta }(x_j))\), for \(x_i, x_j \in \{x_1,\ldots , x_N\}\).
Choice of Covariance Structure
Since the covariance structure controls the correlation scale and smoothness of the stochastic process, careful choice of this function is required to specify a sufficiently informative prior process for the model discrepancy \({\varDelta }\). The smoothness of GP sample paths depends on the way in which the covariance between \({\varDelta }\left( x_i\right) \) and \({\varDelta }\left( x_j\right) \) changes across the input space. One example of a smooth covariance is the squared exponential function,
where \(\sigma ^2\) is a variance parameter and w is the length-scale hyperparameter. A GP with this covariance structure has infinitely differentiable sample paths. Other popular covariance functions include the Matérn [29], whose smoothness can be controlled by the choice of covariance hyperparameter, and the exponential [30], which yields rougher sample paths.
When the GP is used as a prior model, the variance hyperparameter describes prior uncertainty about the unknown function by controlling the pointwise variability of resulting sample paths. A large variance hyperparameter corresponds to a diffuse GP prior distribution, while a smaller variance produces a tighter distribution appropriate when more prior information is available. The length-scale parameter controls how dependence scales with the distance between nearby points. As with all prior hyperparameters, these must either be set by the user or be treated as unknown and estimated from the data within a hierarchical model. Figure 4 illustrates the GP sample paths under a selection of different covariance hyperparameter choices.
A stationary covariance function is one that depends only on the distance between the inputs and is independent of their relative location. Non-stationary covariance models, on the other hand, allow for changes to the dependence structure across the input domain. Figure 3 illustrates the difference between the behaviour of sample paths from a GP with a stationary (left) and a non-stationary (right) covariance. The samples in Fig. 3b initially have stronger dependence, which decreases moving away from 0.
Toy Example
As a simple example, suppose we have measured the mass of an ice block (100 kg), apply an unknown force to it, and then collect observations of the position of the block at different time increments. The ice block accelerates from an initial velocity and position of zero, and we wish to infer from the observations the force which was applied using the fundamental equation,
where the unknown force applied is denoted by \(\theta \). However, suppose that in reality the ice block is melting and the mass is changing with time, and the position of the block under a constant applied force follows the relationship,
where mass indicates the initial mass of the block and \(\lambda \) is a factor controlling the rate of the melt. In this example, there is knowledge missing from the model of the physical system (19), the changing mass of the ice block, which introduces discrepancy between the true force being applied and the model. A comparison of the true behavior, our simplified model of the behavior, and the observations is provided in Fig. 5.Footnote 1 The magnitude of the discrepancy between the imperfect model of the system and its true behavior begins at zero and grows over time. Analyses conducted with and without considering model discrepancy reveal the importance of including this component in the statistical model.
Figure 6 shows marginal histograms from two different analyses performed to estimate the unknown model parameter (\(\theta \), which is the applied force) given the observed data without and with taking into account model discrepancy. A comparison of the two illustrates the need to consider discrepancy in the statistical model. Figure 6a illustrates that how posterior inference under an imperfect model suffers from both bias (a systematic difference between the estimator and the true value) and under-coverage (over-confidence in the results as measured by the amount of spread in the posterior distribution over \(\theta \)). In comparison, Fig. 6b shows results from an analysis in which discrepancy was incorporated through an additive function (as in Eq. 11).
In this case, inference about \(\theta \), as conveyed by the location and spread of the posterior density, is much closer to the ground truth, which is now covered by the central 95% credible interval. It is interesting to note that this bias-variance trade-off [31] is a commonly observed phenomenon, whereby a predictive model with lower bias has a higher variance and vice versa. This dilemma is brought on by the conflict of trying to simultaneously reduce both sources of error (bias and variance) and finding a balance between over-fitting and under-fitting the training data.
With a clear understanding on the importance of considering model misfit in the inference problem, we now proceed to discuss the materials application to which we will be applying these techniques.
Case Study
Reduced-order, phenomenological and homogenized models are well-suited for Bayesian inferential analyses using MCMC simulations due to their low computational cost. In exchange for their simplified formulations and reduced computational cost (when compared to 3D, physics-based and full-field models), these models may leave out certain physics affecting the material behavior or process of interest or may make other simplifying assumptions. It may also be the case that a model of higher fidelity may not be available for a particular application. The bottom line is that while these models may be very reliable over a subset of the domain in their representation of the process or behavior, they may not be reliable over the entire domain (e.g., a model which captures plastic deformation well but does not consider elastic deformation). Consequently, this may introduce a systematic discrepancy over a redistricted domain, or a systematic discrepancy from the true process over the entire domain. However, this discrepancy need not prevent the use of reduced-order models as reliable tools in the prediction of material behavior. With the incorporation of model-form error into the statistical inference problem, their parameters can be reliably recovered without the need the restrict the domain of the model.
Take, for example, the phenomenological visco-plastic self-consistent (VPSC) crystal plasticity model developed by Tomé and Lebensohn at Los Alamos National Laboratory [32]. This model is commonly used to understand deformation and texture evolution at large plastic strains, however it does not include physics to consider the elastic regime of deformation. Because of this, a systematic discrepancy is present between the model and experimental observations of stress-strain response as they are collected. Reliable calibration of this model without restricting the domain requires the consideration of model-form error.
The VPSC crystal plasticity model uses the Voce hardening law [33, 34] to describe the evolution of the critical resolved shear stress at the level of the grain. This phenomenological relationship evolves the critical resolved shear stress (CRSS) as a function of the accumulated shear across all slip systems, \(\alpha \), in the grain,
Here, the incremental shear is denoted by \(\mathrm{d}{\varGamma }\), and unknown parameters in the model are the initial critical resolved shear stress (CRSS), \(\tau _{0}\), the asymptotic CRSS increase \(\tau _{1}\), the initial hardening rate \(\xi _{0}\) and the asymptotic hardening rate \(\xi _{1}\).Footnote 2 Accurate description of hardening in the model is dependent on reliable parameter estimation within the law, which are not based on physics but do have physical meaning and whose values can be extrapolated outside the bounds of calibration to consider various cases of deformation. The VPSC model produces the final texture evolution and the stress–strain response of a material undergoing deformation by homogenizing the local response to obtain the macroscopic response of the material.
One of the two case studies presented in Ricciardi et al. [16] considers inference on the unknown Voce hardening parameters, and propagates this uncertainty to induce a predictive distribution in the stress–strain response enabling prediction for experimental data not yet observed. Observational error as well as sample-to-sample variability were considered in performing inference on the parameters while conditioning on observations from only the plastic regime of deformation. Conditioning on this restricted domain allowed the reasonable assumption that the VSPC model was correct despite its drawbacks in the elastic regime of deformation.
However, in general, we wish to employ the full experimental data collected without restricting the observation domain (i.e., with elastic and plastic data). As we have seen, this risks introducing bias and under-coverage in any parameter estimates when model misfit is not explicitly considered. The missing physics in the VPSC model produces a systematic discrepancy between the model and observations. Therefore, it can no longer be calibrated under the assumption that it forms the mean of the observation process across both elastic and plastic regions. Including model misfit in the analysis will account for this discrepancy, reducing bias in the estimated parameters and propagated model output.
To demonstrate the necessity of including the model discrepancy in the inference problem, an analysis was performed by conditioning the VPSC model on both the elastic and plastic regimes of deformation without consideration of model misfit. As pointed out by Brynjardóttir [17], the posterior distribution of \(\theta \) centers around a ‘posterior best fit’ value which minimizes the mean residual error between the model output and the observations. Thus, ignoring model misfit introduces bias in the parameter estimates and consequently the model trajectory, as illustrated in Fig. 7. The observations of the state are in red and in black are 200 samples drawn from the posterior distribution and propagated through the model to induce a distribution on the true stress–strain behavior given the data. The evident bias is particularly problematic in cases such as this where model parameters are not simply tuning parameters but whose values are of intrinsic interest and necessary for reliable prediction, in both cases of interpolation and extrapolation.
Statistical Model
We assume the true underlying behavior of the system is represented by overall effect \(\theta = (\theta _{1},\ldots ,\theta _{D})^\top \), which is comprised of a D-dimensional vector of unknown model parameters. Yet, due to the random nature of microstructural heterogeneity, observations from each sample are generated under slightly different states which are therefore modeled as random effects. Model parameters corresponding to sample \(s \in \{1,\ldots , S\}\) (where S is the total number of samples) are denoted by \(\theta ^{[s]} = \left( \theta ^{[s]}_{1},\ldots ,\theta ^{[s]}_{D}\right) ^\top \). A D-dimensional Multivariate Normal distribution describes the variation of the random effects around the overall effect,
The decomposition of the precision matrix \({\varLambda } = \text{ diag }(t)\, R \; \text{ diag }(t)\), allows us to define a flexible prior model separately on the correlation matrix, R, and the standard deviations, \(t = (t_{1},\ldots , t_{D})^\top \). Physical constraints on the model parameters are introduced through the indicator function \({\mathbb {I}}\{{\mathscr {C}}(\theta )>0\}\), where positive \({\mathscr {C}}(\theta ) = (\tau _{0},\tau _{1},\theta _{0}-\theta _{1})^\top \) ensures that there is no softening in the material, and the asymptotic hardening rate reaches a limiting value.
The model for the observations follows Eq. (11) and we assume i.i.d. Gaussian error at each variable input represented by \(\delta ^{2}\) such that, \({\varPsi } = \delta ^{2} {\mathbb {I}}_{N}\). In other words, we assume the observational error contribution is the same across all variable inputs. The model for the observations becomes,
where \({\varDelta }\) is an unknown discrepancy function. A Multivariate Normal distribution is assumed for the likelihood of the observations centered at the sum of the model plus the discrepancy with covariance matrix \({\varPsi }^{-1}\).
Diffuse priors are assigned to the overall effective parameters in \(\theta \), reflecting our prior uncertainty about the spread of the sample parameters from their center. The priors on \(\tau _{0}\) and \(\tau _{1}\) were chosen to be Gamma distributed with hyperparameters specifying a mean of 70 for both and a large variance,
Priors on \(\xi _{0}\) and \(\xi _{1}\) were modeled via diffuse Normal distributions,
Prior means were chosen based on previous studies. The error precision, \(\delta ^{2}\) is modeled with a Gamma distribution in order to ensure that only positive real values are possible. A Wishart prior is chosen for the correlation matrix, R, since it is a symmetric positive-definite matrix and the variances, \(t^{2}_{d}, \; d=1,\ldots ,D\) are assigned Gamma distributions. The prior on the unknown discrepancy function, \({\varDelta }\), is a zero-mean Gaussian process with a custom covariance structure, which will be discussed below and shown in Fig. 8. Since the likelihood model (24) only requires \({\varDelta }\) to be evaluated at N discrete points, this prior becomes a Multivariate Normal distribution centered at zero with covariance matrix \({\varGamma }\), obtained by evaluating the GP covariance function at the vector of observation locations \((x_1,\ldots , x_N)^\top \). Summaries of the prior distributions are provided in Table 1.
Since the VPSC model uses a constitutive equation that only accounts for plastic deformation and does not consider the elastic regime of deformation, a large discrepancy is expected in the elastic region, decreasing as we get closer to the elastic–plastic transition. Furthermore the model is expected to have little to no systematic misfit in the plastic region. Because of the contrast in the expected structure of the discrepancy for these two regimes, we select a piece-wise formulation for the prior covariance. The strain at the elastic–plastic transition also marks the transition in the piece-wise covariance function,
We denote the index of the strain at the elastic–plastic transition by t. The strain at the transition is traditionally taken to correspond to the yield stress at .2% offset. While this value will differ between samples, we take this value to be fixed at .0025. In Eq. (27), \({\mathbf {N}}_{el} = \{1,\ldots ,t\}\) is the set of indices within the elastic regime, and \({\mathbf {N}}_{pl} = \{t+1,\ldots ,N\}\) is the set of indices of plastic strain.
For \(\text{ Cov }_{el}\) we choose a non-stationary smooth covariance function since the greatest discrepancy is expected to occur at \(x = 0\) and to taper to zero at \(x_{t}\). While the length-scale and variance hyperparameters of the covariance function can be inferred within the hierarchical model, here we assign values \(w_{el} = 1.4\times 10^{-4}\) and \(\sigma ^{2}_{el} = 1.0 \times 10^{14}\).
A stationary covariance structure is chosen for the plastic region with a length scale of \(w_{pl} = .02\) and a variance of \(\sigma ^{2}_{pl} = 1.0 \times 10^{-4}\). A small prior variance is chosen since we expect very little misfit in the plastic regime. Essentially, the prior penalizes deviations of the model from the observations in this region. Figure 8 shows multiple draws from the prior over \({\varDelta }\) for both the elastic and plastic regions as well as over the full domain.
Random draws over the full domain in a with an inset showing the elastic–plastic transition. Note, the relative prior variances between the elastic and plastic regions are so different that some parts appear to be flat when plotted together. Random draws from the prior of the discrepancy in the elastic region is shown in b and the plastic region in c. Note the difference in the scale of the vertical axes
Results
The MCMC algorithm detailed in [16] was used for this work with appropriate adjustments (i.e., the decomposition of the random effects precision \({\varLambda }\) as well as the inclusion of the model discrepancy \({\varDelta }\)) and can be found in “Appendix 1”.Footnote 3 A total of \(1.25 \times 10^{5}\) Markov chain Monte Carlo samples were simulated targeting the posterior (12) and posterior predictive (14) distributions with the first \(2.5 \times 10^{3}\) samples being discarded as burn-in. Trace plots as well as correlation plots were used to monitor convergence.
Marginal posterior histograms as well as bivariate kernel density estimates of marginal posterior contours are shown in Fig. 9 for a representative random effect and the overall effect. In Fig. 9b the red lines show the marginal as well as bivariate marginal priors placed on the overall effect parameters for comparison. Notably, there is an almost perfect positive posterior correlation between random effect parameters \(\tau _{1}\) and \(\xi _{1}\), also provided in Table 3 in “Appendix 3”. Likewise, there is moderate posterior correlation between the overall parameters. A bivariate kernel density estimate of the marginal posterior over \((\tau _{1},\xi _{1})\) is shown in Fig. 10a, with the overall effect shown as a dashed line. Figure 10b shows a bivariate kernel density estimate of the marginal posterior over \((\tau _{0},\xi _{0})\), with moderate correlation in both the random effects as well as the overall effect. Here, the reader is reminded that correlations between parameters are features of the posterior distribution and not directly of the physical system. Parameter combinations on a given contour of the posterior distribution simply have the same posterior probability given the observed data; a more thorough discussion can be found in [16].
Posterior expectations for the model parameters are provided in Table 2, along with posterior uncertainty summaries, including posterior variances and 95% highest posterior density intervals (HPIs).
Parameter uncertainty was propagated by sampling from the posterior distribution over the model parameters to generate posterior and posterior predictive distributions over the underlying stress–strain response and any unobserved data, respectively. 200 representative samples are shown in Fig. 11b, c, respectively. In both plots, 95% HPIs of the stress–strain response are represented with red dashed lines and the mean stress–strain response is shown with a red line. Here, we take a moment to remind the reader of the subtle difference in the interpretation of these two distributions. While the posterior distribution conveys our subjective belief about the true underlying property, the posterior predictive distribution is our belief about how future experiments will behave, thus resulting in a property distribution which is more diffuse. Figure 11a shows the pointwise posterior mean as well as its decomposition into the VPSC model evaluated at the expected value of the parameters and the mean discrepancy. Also included is an inset of the elastic region.
The pointwise posterior mean (blue), the VPSC model evaluated at the posterior mean parameter values (red) and the mean discrepancy (green) are shown in a with an inset showing the elastic region. 200 draws from the a posterior and b posterior predictive distributions are shown with the mean and 95% highest posterior density intervals shown
An informative comparison can be made between the analyses operated under the assumption that discrepancy is present (Fig. 11b), and the assumption that it is not (Fig. 7). The strong bias evident in the ‘no discrepancy’ analysis is not present when model misfit is accounted for.
Discussion
When to Include Discrepancy While the incorporation of model discrepancy was appropriate and necessary for the study presented here, its inclusion is highly application dependent. In cases such as this where (1) The model is missing physics, resulting in systematic discrepancy between the model output and the observations and (2) There is strong prior knowledge about the misfit between the model and true process (details on this in the following paragraph), then it is appropriate to incorporate discrepancy for inference and prediction. On the other hand, with acknowledgement that no model is perfect, it may be reasonable to exclude discrepancy from the analysis if the model is flexible enough to fit the data and the model is empirical in nature, where parameters do not have physical significance. The condition on the empirical nature is important since even if a model is flexible enough to fit the data (as in the toy example), the parameter estimates will be strongly biased while HPIs may not cover the true parameter values.
Identifiability and Sensitivity of Modeling Discrepancy The sensitivity of modeling discrepancy stems from the property of indentifiability. This describes a desirable property of parameters in statistical models by which their true values can be inferred from the data. The ability to identify model components depends on both the model structure and the type and quantity of data that is available. Parameters which are structurally non-identifiable are not independent of each other, and cannot be uniquely decoupled even with an infinite amount of calibration data In the paper by Brynjarsdóttir [17], this concept is succinctly expressed by rewriting the model for the observations with discrepancy in (10) as,
Note that, even if the true process \(\zeta \) is known perfectly, for every value of \(\theta \), there is a corresponding \({\varDelta }\) that satisfies the condition in (28). Therefore, \(\theta \) and \({\varDelta }\) are not identifiable from the observations.
As discussed in [17], the key to reliable learning about the parameters is to incorporate as much prior information about the discrepancy (and \(\theta \)) as possible. The more realistic the prior information is (i.e., assigning higher prior probability the true \(\theta \) and \({\varDelta }\)), the more reliable the inference will be [17]. Making stronger (but still realistic) prior assumptions typically translates to a reduction in posterior uncertainty over \(\theta \) and \({\varDelta }\). Conversely, lack of understanding of the missing physics may not result in reliable estimates even a discrepancy model is incorporated into the statistical model.
Discrepancy in Applications for ICME A question which arises from the importance of prior modeling is perhaps this: If reliable learning of the parameters requires such careful modeling of the discrepancy prior, why not just improve the constitutive model to include the physics and thus eliminate the need to account for the discrepancy? Often accounting for model misfit is both computationally and philosophically easier than improving a model. As an example, take the case study presented in this paper. Inference was performed on the VPSC model while accounting for discrepancy since the model does not include physics for the elastic regime of deformation. As demonstrated, the misfit due to a visco-plastic model can be handled from a statistical perspective with some basic prior knowledge related to the applicable domain and effective magnitude of the elastic effect. Incorporation of an elastic component to the model, on the other hand, would require a single-crystal elastic constitutive law, numerical values of the components of the 4th order elasticity tensor, a schema for partitioning the total deformation into the elastic and plastic contributions (typically \({\mathbf {F}}={\mathbf {F}}^e\cdot {\mathbf {F}}^p\)), and significant updates to the numerical framework.
Even supposing a more complete model is available, it may still be beneficial to work with a simpler but discrepant model. As an alternative to VPSC, inference could be performed on its more complete counterpart, the elaso-viscoplastic self-consistent model [35] or alternatively full-field elasto-viscoplastic fast Fourier transform [36,37,38] or finite element simulations [39, 40], which do account for elastic deformation. However, the computational cost of these models is such that a technique such as MCMC for direct inference would be too expensive. Still, in many cases, a more complete model is not available for the application due to truly unknown effects, in which case the discrepant model offers the best representation of the quantity of interest. As a result, if a model is very good at representing a material process or behavior on a restricted domain, but not the entire domain (resulting in systematic discrepancy), the consideration of model-form error may still allow reliable inference and prediction on the unknown modeling components.
In a broad sense, while 3D, physics-based and full-field models may have a higher fidelity as a result of their complex formulations, their high parameter dimensionality makes them very expensive and difficult to calibrate. In effect, MCMC simulations are not a feasible option for this class of models where a great number of evaluations are necessary in order to achieve convergence of the Markov chain to the target posterior. Of course, these high-fidelity models can make use of MCMC simulations in tandem with the adoption of a surrogate model or other techniques such as multi-fidelity optimization [41] to recover information on unknown modeling components. However, the framework we present in this work is intended for the analysis of models for which MCMC is not prohibitively expensive (although, it can be extended to include surrogate models). While a comparison of the techniques of emulation versus discrepancy modeling is outside the scope of this paper, it is worthwhile to note that emulation and surrogate inference come with its own set of challenges which may yet lead to difficulty in learning the true model parameters.
‘Lower’-Fidelity Models for Design The inclusion of model misfit into the inferential and predictive problem has important implications for reduced-order, homogenized, and phenomenological models. Accounting for model discrepancy allows uncertainty propagation for reliable posterior predictions even when the model is incomplete or our physical understanding is not perfect. This has great value for design and engineering problems since it opens the door for the reliable use of these predictions in design and optimization applications and is a main focus of ongoing work by the authors. Furthermore, the repercussion of biased parameter estimates compounds with long-term behavior and property predictions. For example, although the VPSC model is primarily utilized to understand plastic behavior at large strain, unaccounted-for bias in the elastic region would have a strong affect on these long-term predictions, further underlying the importance of accounting for model discrepancy.
Conclusion
There is a great significance to the goals of ICME in accounting for model misfit in uncertainty analyses. Two primary goals of ICME are (1) The linking of models across vast scales in order to account for mechanisms and phenomena affecting the material behavior and performance on many different levels, and (2) A reduction in the volume of experiments needed for design. This relies on being able to first quantify and then propagate uncertainty in and between models in the modeling chain in order to obtain a reliable processing \(\leftrightarrow \) performance predictions and to establish a design confidence. However, many models which are used for design purposes are computationally intensive, with run times exceeding resources typically available to perform advanced UQ techniques without emulation. Being able to incorporate model misfit in to the UQ analysis opens the door to the use of reduced-order, phenomenological, or homogenized models in reliable design work.
In this work, a UQ analysis was performed on the phenomenological VPSC model under the framework presented in [16], with appropriate adjustments to incorporate model misfit. The VPSC model, which accounts only for the plastic regime of deformation, was calibrated with data from both the elastic and plastic region. Uncertainties about unknown model parameters were established and propagated through the model for inference and prediction on the stress–strain response. Posterior summaries such as the MAP and HPI parameters and model evaluations were used to summarize the analysis.
Notes
The observant reader might notice some observations indicate a negative position due to the noise; while this example is not perfect, it serves the purpose of emphasizing the importance of considering model misfit in the analysis.
The hardening rates are traditionally represented by \(\theta _{0}\) and \(\theta _{1}\) in the Voce hardening law. However, to avoid confusion with parameters in the statistical model, this notation is adopted as in [16].
The code and required data to reproduce these results is made available on GitHub at https://github.com/mesoOSU/UQ.
References
Honarmandi P, Paulson N, Arroyave R, Stan M (2019) Uncertainty quantification and propagation in calphad modelling. Model Simul Mater Sci Eng. https://doi.org/10.1088/1361-651X/ab08c3
Paulson NH, Bocklund BJ, Otis RA, Liu Z-K, Stan M (2019) Quantified uncertainty in thermodynamic modeling for materials design. Acta Mater 174:9–15
Olbricht W, Chatterjee ND, Miller K (1994) Bayes estimation: a novel approach to derivation of internally consistent thermodynamic data for minerals, their uncertainties, and correlations. Part I: theory. Phys Chem Miner 21(1–2):36–49
Königsberger E (1991) Improvement of excess parameters from thermodynamic and phase diagram data by a sequential bayes algorithm. Calphad 15(1):69–78
Malakhov DV (1997) Confidence intervals of calculated phase boundaries. Calphad 21(3):391–400
Otis RA, Liu Z-K (2017) High-throughput thermodynamic modeling and uncertainty quantification for ICME. JOM 69(5):886–892
Stan M, Reardon BJ (2003) A bayesian approach to evaluating the uncertainty of thermodynamic data and phase diagrams. Calphad 27(3):319–323
Kouchmeshky B, Zabaras N (2009) The effect of multiple sources of uncertainty on the convex hull of material properties of polycrystals. Comput Mater Sci 47(2):342–352
Yeratapally SR, Glavicic MG, Argyrakis C, Sangid MD (2017) Bayesian uncertainty quantification and propagation for validation of a microstructure sensitive model for prediction of fatigue crack initiation. Reliab Eng Syst Saf 164:110–123
Kouchmeshky B, Zabaras N (2010) Microstructure model reduction and uncertainty quantification in multiscale deformation processes. Comput Mater Sci 48(2):213–227
Koslowski M, Strachan A (2011) Uncertainty propagation in a multiscale model of nanocrystalline plasticity. Reliab Eng Syst Saf 96(9):1161–1170
Fezi K, Krane MJM (2016) Uncertainty quantification of modelling of equiaxed solidification. In: IOP conference series: materials science and engineering, vol 143, p 012028. IOP Publishing
Plotkowski A, Krane MJM (2017) Quantification of epistemic uncertainty in grain attachment models for equiaxed solidification. Metall Mater Trans B 48(3):1636–1651
Sun G, Li G, Zhou S, Wei X, Yang X, Li Q (2011) Multi-fidelity optimization for sheet metal forming process. Struct Multidiscipl Optim 44(1):111–124
Talapatra A, Boluki S, Duong T, Qian X, Dougherty E, Arróyave R (2018) Autonomous efficient experiment design for materials discovery with bayesian model averaging. Phys Rev Mater 2(11):113803
Ricciardi DE, Chkrebtii OA, Niezgoda SR (2019) Uncertainty quantification for parameter estimation and response prediction. Integr Mater Manuf Innov 8(3):273–293
Brynjarsdóttir J, O’Hagan A (2014) Learning about physical parameters: the importance of model discrepancy. Inverse Prob 30(11):114007
Gelfand AE, Smith AFM (1990) Sampling-based approaches to calculating marginal densities. J Am Stat Assoc 85(410):398–409
Gilks WR, Richardson S, Spiegelhalter D (1995) Markov chain Monte Carlo in practice. Chapman and Hall/CRC, London
Brooks S, Gelman A, Jones G, Meng X-L (2011) Handbook of markov chain monte carlo. CRC Press, London
Robert C, Casella G (2013) Monte Carlo statistical methods. Springer, Berlin
Metropolis N, Rosenbluth AW, Rosenbluth MN, Teller AH, Teller E (1953) Equation of state calculations by fast computing machines. J Chem Phys 21(6):1087–1092
Hastings WK (1970) Monte carlo sampling methods using markov chains and their applications. Biometrika. https://doi.org/10.2307/2334940
Geman S, Geman D (1984) Stochastic relaxation, gibbs distributions, and the Bayesian restoration of images. IEEE Trans Pattern Anal Mach Intell 6:721–741
Barnard J, McCulloch R, Meng X-L (2000) Modeling covariance matrices in terms of standard deviations and correlations, with application to shrinkage. Stat Sin 10:1281–1311
O’Malley AJ, Zaslavsky AM (2008) Domain-level covariance analysis for multilevel survey data with structured nonresponse. Journal of the American Statistical Association 103(484):1405–1418
Kennedy MC, O’Hagan A (2001) Bayesian calibration of computer models. J R Stat Soc Ser B (Stat Methodol) 63(3):425–464
Rasmussen CE (2003) Gaussian processes in machine learning. In: Summer school on machine learning. Springer, pp 63–71
Matérn B (1960) Spatial variation, vol 36 of. Lecture notes in statistics
Bjørnstad ON, Falck W (2001) Nonparametric spatial covariance functions: estimation and testing. Environ Ecol Stat 8(1):53–70
Fortmann RS (2012) Understanding the bias-variance tradeoff. I. http://scott.fortmann-roe.com/docs/BiasVariance.html (hämtad 2017-09-29)
Tomé CN, Lebensohn RA (2009) Manual for code visco-plastic self-consistent (VPSC). Los Alamos National Laboratory, New Mexico
Voce E (1948) The relationship between stress and strain for homogeneous deformation. J Inst Metals 74:537–562
Voce E (1955) A practical strain hardening function. Metallurgia 51:219–226
Wang H, Wu PD, Tomé CN, Huang Y (2010) A finite strain elastic–viscoplastic self-consistent model for polycrystalline materials. J Mech Phys Solids 58(4):594–612
Lebensohn RA, Kanjarla AK, Eisenlohr P (2012) An elasto-viscoplastic formulation based on fast fourier transforms for the prediction of micromechanical fields in polycrystalline materials. Int J Plast 32:59–69
Lebensohn RA, Castañeda PP, Brenner R, Castelnau O (2011) Full-field vs. homogenization methods to predict microstructure–property relations for polycrystalline materials. In: Computational methods for microstructure-property relationships. Springer, pp 393–441
Zhao P, Low TSE, Wang Y, Niezgoda SR (2016) An integrated full-field model of concurrent plastic deformation and microstructure evolution: application to 3d simulation of dynamic recrystallization in polycrystalline copper. Int J Plast 80:38–55
Roters F, Eisenlohr P, Kords C, Tjahjanto DD, Diehl M, Raabe D (2012) Damask: the düsseldorf advanced material simulation kit for studying crystal plasticity using an fe based or a spectral numerical solver. Procedia Iutam 3:3–10
Roters F, Eisenlohr P, Hantcherli L, Tjahjanto DD, Bieler TR, Raabe D (2010) Overview of constitutive laws, kinematics, homogenization and multiscale methods in crystal plasticity finite-element modeling: Theory, experiments, applications. Acta Mater 58(4):1152–1211
Forrester AIJ, Sóbester A, Keane AJ (2007) Multi-fidelity optimization via surrogate modelling. Proc R Soc Math Phys Eng Sci 463(2088):3251–3269
Acknowledgements
S.R.N and D.E.R. received support from The Air Force Research Laboratory under Award FA8650-17-1-5277 “Ensemble predictions of material behavior for ICMSE for additive structures.” S.R.N. and O.A.C. received seed funding for this work through the Center for Emergent Materials: An NSF MRSEC (Chris Hammel PI, NSF Award DMR-0820414). Computing resources for the Ohio Supercomputer Center were provided through the Simulation Innovation and Modeling Center (SIMCenter) at Ohio State.
Author information
Authors and Affiliations
Corresponding author
Appendices
Appendix 1: MCMC Sampler Implementation

Appendix 2: Derivation of Full-Conditionals
Derivation of full conditional distributions over \(\delta ^{2}, {\varPsi }, R\) and \({\varDelta }\).
Independent Error Precision,\(\delta ^{s}\)
Full Error Precision Matrix,\({\varPsi }\)
Random Effects Precision (\({\varLambda }\)) Decomposed Correlation, R
Discrepancy,\({\varDelta }\)
Appendix 3: Parameter Correlation
Appendix 4: Additional Figures
Marginal posterior distributions: Fig. 12 compares the distributions between the overall (dashed lines) and random effects (solid lines).
Rights and permissions
About this article
Cite this article
Ricciardi, D.E., Chkrebtii, O.A. & Niezgoda, S.R. Uncertainty Quantification Accounting for Model Discrepancy Within a Random Effects Bayesian Framework. Integr Mater Manuf Innov 9, 181–198 (2020). https://doi.org/10.1007/s40192-020-00176-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40192-020-00176-2