Abstract
The present study was focused on the biodegradation of monocrotophos (MCP) by indigenous bacteria isolated from sugarcane cultivable soil. The isolates were tested for pesticide tolerance, and the strain VITNNDJ5 showed maximum MCP tolerance and highest degradation potential. The presence of candidate gene for pesticide degradation and plant growth promoting rhizobacterial traits confirmed VITNNDJ5 to be the effective strain. Biodegradation of MCP (1000 mg L−1) was monitored using UV spectrophotometer and HPLC; the degradation products were identified by GCMS. The degradation kinetics was analyzed, and the rate constant (k) and half-life (t1/2) were calculated. Maximum degradation of up to 93% was observed within 5 days. The 16S rRNA gene sequencing revealed VITNNDJ5 to be the closest neighbor of Bacillus aryabhattai (GenBank accession number—KU598848). The augmentation of VITNNDJ5 to the rhizosphere of Liriope muscari enhanced the plant growth and degradation of MCP in soil. The fate of MCP in soil was analyzed, and a metabolic pathway was proposed.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Agriculture plays a pivotal role in the Indian economy, and according to the ministry of agriculture and farmers welfare, Government of India approximately 58% of the Indian population depends on agriculture for livelihood. Reports have shown that about 15–20% of the annual crop production is being destroyed by parasites such as pests, insects and herbs (Bhalerao and Puranik 2009). Pesticides are synthetic chemicals widely used in agriculture to prevent infestation by pests, thereby increasing productivity. The indiscriminate application of pesticide leads to the common occurrence of its residue in several biological and non-biological entities posing serious threats to humans and the environment. The physical and chemical methods implemented for the decomposition of pesticides results in the production of toxic intermediates causing more harm to the environment. Therefore, the removal of pesticides through a biological process can be very effective (Al-Qurainy and Abdel-Megeed 2009).
Organophosphorus pesticides (OPPs) are the most widely used insecticides representing 34% of worldwide insecticide consumption (Singh and Walker 2006). OPPs are known to inhibit the cholinesterase neurotransmitters, including acetylcholinesterase, butyrylcholinesterase and pseudocholinesterase (Abraham et al. 2014). The irreversible inhibition of these enzymes leads to the disruption of endocrine functions, failure of the nervous system, defect in childbirth, impotency and disruption of growth, development and reproduction in animals, birds and humans. The non-specific mode of interaction of OPPs with lower and higher life forms leads to biomagnifications thereby raising the toxicity in humans (Ali et al. 2012). Monocrotophos (MCP) also known as dimethyl-(E)-1-methyl-2-(methylcarbamoyl) vinyl phosphate is an organophosphate insecticide widely used for its high efficiency, availability and low cost. MCP is moderately persistent in nature with a half-life of 7–10 days in soil under direct sunlight, 25–30 days under shaded condition and under controlled environmental conditions with 25 °C temperature and alkaline pH (8–9); the half-life is 20 days (Lee et al. 1990; Guth 1994; Gundi and Reddy 2006). MCP can be degraded by various physical, chemical and biological processes.
Biological detoxification is eco-friendly, cost-effective and has a broad range of substrate specificity. Microorganisms play an effective role in the biodegradation of pesticides and reducing their toxicity (Bhalerao and Puranik 2009; Singh and Walker 2006). There are several reports on the biodegradation of MCP using indigenous soil bacteria. Acharya et al. (2015) reported a Bacillus subtilis KPA-1 strain showing 94% MCP biodegradation potential. Similarly, Pseudomonas fluorescens and Klebsiella sp. were also reported for their ability in degrading MCP (KaviKarunya 2012). Sun et al. (2016) have reported 100% degradation of 0.2 mM MCP within 36 h by Starkeya novella YW6 strain. A study on plasmid-mediated biodegradation of MCP using Pseudomonas mendocina strain was reported by Bhadbhade et al. (2002a). B. megaterium MCM B-423 and Arthrobacter atrocyaneus MCM B-425 strains were also reported for biomineralization of MCP up to 93% and 83% within 8 days (Bhadbhade et al. 2002b). Similarly, rapid biodegradation of MCP was observed in the culture medium and soil by Paracoccus sp. M-1 isolated from wastewater sludge (Jia et al. 2006). In the contaminated soil, MCP acts as a sole source of carbon and phosphorus for many indigenous bacteria since they synthesize enzymes like organophosphate hydrolase (OPH) encoded by opdA gene which degrades MCP and other organophosphorus pesticides (Horne et al. 2002). The phosphatase and esterase enzymes have a direct influence on the biodegradation of MCP (Subhas and Singh 2003; Chaudhry et al. 1988; Jain and Garg 2013). The metabolism of MCP involves the production of O-desmethyl monocrotophos, monomethyl phosphate, dimethyl phosphate, N-methylacetoacetamide, N-methylbutyramide, CO2, methylamine, ammonia, etc. (Bhadbhade et al. 2002b).
Phytoremediation process has proven to be cost-effective, eco-friendly and efficient in eliminating a wide range of organic and inorganic pollutants from the environment. Studies on pesticide degradation in soil have revealed that plant species such as Ricinus communis L., Avena sativa L. and Cucurbita pepo are capable of playing a predominant role in the remediation of pesticides and other environmental pollutants (Rissato et al. 2015; Truu et al. 2015). Similarly, aquatic plants Myriophyllum aquaticum, Spirodela oligorrhiza L. and Elodea canadensis are reported for enzymatic degradation of OPPs in water (Gao et al. 2000). In addition to their role in pesticide degradation, plants can also influence the growth of indigenous microbes by releasing catabolic enzymes, carbohydrates, sugars, amino acids, alcohols, organic acids, etc. (Liu et al. 2018). The mechanism of phytoremediation involves several remediation strategies like phytodegradation, rhizodegradation, phytoextraction, phytotransformation, phytostabilization rhizofiltration. Carboxylesterase, cytochrome P450 and glutathione S-transferase are the key enzymes involved in phytoremediation of pesticides (Dubey and Fulekar 2013; Velázquez-Fernández et al. 2012). Hence, the present study was focused on the integrated application of plant and indigenous bacterial strain for the biodegradation of MCP in soil and the research was carried out during 2016–2017 at Vellore Institute of Technology, Tamil Nadu, India.
Materials and methods
Soil sampling and chemical reagents
Soil samples for the isolation of bacteria were collected from the sugarcane fields by random sampling procedure near Brahmapuram, Vellore, Tamil Nadu, India (latitude—12°58′15″N, longitude—79°11′1″E). Rhizosphere soil (5–10 cm depth) from five different locations with approximately 100 m distance was collected by tapping the roots so as to obtain the soil adhering to the rhizoplane region and was transferred into sterile polyethylene bags. Samples were transported to the laboratory for isolation of indigenous bacteria (Barillot et al. 2013). The soil was clay loamy in texture with a pH of 6.9.
Commercial grade monocrotophos (monocrotophos 53% and cyclohexanone 47% w/w) was obtained from agrochemical shop in Vellore. Analytical grade ethyl acetate, acetonitrile and methanol were obtained from Sisco Research Laboratories (SRL), and p-nitrophenyl phosphate (p-NPP), methylamine, p-nitrophenol were procured from Sigma-Aldrich. Minimal salt medium (MSM) contained (Na2HPO4, 5.8; KH2PO4, 3.0; NaCl, 0.5; NH4Cl, 1; MgSO4, 0.25 in gL−1 and pH 6.8–7.0) (Abraham et al. 2014), Davis Mingioli’s synthetic medium (K2HPO4, 3; KH2PO4, 7; MgSO4, 0·1; and (NH4)2SO4, 1 in gL−1) (Bhalerao and Puranik 2009).
Isolation of bacteria and screening for MCP degradation
Soil samples were mixed thoroughly, 10 g was added in 100 mL of MSM supplemented with 50 mg L−1 MCP, and the flask was incubated in an orbital shaker (120 rpm, 28 ± 2 °C) for 48 h. The enriched sample was serially diluted and was plated onto MSM agar medium. Morphologically distinct colonies were purified and were maintained in glycerol stock. The isolates obtained were screened for their tolerance against MCP (100–4500 mg L−1) in MSM using agar plate and broth assays (Bhalerao and Puranik 2007). For the detection of opdA gene, the bacterial genomic DNA was isolated by phenol–chloroform method (He 2011) and was used as DNA template for the amplification of opdA gene using the Opda-F 5′ TGTTCCGGTAACCACTCACA 3′ and Opda-R 5′ CACTCTCAGAGGGACGAAGG3′ forward and reverse primers as described by Ali et al. (2012).
Analysis of plant growth promoting traits of VITNNDJ5
Production of ammonia was assessed in freshly prepared peptone water inoculated with VITNNDJ5 strain and incubated in an orbital shaker at 120 rpm for 72 h (Cappuccino and Sherman 1992). The ability of the isolate VITNNDJ5 in solubilizing insoluble phosphate was tested using the standard National Botanical Research Institute’s phosphate (NBRIP) growth medium. The isolate was spot inoculated onto the NBRIP medium and was incubated for 7 days at 28 ± 2 °C (Nautiyal 1999).
The solubilization efficiency (SE) was determined using the formula (Eq. 1),
For the production of indole acetic acid (IAA), VITNNDJ5 was inoculated in MSM supplemented with 5 mM tryptophan and was incubated at 28 ± 2 °C for 48 h in an orbital shaker at 120 rpm. Culture broth (2 mL) was centrifuged at 12,000 rpm, and Salkowski’s reagent was added to the supernatant. Tubes were kept undisturbed in dark for 15–25 min to allow the reaction to take place. The IAA produced was quantified at 530 nm (Gordon and Weber 1951; Das et al. 2014). FISS minimal medium supplemented with 0.5 µmol iron was inoculated with VITNNDJ5 and was incubated for 24 h on an orbital shaker at 30 °C. The broth was centrifuged at 10,000 rpm for 10 min at 4 °C, and the supernatant was added into the wells on chrome azurol S (CAS) plates. The presence of orange halo zone around the well indicated the production of siderophore (Schwyn and Neilands 1987).
Enzymatic assay
Detection and estimation of Organophosphate hydrolase
Enzyme assay was performed according to the protocol reported by Jia et al. (2006) with some modification where VITNNDJ5 was inoculated into MSM supplemented with 500 mg L−1 of MCP and was incubated in an orbital shaker at 120 rpm for 48 h. Cells were recovered at the mid-log phase by centrifugation (10,000 rpm for 10 min) and were tested for their ability to hydrolyze MCP. The pellet was washed and suspended in three volumes of Tris–HCl buffer (0.01 M, pH 8.1) and was stored at 4 °C. Further, the cells were ruptured by sonication (at 1.4 amp with 3-s pulse for 4 min with a 30 s interval in between) and were centrifuged at 10,000 rpm for 10 min for the removal of cell debris and the supernatant was stored at − 20 °C (Subhas and Singh 2003). The cell-free extract (crude enzyme, 50 µL), along with MCP (substrate) and BSA standards were used to for the estimation of total protein and determination of enzyme activity (Lowry et al. 1951; Ali et al. 2012).
Detection and estimation of phosphatase
Cell suspension (108 cfu mL−1) was inoculated into 250 ml Erlenmeyer flask containing 100 mL MSM (1000 mg L−1 MCP) and was incubated at 28 ± 2 °C for 48 h under shaking condition. Culture media was centrifuged, and the cell-free supernatant (CFS) was used for the detection of phosphatase enzyme by King–Armstrong method using p-nitrophenyl phosphate (p-NPP) and MCP as substrates (King et al. 1951). The amount of p-nitrophenol was estimated by measuring the absorbance at 400 nm. Degradation of MCP was measured by estimating the amount of phosphate released using KH2PO4 as standard. The enzyme activity was analyzed by calculating the amount of p-nitrophenol and phosphate (in µg mL−1) released in unit time (Bhadbhade et al. 2002b).
Identification of VITNNDJ5
The isolate VITNNDJ5 was identified based on various morphological and biochemical characteristics using the standard procedures described by Cappuccino and Sherman (1992). Overnight grown cell suspension with and without pesticide treatments was fixed using 3% glutaraldehyde followed by a series of dehydration steps using ethanol (30–100%). The fixed samples were attached to a metal mount using carbon tape and were sputter coated with gold under vacuum condition in argon atmosphere. The cell surface morphology of VITNNDJ5 was analyzed by scanning electron microscope (SEM) (model ZEISS EVO 18). The molecular characterization of bacterial strain was performed by 16S rRNA gene sequencing using the universal primers 27F (5′ AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′). The sequences were compared using BLAST, and related sequences were obtained from the sequence database for comparison. Multiple alignments were performed, and the phylogenetic tree was constructed by neighbor joining method using the default parameters of MEGA4 (Tamura et al. 2007) and bootstrapped using 1000 bootstrap trials.
Biodegradation of MCP in MSM
The overnight culture of VITNNDJ5 prepared in MSM was centrifuged at 4600 rpm for 5 min at 4 °C. The pellet obtained was washed twice and was resuspended in N-saline to obtain an OD of 0.7. Seed culture (2% v/v) was inoculated into Erlenmeyer flasks containing 150 mL MSM supplemented with 1000 mg L−1 MCP. The effects of additional carbon sources (glucose, sucrose, fructose and cellulose) and pH (6, 7, 8 and 9) on MCP biodegradation were studied (Acharya et al. 2015; Tang and You 2012). The desired pH was obtained using 0.1 N NaOH and 0.1 N HCl solutions. MSM without pesticide supplementation was used to compare the growth pattern and the uninoculated media served as control. Degradation study was carried out for 5 days, and samples were collected at an interval of 24 h followed by measurement of pH and cell density (OD620 nm). The recovered samples were harvested at 7200 rpm for 10 min, and the cell-free supernatants (CFS) were used for the detection of MCP. The concentration of MCP in the test and control samples was assessed by UV/Vis spectrophotometer (AU-2603) at 214 nm. Further, the % degradation was calculated using the following formula (Eq. 2),
The CFS was extracted with an equal volume of ethyl acetate and was filtered twice through 0.22 µm syringe filter. MCP biodegradation was analyzed by HPLC (Waters 1525 binary HPLC pump, Milford, USA) using Symmetry C18 reverse phase column (Waters 5 mm, 4.6 mm × 150 mm). The sample volume of 20 µL was injected with an isocratic mobile phase composed of acetonitrile and water (70:30, V/V), which was pumped through the column at a flow rate of 1 mL min−1, for a duration of 15 min, and residual MCP was detected at 214 nm. The retention time of MCP in the control sample was 2.54 min.
Identification of the degraded products upon MCP metabolism
MCP and its degradation products were identified by GCMS (Clarus 680, Perkin Elmer) with Elite-5MS column (30.0 m length, 0.25 mm internal diameter (ID) and 250 μm film thickness). The components were separated using Helium as carrier gas at a constant flow rate of 1 mL min−1. The injector temperature was set at 260 °C during the chromatographic run. The extracted residue (1 μL) was injected into the instrument with the initial oven temperature of 60 °C for 2 min followed by 300 °C at the rate of 10 °C min−1, and 300 °C was held for 6 min. The conformational change in the original MCP structure and the occurrence of new chemical bonds were investigated by FTIR analysis using a Nicolet FTIR (AVATAR-330) model with 16 scan speed and resolution of 4 cm−1 in the mid-IR region of 400–4000 cm−1 as described by Abraham et al. (2014).
Biodegradation of MCP in artificially contaminated soil
Two experimental setups for phyto- and rhizoremediation of MCP were designed, and the study was conducted in greenhouse condition. Red loamy soil was collected from 0 to 20 cm depth from the nursery of Vellore Institute of Technology, Tamil Nadu. The physicochemical properties of soil are mentioned in online resource Table 1. The soil was air-dried, sieved with 2-mm pore-sized mesh to obtain even sized particles (1 kg, treated with 50, 100 and 150 mg L−1 of MCP) and was transferred into low-density polyethylene (LDPE) bags (Truu et al. 2015). Liriope muscari commonly known as “silver dragon lilyturf” plants were collected from VIT nursery. Plants were cleaned in running tap water, the shoots were cut into 10 cm height, and pre-germinated roots were removed from the tuber. Plants were allowed to acclimatize for 10 days. Bacterial inoculum (15 mL per pot) with 0.7 OD620mn was augmented to the rhizoremediation soil. The study was carried out for 80 days, and the treated and untreated samples were collected at a regular interval of 20 days. The available bacterial load in both phyto- and rhizoremediation soil was estimated regularly by serial dilution and spread plate technique. Bacterial colonies with similar morphology as VITNNDJ5 were enumerated and characterized using morphological and biochemical methods (Cappuccino and Sherman 1992).
Analysis of MCP biodegradation and identification of metabolites to propose the biodegradation pathway
The air-dried soil sample (20 g) was extracted twice using 100 mL of ethyl acetate with anhydrous sodium sulfate (Sharma and Rajput 2012). Ethyl acetate extract was filtered through a 0.22-mm syringe filter and was analyzed for MCP residues by UV/Vis spectrophotometer (AU-2603) at 214 nm. Residual MCP and the degradation products in the soil were identified by GCMS as described earlier. A biodegradation pathway for MCP was proposed using PathPred: Pathway Prediction server—GenomeNet and KEGG PATHWAY database—GenomeNet (Moriya et al. 2010).
MCP biodegradation kinetics
The dissipation pattern of MCP from both MSM and contaminated soil was found to be well fitted to the first-order kinetic model (Eqs. 3, 4).
upon linear arrangement it produced,
where C0 is the initial concentration of MCP supplemented, followed by Ct, the concentration of MCP at time t (in days), and k represents the degradation rate constant. The time (half-life, t1/2) at which half of the initial MCP concentration was degraded from the media and soil was calculated using the linear regression equation.
Assessment of plant growth parameters
Plant samples were collected from each experimental setup at regular intervals and were assessed for MCP uptake, root length, shoot height and chlorophyll content. Fresh leaves were washed thoroughly in tap water followed by distilled water and were kept for drying in room temperature. The leaf sample (0.5 g) was homogenized with 10 ml of 80% and 100% acetone for the extraction of pigments (Lichtenthaler 1987). The homogenized mixture was centrifuged at 10,000 rpm for 15 min at 4 °C, and 0.5 mL of the supernatant was transferred to a fresh tube containing 4.5 mL of 80% acetone. The presence of chlorophyll a and chlorophyll b was determined by measuring the absorbance at 663 and 645 nm.
The total chlorophyll content was calculated using Eq. 5 (Arnon 1949),
The plant root and shoot samples were dried by heating intermittently in a hot air oven at 60 °C for 12 h, and 5 g of plant sample was macerated and extracted twice with 5 mL of ethyl acetate and was analyzed for the presence of MCP by GCMS (Sharma and Rajput 2012).
Result and discussion
Isolation and screening for MCP degrading bacteria
A total of 15 MCP resistant morphologically distinct bacterial colonies were isolated on MSM agar plates supplemented with 200 mg L−1 of MCP. MTC plate assay and broth assay confirmed the isolate VITNNDJ5 to be effective as it was tolerant up to 4500 mg L−1of MCP (Fig. 1a). Indigenous microbes are known to tolerate pesticides and degrade monocrotophos as well as other OPPs in various environmental compartments due to continuous exposure (Rangaswamy and Venkateswarlu 1992; Chanika et al. 2011; Shafiani and Malik 2003). Bhadbhade et al. (2002b) elucidated the ability of Bacillus sp. and Arthrobacter sp. to tolerate and efficiently degrade MCP up to 1000 mg L−1. Bhalerao and Puranik (2009) also reported a series of fungal strain capable of tolerating and degrading various concentrations of MCP. The presence of opdA gene was confirmed by the appearance of a 380 bp sized amplicon on an agarose gel as shown in lane 1 (Fig. 1b). Similarly, the presence of opdA gene in Bacillus sp. isolated from agricultural soil was reported by Ali et al. (2012) and Acharya et al. (2015).
Analysis of plant growth promoting traits of VITNNDJ5
The strain VITNNDJ5 was capable of producing indole acetic acid (IAA), ammonia and solubilizes insoluble phosphate. Many root-associated bacteria are known to possess one or more plant growth promoting (PGP) traits attributing to their ability in degrading pesticides and other toxic contaminants (Das et al. 2014). IAA, a phytohormone (auxin) was produced by the strain VITNNDJ5 upon bioconversion of tryptophan into IAA by the enzyme tryptophanase. Production of IAA via indole-3 pyruvic acid and indole-3-acetic aldehyde has been observed in several bacterial genera like Agrobacterium, Pseudomonas, Azospirillum, Rhizobium, Klebsiella, etc. It has been reported that Pseudomonas, Azospirillum, Cyanobacteria and Bacillus genera are the tryptophan-dependent IAA producers (Ahemad and Kibret 2014). The isolate VITNNDJ5 was capable of producing 161.99 µg mL−1 of IAA in media supplemented with l-tryptophan. The formation of a clear halo zone around the bacterial colony in NBRIP agar plates confirmed the ability of the isolate in solubilizing insoluble phosphate with an efficiency of 57%. The isolate showed a positive result for ammonia production which correlated with the study reported by Mishra et al. (2010) and Ghosh et al. (2018). However, siderophore production was found to be negative as no halo zone was observed (Table 1). Bacillus sp. are known for their active participation in plant growth promotion under toxic stress. Bacillus aryabhattai is one of the newly isolated strains reported to have efficient PGP characteristics (Ghosh et al. 2018; Ramesh et al. 2014)
Enzyme Assay
The strain VITNNDJ5 showed an OPH enzyme activity of 71.49 U mL−1 after 48 h of incubation. The data obtained were similar to that of the previous reports on the production of OPH enzyme by Bacillus sp. and Paracoccus sp. (Ali et al. 2012; Jia et al. 2006). Phosphatase was detected and estimated with the production of inorganic phosphates from MCP and p-nitrophenol from p-NPP (positive control). The activity of phosphatase was found to be 62 U mL−1 with MCP as substrate and 95.67 U mL−1 with p-NPP as substrate (Table 1), which confirms the ability of the VITNNDJ5 in degrading MCP upon the production of phosphatase. Bhadbhade et al. (2002b) also suggested a similar report on the involvement of bacterial enzyme phosphatase in biodegradation of MCP. OPH and phosphatase involved in the process are responsible for the cleavage of P–S bond in phosphorodithioates and phosphorothioates and P–O bond cleavage in phosphorothioate type of OPP. However, in MCP the hydrolysis of P–O alkyl and P–O aryl bonds plays a significant role in the detoxification process. Production of OP hydrolyzing enzymes by various bacterial and fungal strains such as Pseudomonas sp., E. coli, Agrobacterium sp., Bacillus sp., P. aculeatum and A. niger, has been reported by Ali et al. (2012) and Jain and Garg (2013).
Identification of VITNNDJ5
The colony morphology of effective isolate VITNNDJ5 was observed to be large (3–4 mm), round and creamy white in color when grown on MSM. The isolate was a gram-positive, motile bacterium, and SEM micrograph showed rod-shaped bacterial structure (Fig. 2a). The cells were found to be pleomorphic which could be due to the supplementation of MCP (Fig. 2b). VITNNDJ5 was positive for MR-VP, oxidase, catalase, amino acid decarboxylase (lysine and ornithine), TSI and gelatine liquefaction tests, whereas it showed negative results for indole production and citrate utilization tests. The 16S rRNA gene sequencing and phylogenetic tree identified the isolate to be the closest neighbor of B. aryabhattai (Fig. 2c). The nucleotide sequence was submitted to GenBank with accession number KU598848. Similar morphological and biochemical characteristics were reported by Shivaji et al. (2009) for B. aryabhattai. Bacillus is the predominant genus in the environment and hence is involved in the degradation of xenobiotics. There are reports on various environmental strains of Bacillus sp. that play a pivotal role in the degradation of pesticide upon expression of OP degrading gene (Ali et al. 2012; KaviKarunya 2012; Pailan et al. 2015).
Kinetics of MCP biodegradation in MSM and optimization of carbon source and pH
Bacterial growth kinetics and degradation dynamics of MCP in MSM with and without supplementation of glucose are demonstrated in Fig. 3a. The strain VITNNDJ5 was found to utilize all the four carbon sources. However, the optimization of carbon source revealed that the bacterial growth and MCP biodegradation rate were comparatively higher in the media supplemented with glucose, hence was considered as the suitable carbon source (Fig. 3b). The growth of VITNNDJ5 in MSM (with and without glucose) was minimal during the initial 24 h. Nevertheless, in MSM with glucose, the growth was further accelerated due to the presence of an additional carbon source. Utilization of glucose as a co-substrate during MCP biodegradation was also reported by Acharya et al. (2015). No significant difference in the biodegradation of MCP was observed at various pHs during the initial growth phase. With an increase in the incubation time, maximum biodegradation of MCP was achieved in the media with pH—8 (Fig. 3c). Reports have shown the significance of neutral to slightly alkaline pH in the enhancement of OPP metabolism (Tang and You 2012; Acharya et al. 2015). In addition, the decrease in initial pH from 7.6 to 3.8 after 5 days could be due to the production of organic acids as a result of MCP biodegradation. Significant reduction in pH during pesticide biodegradation by fungal and bacterial strains has been reported by Siddique et al. (2003). The biodegradation process without an additional carbon source (MSM + MCP) followed first-order kinetics with a rate constant of 0.166 day−1 resulting in extended half-life (4.6 days). However, the presence of glucose in MSM as a co-substrate accelerated the dissipation rate (0.562 day−1) and achieved a shorter half-life of 1.7 days (Table 2). The isolate VITNNDJ5 could achieve 93% and 52% degradation in MSM after 5 days with and without supplementation of glucose (Fig. 3b). It was evident that VITNNDJ5 was unable to degrade MCP effectively when glucose was not supplemented and thereby lead to a stress condition, whereas rapid multiplication of cells and expression of opdA gene led to enhanced biodegradation of MCP upon addition of carbon source to the media. Similarly, under optimized conditions, 94.2% degradation of MCP within 144 h by an environmental B. subtilis strain was previously reported (Acharya et al. 2015). Bhalerao and Puranik (2009), in their study on biodegradation of MCP by a fungal strain Aspergillus oryzae, reported that the biodegradation process followed first-order kinetic model.
MCP biodegradation by VITNNDJ 5 and the parameters effecting: a MCP degradation and bacterial growth kinetics with and without glucose, b biodegradation of MCP (1000 mg L−1) with and without supplementation of carbon source (CS), c optimization of pH for MCP (1000 mg L−1) biodegradation. MCPC: MCP concentration in mg L−1, MCPC (MSM + MCP + Glucose): MSM supplemented with MCP and glucose, MCPC (MSM + MCP): MSM without glucose inoculate with VITNNDJ5, BG: bacterial growth. The values represented in the figures are the means of triplicate with a standard deviation within 5% of the mean value
Detection of MCP biodegradation and identification of metabolites
The MCPC (MSM + MCP) sample showed a peak at the retention time of 2.54 min for MCP in HPLC (Fig. 4b). MSM without pesticide was analyzed for eliminating the interfering peaks (Fig. 4a). The biodegradation of MCP was monitored by the gradual disappearance of the MCP peak (Fig. 4c). Further FTIR spectra of the control sample (MCPC) showed a sharp peak at 1028 wavenumber which indicated the C-O link that connects the organophosphate moiety to the methyl acetoacetamide compound (Fig. 4d). In the treated sample (MSM + MCP + Isolate), a drastic reduction in the 1028 peak height could explain the breakage of C-O bond leaving both organophosphate and methyl acetoacetamide groups vulnerable to undergo enzymatic degradation. Shifting of existing peaks and appearance of new peaks represented the breakdown or conformational changes in the original structure and formation of new metabolites (Fig. 4e). GCMS analysis was carried out to identify the degradation product of MCP based on the m/z values in the mass spectra. The presence of MCP was confirmed with an m/z ratio of 223 (M = 223.0610) (Fig. 5a). Formation of intermediate products such as monomethyl phosphate [m/z: 113, (M + 1H)+, where M = 112.0210] and dimethyl phosphate [m/z: 127, (M + 1H)+, where M = 126.0480] (Fig. 5b, c) was observed. Formation of dimethyl phosphate and monomethyl phosphate during the biodegradation of MCP was also reported by Abraham and Silambarasan (2015). Acetic acid derivatives, organic acids and aldehydes were also identified as the end products of MCP biodegradation (Table 3). Reports have shown the production of beneficial and metabolically active by-products during the biodegradation of pesticides; for instance, the production of aldehydes and organic acids upon bacterial degradation of MCP was reported by Buvaneswari et al. (2018) and Abraham et al. (2014).
Analysis of MCP biodegradation by HPLC and FTIR: a HPLC chromatogram for MSM without MCP (control 1), b MCPC (MSM + MCP) (control 2), c (MSM + MCPC + Isolate) degraded sample after 5 days of incubation, d, e FTIR spectra representing the peaks for compounds in MCPC (MSM + MCP) (control 2) and (MSM + MCPC + Isolate), respectively
Biodegradation of MCP in artificially contaminated soil
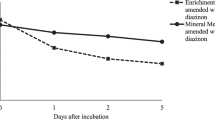
The phyto- and rhizoremediation studies revealed the rate of MCP biodegradation in the soil to be rapid at the initial 20 days as compared to that of the consecutive intervals, which could be due to the bioavailability of substrate. However, the process was found to be decelerated after 20 days of treatment, leading to a final degradation up to 99.2% for 50 mg kg−1 and 98% for 100 and 150 mg kg−1 setups (Fig. 6a). The presence of MCP in the soil after 20 days of phytoremediation treatment proved the process to be slower. In the rhizoremediation process, a symbiotic association between the PGPR strain VITNNDJ5 and plant L. muscari led to cometabolism, thereby reducing the half-life of MCP. Within 40 days of treatment, 99% degradation was noticed for all the tested concentrations (Fig. 6a). The microbial load in rhizoremediation soil was found to be higher as compared to phytoremediation soil, which could be due to the augmentation of effective indigenous bacterial strain VITNNDJ5 to the rhizosphere. A gradual increase in the population of VITNNDJ5 strain was noticed at each interval. The natural attenuation was not efficient as traces of MCP were prominently found in the soil, which could be due to the inadequacy of the indigenous microbial population to degrade increased concentrations of MCP. No trace of MCP was detected in the plant parts which can be due to rapid cometabolism of MCP by plant and bacterial system. PGPR bacteria have also been reported for their role in accelerating pesticide degradation in plant bodies (Feng et al. 2017). Hence, the process of biodegradation adapted by L. muscari could be phytodegradation. Dubey and Fulekar (2013) reported the interaction between plant and bacteria in the cometabolism of pesticide.
Identification of metabolites in the soil and analysis of potential biodegradation pathway
Organic acids, amines, aldehydes, amino acids, carbohydrates and their conjugates were obtained as end products in the soil (Table 3). Plants are known for the production of various low molecular weight and high molecular weight organic acids and amino acids under pesticide stress conditions (Singer et al. 2003). GCMS data confirmed the presence of carbohydrates, amino acids and protein building blocks in the rhizoremediation soil. Cyclohexanone, a component of MCP formulation, was detected in the soil during the initial 20 days of MCP biodegradation (Online resource Fig. 1a). Further, the degradation of cyclohexanone was confirmed as it disappeared in consecutive intervals. Dhanya et al. (2016) reported biotic and abiotic decomposition of cyclohexanone using a mixed culture of Pseudomonas and production of phenol and its derivatives as major intermediates, and a novel degradation pathway of cyclohexanone was proposed by Yi et al. (2011). Figure 6b elucidates the bioconversion of MCP into non-toxic end products via several degradation pathways (benzoate degradation, styrene degradation, propanoate and glyoxylate metabolism pathway) which could allow them to be further utilized in energy generation through glycolysis/gluconeogenesis, TCA cycle, etc., by living organisms. The pathway suggested a possible breakdown of MCP into dimethyl phosphate via hydrolysis. The dimethyl phosphate group further led to the formation of phosphoric acid and acetic acid esters by hydrolase and monooxygenase enzymes. A similar report on the formation of dimethyl phosphate, methylacetoacetamide and phosphoric acid during MCP biodegradation was shown by Abraham and Silambarasan (2015). The formation of an amide compound, N-(hydroxymethyl)acetamide, was facilitated by the demethylation of MCP which further biotransformed into acetamide followed by acetic acid liberating molecules of H2O and CO2 in the process (Online resource Fig. 1). Eadsforth (1983) also reported the formation N-hydroxymethyl derivatives upon demethylation of MCP. Xiong et al. (2018) in a recent study on degradation of N-hydroxymethyl acetamide reported a similar pattern of degradation. The breakdown of MCP into orthophosphoric acid and acetic acid could also be achieved from a phosphonoacetate intermediate via phosphonate degradation pathway. Reports have shown the breakdown of phosphonoacetate into orthophosphoric acid and acetic acid by bacterial phosphonoacetate hydrolase enzyme (McMullan et al. 1992; Panas et al. 2006).
Assessment of plant growth parameters
The plants were capable of surviving at all the concentrations of MCP without accumulation of pesticide. Increase in the root and shoot lengths was observed in both phyto- and rhizoremediation setups, but the growth rate was significantly higher in the rhizoremediation process which could be due to the augmentation of indigenous PGP strain VITNNDJ5 (Fig. 7a). Reports have shown the interaction of plant root exudates and bacterial enzymes to be beneficial for plant growth and metabolism (Singer et al. 2003; Dubey and Fulekar 2013). The decrease in the concentration of MCP was directly proportional to the plant growth and availability of non-toxic by-products such as carbohydrates, amino acids, organic acids, etc., in soil (Table 3). The study also revealed the production of chlorophyll to be 0.01 µgml−1 in phyto and 0.012 µgml−1 in rhizoremediation setup (Fig. 7b), which shows the ability of L. muscari in stabilizing its growth and metabolism even at higher concentrations of pesticide. Several plant species have been reported for their ability to tolerate and degrade agrochemical compounds (Al-Qurainy and Abdel-Megeed 2009).
Conclusion
The study highlighted the ability of an effective indigenous strain VITNNDJ5 in degrading MCP effectively (93%). The molecular characterization revealed the isolate to be the closest neighbor of B. aryabhattai. The strain VITNNDJ5 was identified to be a PGPR capable of degrading MCP efficiently in an integrated bio-system of the plant (L. muscari) and bacteria. A biodegradation pathway illustrated the bioconversion of MCP into non-hazardous end products. The possibility of the strain in degrading other classes of pesticides and environmental contaminants can be studied further. The isolate can further be assessed for large-scale pesticide biodegradation under field conditions.
References
Abraham J, Silambarasan S (2015) Bacterial degradation of monocrotophos and phyto-and cyto-toxicological evaluation of metabolites. Toxicol Environ Chem 97(9):1202–1216. https://doi.org/10.1080/02772248.2015.1092541
Abraham J, Silambarasan S, Logeswari P (2014) Simultaneous degradation of organophosphorus and organochlorine pesticides by bacterial consortium. J Taiwan Inst Chem Eng 45(5):2590–2596. https://doi.org/10.1016/j.jtice.2014.06.014
Acharya KP, Shilpkar P, Shah MC, Chellapandi P (2015) Biodegradation of insecticide monocrotophos by Bacillus subtilis KPA-1, isolated from agriculture soils. Appl Biochem Biotechnol 175(4):1789–1804. https://doi.org/10.1007/s12010-014-1401-5
Ahemad M, Kibret M (2014) Mechanisms and applications of plant growth promoting rhizobacteria: current perspective. J King Saud Univ Sci 26(1):1–20. https://doi.org/10.1016/j.jksus.2013.05.001
Ali M, Naqvi TA, Kanwal M, Rasheed F, Hameed A, Ahmed S (2012) Detection of the organophosphate degrading gene opdA in the newly isolated bacterial strain Bacillus pumilus W1. Ann Microbiol 62(1):233–239. https://doi.org/10.1007/s13213-011-0251-4
Al-Qurainy F, Abdel-Megeed A (2009) Phytoremediation and detoxification of two organophosphorous pesticides residues in Riyadh area. World Appl Sci 6:987–998
Arnon DI (1949) Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. World Appl Sci 24(1):1–15
Barillot CD, Sarde CO, Bert V et al (2013) A standardized method for the sampling of rhizosphere and rhizoplan soil bacteria associated to a herbaceous root system. Ann Microbiol 63:471. https://doi.org/10.1007/s13213-012-0491-y
Bhadbhade B, Dhakephalkar P, Sarnaik S et al (2002a) Plasmid-associated biodegradation of an organophosphorus pesticide, Monocrotophos, by Pseudomonas mendocina. Biotechnol Lett 24:647. https://doi.org/10.1023/A:1015099409563
Bhadbhade BJ, Sarnaik SS, Kanekar PP (2002b) Biomineralization of an organophosphorus pesticide, Monocrotophos, by soil bacteria. J Appl Microbiol 93(2):224–234. https://doi.org/10.1046/j.1365-2672.2002.01680.x
Bhalerao TS, Puranik PR (2007) Biodegradation of organochlorine pesticide, endosulfan, by a fungal soil isolate, Aspergillus niger. Int Biodeterior Biodegrad 59(4):315–321. https://doi.org/10.1016/j.ibiod.2006.09.002
Bhalerao TS, Puranik PR (2009) Microbial degradation of monocrotophos by Aspergillus oryzae. Int Biodeterior Biodegrad 63(4):503–508. https://doi.org/10.1016/j.ibiod.2008.11.011
Buvaneswari G, Thenmozhi R, Nagasathya A, Thajuddin N, Kumar P (2018) GC–MS and molecular analyses of monocrotophos biodegradation by selected bacterial isolates. Afr J Microbiol Res 12(3):52–61. https://doi.org/10.5897/AJMR2017.8696
Cappuccino JG, Sherman N (1992) Biochemical activities of microorganisms. Microbiology, a laboratory manual, 1st edn. The Benjamin/Cummings Publishing Co, Menlo Park, pp 105–300
Chanika E, Georgiadou D, Soueref E et al (2011) Isolation of soil bacteria able to hydrolyze both organophosphate and carbamate pesticides. Bioresour Technol 102(3):3184–3192. https://doi.org/10.1016/j.biortech.2010.10.145
Chaudhry GR, Ali AN, Wheeler WB (1988) Isolation of a methyl parathion-degrading Pseudomonas sp. that possesses DNA homologous to the opd gene from a Flavobacterium sp. Appl Environ Microbiol 54(2):288–293
Das S, Jean JS, Kar S, Chou ML, Chen CY (2014) Screening of plant growth-promoting traits in arsenic-resistant bacteria isolated from agricultural soil and their potential implication for arsenic bioremediation. J Hazard Mater 272:112–120. https://doi.org/10.1016/j.jhazmat.2014.03.012
Dhanya V, Usharani MV, Jayadev P (2016) Biodegradation of cyclohexanol and cyclohexanone using mixed culture of Pseudomonas in activated sludge process. J Environ Res Dev 11(2):324
Dubey KK, Fulekar MH (2013) Rhizoremediation of pesticides: mechanism of microbial interaction in mycorrhizosphere. Int J Adv Res Technol 2:193–210
Eadsforth CV (1983) Investigation of pesticide metabolites in the human. In: Reid E, Leppard JP (eds) Drug metabolite isolation and determination. Springer, Boston, pp 119–133. https://doi.org/10.1007/978-1-4684-4484-1_16
Feng F, Ge J, Li Y, He S, Zhong J, Liu X, Yu X (2017) Enhanced degradation of chlorpyrifos in rice (Oryza sativa L.) by five strains of endophytic bacteria and their plant growth promotional ability. Chemosphere 184:505–513. https://doi.org/10.1016/j.chemosphere.2017.05.178
Gao J, Garrison AW, Hoehamer C, Mazur CS, Wolfe NL (2000) Uptake and phytotransformation of organophosphorus pesticides by axenically cultivated aquatic plants. J Agric Food Chem 48(12):6114–6120. https://doi.org/10.1021/jf9904968
Ghosh PK, Maiti TK, Pramanik K, Ghosh SK, Mitra S, De TK (2018) The role of arsenic resistant Bacillus aryabhattai MCC3374 in promotion of rice seedlings growth and alleviation of arsenic phytotoxicity. Chemosphere 211:407–419. https://doi.org/10.1016/j.chemosphere.2018.07.148
Gordon SA, Weber RP (1951) Colorimetric estimation of indoleacetic acid. Plant Physiol 26(1):192–195
Gundi VA, Reddy BR (2006) Degradation of monocrotophos in soils. Chemosphere 62(3):396–403. https://doi.org/10.1016/j.chemosphere.2005.04.076
Guth JA (1994) Monocrotophos—environmental fate and toxicity. In: Ware GW (ed) Reviews of environmental contamination and toxicology. Reviews of environmental contamination and toxicology, vol 139. Springer, New York. https://doi.org/10.1007/978-1-4684-7071-0_7
He F (2011) E. coli genomic DNA extraction. Bio-protocol 1:e97. https://doi.org/10.21769/BioProtoc.97
Horne I, Sutherland TD, Harcourt RL, Russell RJ, Oakeshott JG (2002) Identification of an opd (organophosphate degradation) gene in an Agrobacterium isolates. Appl Environ Microbiol 68(7):3371–3376. https://doi.org/10.1128/aem.68.7.3371-3376.2002
Jain R, Garg V (2013) Enzymatic degradation of monocrotophos by extracellular fungal OP hydrolases. Appl Biochem Biotechnol 171(6):1473–1486. https://doi.org/10.1007/s12010-013-0438-1
Jia KZ, Cui ZL, He J, GuoP Li SP (2006) Isolation and characterization of a denitrifying monocrotophos-degrading Paracoccus sp. M-1. FEMS Microbiol Lett 263(2):155–162. https://doi.org/10.1111/j.1574-6968.2006.00389.x
KaviKarunya S (2012) Biological degradation of chlorpyrifos and monocrotophos by bacterial isolates. Int J Pharm Biol Arch 3(3):685–691
King EJ, Abul-Fadl MAM, Walker PG (1951) King-Armstrong phosphatase estimation by the determination of liberated phosphate. J Clin Pathol 4(1):85
Lee PW, Fukuto JM, Hernandez H, Stearns SM (1990) Fate of monocrotophos in the environment. J Agric Food Chem 38:567–573
Lichtenthaler HK (1987) Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. In: Packer L, Douce R (eds) Methods in enzymology, vol 148. Academic Press, pp 350–382. https://doi.org/10.1016/0076-6879(87)48036-1
Liu T, Xu S, Lu S, Qin P, Bi B, Ding H, Liu Y, Guo X, Liu X (2018) A review on removal of organophosphorus pesticides in constructed wetland: performance, mechanism and influencing factors. Sci Total Environ 651(2):2247–2268. https://doi.org/10.1016/j.scitotenv.2018.10.087
Lowry OH, Rosebrough NJ, Farr AL, Randall RJ (1951) Protein measurement with the folin phenol reagent. J Biol Chem 193:265–275
McMullan G, Harrington F, Quinn JP (1992) Metabolism of phosphonoacetate as the sole carbon and phosphorus source by an environmental bacterial isolate. Appl Environ Microbiol 58(4):1364–1366
Mishra RK, Prakash O, Alam M, Dikshit A (2010) Influence of plant growth promoting rhizobacteria (PGPR) on the productivity of Pelargonium Graveolens l. herit. Rec Res Sci Technol 2(5):53–57
Moriya Y, Shigemizu D, Hattori M, Tokimatsu T, Kotera M, Goto S, Kanehisa M (2010) PathPred: an enzyme-catalyzed metabolic pathway prediction server. Nucleic Acids Res 38(Issue suppl 2):W138–W143. https://doi.org/10.1093/nar/gkq318
Nautiyal CS (1999) An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiol Lett 170(1):265–270. https://doi.org/10.1111/j.1574-6968.1999.tb13383.x
Pailan S, Gupta D, Apte S, Krishnamurthi S, Saha P (2015) Degradation of organophosphate insecticide by a novel Bacillus aryabhattai strain SanPS1, isolated from soil of agricultural field in Burdwan, West Bengal, India. Int Biodeterior Biodegrad 103:191–195. https://doi.org/10.1016/j.ibiod.2015.05.006
Panas P, Ternan NG, Dooley JS, McMullan G (2006) Detection of phosphonoacetate degradation and phnA genes in soil bacteria from distinct geographical origins suggest its possible biogenic origin. Environ Microbiol 8(5):939–945. https://doi.org/10.1111/j.1462-2920.2005.00974.x
Ramesh A, Sharma SK, Sharma MP, Yadav N, Joshi OP (2014) Inoculation of zinc solubilizing Bacillus aryabhattai strains for improved growth, mobilization and biofortification of zinc in soybean and wheat cultivated in Vertisols of central India. Appl Soil Ecol 73:87–96. https://doi.org/10.1016/j.apsoil.2013.08.009
Rangaswamy V, Venkateswarlu K (1992) Degradation of selected insecticides by bacteria isolated from soil. Bull Environ Contam Toxicol 49(6):797–804
Rissato SR, Galhiane MS, Fernandes JR, Gerenutti M, Gomes HM, Ribeiro R, Almeida MVD (2015) Evaluation of Ricinus communis L. for the phytoremediation of polluted soil with organochlorine pesticides. Biomed Res Int 2015:1–8. https://doi.org/10.1155/2015/549863
Schwyn B, Neilands JB (1987) Universal chemical assay for the detection and determination of siderophores. Anal Biochem 160(1):47–56. https://doi.org/10.1016/0003-2697(87)90612-9
Shafiani S, Malik A (2003) Tolerance of pesticides and antibiotic resistance in bacteria isolated from wastewater-irrigated soil. World J Microbiol Biotechnol 19(9):897–901. https://doi.org/10.1023/B:WIBI.0000007290.94694.4f
Sharma PK, Rajput SK (2012) A new sensitive and facile spectrophotometric determination of monocrotophos in environmental, biological and agricultural samples. Int J Sci Eng Res 3:10–23
Shivaji S, Chaturvedi P, Begum Z et al (2009) Janibacter hoylei sp. nov., Bacillus isronensis sp. nov. and Bacillus aryabhattai sp. nov., isolated from cryotubes used for collecting air from the upper atmosphere. Int J Syst Evol Microbiol 59(12):2977–2986
Siddique T, Okeke BC, Arshad M, Frankenberger WT (2003) Biodegradation kinetics of endosulfan by Fusarium ventricosum and a Pandoraea species. J Agric Food Chem 51(27):8015–8019. https://doi.org/10.1021/jf030503z
Singer AC, Crowley DE, Thompson IP (2003) Secondary plant metabolites in phytoremediation and biotransformation. Trends Biotechnol 21(3):123–130. https://doi.org/10.1016/S0167-7799(02)00041-0
Singh BK, Walker A (2006) Microbial degradation of organophosphorus compounds. FEMS Microbiol Rev 30(3):428–471. https://doi.org/10.1111/j.1574-6976.2006.00018.x
Subhas, Singh DK (2003) Utilization of monocrotophos as phosphorus source by Pseudomonas aeruginosa F10B and Clavibacter michiganense subsp. insidiosum SBL 11. Can J Microbiol 49(2):101–109. https://doi.org/10.1139/w03-013
Sun L, Zhu S, Yang Z, Chen Q et al (2016) Degradation of monocrotophos by Starkeya novella YW6 isolated from paddy soil. Environ Sci Pollut Res Int 23(4):3727–3735. https://doi.org/10.1007/s11356-015-5606-0
Tamura K, DudleyJ Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24(8):1596–1599. https://doi.org/10.1093/molbev/msm092
Tang M, You M (2012) Isolation, identification and characterization of a novel triazophos-degrading Bacillus sp. (TAP-1). Microbiol Res 167(5):299–305. https://doi.org/10.1016/j.micres.2011.10.004
Truu J, Truu M, Espenberg M, Nõlvak H, Juhanson J (2015) Phytoremediation and plant-assisted bioremediation in soil and treatment wetlands: a review. Open Biotechnol J 9:85–92. https://doi.org/10.2174/1874070701509010085
Velázquez-Fernández JB, Martínez-Rizo AB, Ramírez-Sandoval M, Domínguez-Ojeda D (2012) Biodegradation and bioremediation of organic pesticides. In: Soundararajan RP (ed) Pesticides-recent trends in pesticide residue assay. IntechOpen, pp 253–272. http://dx.doi.org/10.5772/46845
Xiong Z, Lai B, Yang P (2018) Insight into a highly efficient electrolysis-ozone process for N,N-dimethylacetamide degradation: quantitative analysis of the role of catalytic ozonation, fenton-like and peroxone reactions. Water Res 140:12–23. https://doi.org/10.1016/j.watres.2018.04.030
Yi T, Lee EH, Ahn YG, Hwang GS, Cho KS (2011) Novel biodegradation pathways of cyclohexane by Rhodococcus sp. EC1. J Hazard Mater 191(1–3):393–396. https://doi.org/10.1016/j.jhazmat.2011.04.080
Acknowledgements
The authors would like to thank the management of Vellore Institute of Technology for providing all the resources and technical support aided to carry out the research. Heartfelt thanks to Dr. S. Vino and Dr. S. Sajitha Lulu for providing help for in silico studies. Authors extend their gratitude to DST–FIST cum VIT Funded Scanning Electron Microscope Lab Facility, SIF-VIT for GC–MS and FTIR facilities and TBI–VIT for providing and HPLC facility to carry out analytical studies.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors would like to declare that they have no conflict of interest.
Additional information
Editorial responsibility: Ta Yeong Wu.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Dash, D.M., Osborne, J.W. Biodegradation of monocrotophos by a plant growth promoting Bacillus aryabhattai (VITNNDJ5) strain in artificially contaminated soil. Int. J. Environ. Sci. Technol. 17, 1475–1490 (2020). https://doi.org/10.1007/s13762-019-02432-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13762-019-02432-1