Abstract
The recognition of adverse environmental impact of acid rain is a prerequisite for finding feasible approaches to alleviate such damage to plants. We studied the regulation of nitrogen and phosphorus absorption by plasma membrane H+-ATPase and its expression at transcriptional levels in rice roots exposed to acid rain under hydroponic conditions. At pH 5.0 and 3.5, acid rain increased activity of plasma membrane H+-ATPase by increasing transcriptional levels of OSA1, 4, 5, 6, 7, 8, 9, and 10 and promoted ammonium absorption. However, nitrates and phosphorus contents in roots were decreased by acid rain (pH 3.5) due to a decrease in the H+ gradient. At pH 2.5, acid rain decreased nitrogen and phosphorus content in roots by decreasing plasma membrane H+-ATPase activity and its expression at transcriptional levels (OSA1–OSA10) as well as synthesis of ATP and inhibited the growth. After a 5-day recovery (without acid rain), all the parameters in roots treated with acid rain (pH 5.0 or 3.5) were greater than the data measured during the exposure period. However, all the parameters in roots treated with acid rain (pH 2.5) could not be recovered because heavy acid rain caused irreversible inhibition on plasma membrane H+-ATPase activity. Hence, we concluded that plasma membrane H+-ATPase plays a role in adaptation of rice seedlings to acid rain by regulating the absorption of nitrogen and phosphorus.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Acid rain is one of the major global environmental pollutants and severely affects agricultural and forest production (Menz and Seip 2004; Andonova 2007; Sanchez et al. 2010). Previous studies show that acid rain not only has direct negative effects on plants by inhibiting plant growth, reducing photosynthetic rate, damaging cell membrane intercity, and destroying ultrastructure of chloroplast, but also indirectly influences soil properties and microbial composition (Ling et al. 2010; Wen et al. 2011; Sun et al. 2014; Zhang et al. 2014). When soil pH is lowered to 4, most of nutritional elements such as magnesium, calcium, phosphorus, and soluble nitrogen in plant roots are decreased, resulting in nutrient deficiency and abnormal growth of plants (Sun et al. 2013; Du et al. 2014). In addition, acid rain increases extracellular concentration of H+ and can be a threat to membrane structure by exchanging with positive ions (which is the most important in calcium) in membrane, resulting in membrane permeability and hence increased intracellular leakage. Therefore, loss of mineral elements in the leaves by acid rain precedes change in appearance (DeHayes et al. 1999; Tarhanen et al. 1999). Remarkably, the loss of nutritional elements in plants affects inevitably growth and physiological metabolisms in plants. Theoretically, nutritional elements in plants depend on absorption by roots from soil (Hodge et al. 2009; Farid et al. 2015; Pereira et al. 2015). Hence, it should be informative to know effects of acid rain on the nutrient absorption of plants for clarifying the mechanism of plant tolerance to acid rain. The information will be helpful to find ways to alleviate such damage caused by acid rain.
Plasma membrane H+-ATPase constitutes a family of proton pumps driven by hydrolysis of ATP and is found exclusively in the plasma membrane of plants. The primary role of plasma membrane H+-ATPase is to provide an energy source for nutrient transport into the cell and extrusion of positive charges (H+), thus forming membrane potential (Palmgren 2001). Zhu et al. (2009) found a high regulation of various plasma membrane H+-ATPases in rice roots is adapted to rhizosphere acidification caused by ammonium absorption. Zeng et al. (2012) also found an involvement of plasma membrane H+-ATPase in the stimulated uptake of phosphorus by rice roots supplemented with ammonium. Thus, we presume that the regulation of plasma membrane H+-ATPase on absorption of nutrient could be important to clarify the tolerance of plants to acid rain stress. In our previous study (Liang et al. 2015), we found that plasma membrane H+-ATPase plays a role in adaptation of plants to acid rain by regulating intracellular pH for avoiding the damage to intensity of plasma membrane. However, there is limited literature on the regulation mechanism of plasma membrane H+-ATPase on absorption of nitrogen (ammonium and nitrates) and phosphors in plant roots exposed to acid rain stress. Hence, it would be interesting to reveal the adaptation of plants to acid rain from a new perspective of plasma membrane H+-ATPase regulation during nutrient absorption in plants stressed by acid rain. The exposure of plants to acid rain is a discontinuous process where plants are first exposed to acid rain followed by a restoration period. Therefore, it is essential to clarify the adaptation of plants during exposure and recovery periods being closed to the real situation when acid rain happened. Based on the findings, necessary measures to eliminate the negative effects of acid rain will be possible.
This study aimed at (1) clarifying response of plasma membrane H+-ATPase activity and its expression at transcriptional levels, intracellular H+, and ATP content in rice roots to different pHs during exposure and recovery periods; (2) revealing response of the ammonium, nitrates, and phosphorus content in rice roots to acid rain at different pHs during exposure and recovery periods; the relationship between plasma membrane H+-ATPase activity and nutrient absorption during exposure and recovery periods; and (3) studying response of the biomass and root cap ratio of rice seedlings to acid rain at different pHs during exposure and recovery periods. These results will help us understand the adaptation of plant to acid rain stress and can provide a new direction for finding effective ways to alleviate the damage to plants caused by acid rain.
Materials and methods
Plant material and culture conditions
Seeds of rice “Huaidao 8” (Oryza sativa) (Wuxi Seed CO., Ltd., China) were surface-sterilized with HgCl2 solution (0.1 %, w/v) for 5 min and washed with distilled water several times. After soaking in distilled water for 12 h, the seeds were placed in a dish (90 mm) under-laid with three layers of filter paper and germinated in an incubator at 25 ± 1 °C. After 2 days, the seedlings were transferred to a plastic container with vermiculite. When the first two leaves appeared, rice seedlings were cultivated in routine nutrition solution [150 mM (NH4)2SO4, 24 mM KH2PO4, 42 mM K2SO4, 120 mM CaCl2·H2O, 120 mM MgSO4·7H2O, 60 mM Na2SiO3·9H2O, 1.08 mM MnCl2·4H2O, 2.4 mM H3BO3, 2.40 mM Fe(III)-EDTA, 46.82 μM Na2MoO4·2H2O, 92.39 μM ZnSO4·7H2O, and 38.40 μM CuSO4·5H2O, pH 5.5] prepared by the method provided by the International Rice Research Institute (Zhu et al. 2009) in a growth chamber with a light intensity of 300 mol·m−2·s−1 photosynthetically active radiation, temperature of 25 °C/20 °C (14 h/10 h), relative humidity of 70 %/80 % (day/night), and the nutrient solution was renewed every 5 d to stabilize pH. After 30 days, rice seedlings were used to do experiments.
SAR treatment
Acid rain is formed from SO2 and nitrous oxides (NOx) emitted to the atmosphere. Simulated acid rain (SAR) was prepared by mixing concentrated H2SO4 and HNO3 in a ratio of 3:1 (v/v), similar to the ratio of SO4 2−: NO3 − in ambient rain in South China (Zhang et al. 1996; Liang and Wang 2013). The solution was diluted with deionized water to pH 5.0, 3.5, and 2.5. SAR (pH 5.0, 3.5, and 2.5) was sprayed at 24-h intervals on the leaves of rice seedlings till drops began to fall. At the same time, the pH of nutrient solution was adjusted to 5.0, 3.5, and 2.5. As the control, same amount of deionized water (pH 7.0) was applied to rice leaves. All treatments were done in triplicate. After a 5-day SAR treatment, half of the rice seedlings were collected for analysis. The rest of rice seedlings were cultured for another 5 days without SAR, same with the control conditions, and then were collected for analysis.
Determination of ATP content
ATP was extracted from rice roots according to the method described by Liu et al. (2006). Rice roots (2 g) was rapidly frozen in liquid nitrogen and homogenized into powder. Adenosine phosphates were extracted from the powder with perchloric acid (0.6 mol L−1, 10 mL) in the ice bath for 1 min by the method described by Yang et al. (2002). The mixture was centrifuged at 6000×g for 10 min at 4 °C. Supernatant (6 mL) was quickly neutralized to pH 6.5–6.8 with KOH (1 mol·L−1) and kept on an ice bath for 30 min. The filtrate solution was filtered again through a 0.45-mm filter. The final filtrate solution was made up to 8 mL and then stored at −30 °C prior to the analysis. ATP content was measured by high-performance liquid chromatography. High-performance liquid chromatography separation was achieved using continuous gradient elution. The elution programs were as follows: 0 min 100 % A, 0 % B; 2 min 95 % A, 5 % B; 4 min 80 % A, 20 % B; 5.3 min 75 % A, 25 % B and 6 min 100 % A, 0 % B. Mobile phase A consisted of dipotassium hydrogen phosphate (0.06 mol·L−1) and potassium dihydrogen phosphate (0.04 mol·L−1), whereas mobile phase B consisted of 100 % acetonitrile. Finally, the program took a further 1 min to return to the initial conditions and stabilize. Flow rate of the mobile phase was 1.2 mL·min−1, while the injection volume was 20 mL. The total retention time was about 5 min, and the gradient was run for 6 min to ensure full separation.
Hydrolytic activity of plasma membrane H+-ATPase measurement
Rice roots were ground in ice-cold homogenization buffer with a mortar and pestle. The homogenization buffer contained 250 mM sucrose, 250 mM KI, 2 mM EGTA, 10 % (v/v) glycerol, 0.5 % (w/v) bovine serum albumin, 2 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 5 mM 2-mercaptoethanol, and 50 mM bis–tris propane-2-(N-morpholino) ethanesulfonic acid (pH 7.8). The homogenate (adjusted to a grinding medium–tissue ratio of 4 mL g−1 fresh weight) was filtered through two layers of Miracloth (Calbiochem-Novabiochem Corporation, San Diego, CA) and centrifuged in a swinging bucket rotor at 11,500×g (Optima TM L-80 XP Ultracentrifuge, Beckman Coulter Inc. Fullerton, CA) at 0 °C for 10 min. The supernatants were centrifuged at 87,000 × g for 35 min. The microsomal pellets were resuspended in phosphate buffer that contained 250 mM sucrose, 3 mM KCl, and 5 mM KH2PO4 (pH 7.8). The microsomal membrane preparation was fractionated by two-phase partitioning in aqueous dextran T500 (Sigma, St. Louis, MO) and polyethylene glycol (Sigma) according to the method provided by Larsson et al. (1987) and Klobus and Buczek (1995). Phase separations were carried out in a series of 32-g phase systems that contained 6.1 % (w/w) dextran T500, 6.1 % (w/w) polyethylene glycol 3350, 250 mM sorbitol, 3 mM KCl, and 5 mM bis–tris propane-2-(N-morpholino) ethanesulfonic acid (pH 7.8). The plasma membrane obtained by this procedure was used to determine the hydrolytic activity of plasma membrane H+-ATPase. Protein was measured according to the method described by Bradford (1976).
Hydrolytic activity of plasma membrane H+-ATPase was determined by measuring the Pi amount after 30-min hydrolysis. The reaction reagents (0.5 mL) contained 30 mM bis–tris propane-2-(N-morpholino) ethanesulfonic acid (pH 6.5), 5 mM MgSO4, 50 mM KCl, 50 mM KNO3, 1 mM Na2MoO4, 1 mM NaN3, 0.02 % (w/v) Brij58, 5 mM ATP-Na2, and 30 mL protein (plasma membrane) in the presence and absence of inhibitors: 50 mM KNO3 (for tonoplast H+-ATPases), 1 mM Na2MoO4 (for nonspecific phosphatases), 1 mM NaN3 (for mitochondrial membrane H+-ATPases), or 0.3 mM Na3VO4 (for plasma membrane H+-ATPases). The reaction was performed at 30 °C for 30 min and stopped by adding 1 mL stopping reagent [2 % (v/v) concentrated H2SO4, 5 % (w/v) sodium dodecyl sulfate, and 0.7 % (w/v) (NH4)2MoO4] followed immediately by adding 50 mL 10 % (w/v) ascorbic acid. After 10 min, 1.45 mL arsenite-citrate reagent [2 % (w/v) sodium citrate, 2 % (w/v) sodium m-arsenite, and 2 % (w/v) glacial acetic acid] was added to prevent phosphate from releasing because of H+-ATPase activity from ATP hydrolysis under acidic conditions (Baginski et al. 1967). After 30 min, the absorbance was measured by spectrophotometer at 820 nm. Plasma membrane H+-ATPase activity was calculated as phosphate liberated in excess of boiled-membrane control. By analyzing ATPase hydrolytic activity of the membrane fraction in the presence and absence of inhibitors as described by Wakeel et al. (2010), there were no differences in purity for membranes from the control and SAR-treated plants (Table S1 and Table S2).
RNA extraction and real-time RT-PCR
To evaluate the expression of 10 genes [OSA1 (GenBank accession: AJ439999.1), OSA2 (GenBank accession: AJ440000.1), OSA3 (GenBank accession: AJ440001.1), OSA4 (GenBank accession: AJ440002.1), OSA5 (GenBank accession: AJ440216.1), OSA6 (GenBank accession: AJ440217.1), OSA7 (GenBank accession: AJ440218.1), OSA8 (GenBank accession: AJ440219.1), OSA9 (GenBank accession: AJ440220.1), OSA10 (GenBank accession: AJ440221.1)] encoding plasma membrane H+-ATPase, real-time PCR analysis with specific primers for each gene was performed. For the normalization of expression of each OSA gene, the actin gene (GenBank accession no: NM197297) was used as the internal standard. All primers were designed on the basis of the sequences published in GenBank by Beacon Designer 7.0 (Premier Biosoft, Palo Alto, CA) (Liang et al. 2014), being the same as used in previous study (Liang et al. 2015). Total RNA from roots of rice seedlings was isolated using Tri-Reagent (Sigma-Aldrich, St. Louis) according to the manufacturer’s instructions. Total RNA yield was determined using NanoDrop Spectrophotometer ND-1000 (Thermo Scientific, Wilmington, DE), and the 260/280 nm ratio showed expected values between 1.9 and 2.0. To avoid any DNA contamination, RNA samples were treated with RNase-free DNase I (Thermo Scientific, Wilmington, DE) and then reverse-transcribed into first-strand cDNA with a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Inc., Foster City, CA) according to the manufacturer’s instructions. The cDNA was then used as the template for PCR amplification with SYBRGreen PCR Master Mix kit (Applied Biosystems Inc., Foster City, CA). PCR cycling was performed with Real-Time PCR System (Bio-Rad, Hercules, CA). After an initial denaturation at 95 °C for 2 min, 40 cycles were performed (melting at 94 °C for 10 s and annealing and extension at 60 °C for 40 s). Agarose gel electrophoresis was done to identify the product of PCR. Relative expression abundance was quantified using comparative quantization analysis settings (Optical System software, version 1.0).
Protoplasts isolation and intracellular H+ measurement
Fresh rice roots were cut into pieces (0.5 mm length) and put into a test tube. The enzyme solution containing 2 % cellulase (lyophilized, 10.0 U mg−1), 0.5 % macerase (lyophilized, 0.5 U mg−1), 0.1 % (w/v) pectolyase (Y-23), 5 mmol L−1 MES, and 0.45 mmol L−1 mannitol was added into tubes with pieces of roots. Tubes were shaken at 40 rpm at 30 °C in the dark for 3 h. Then, the reaction mixture was filtered with plug (400 meshes). The filtrate was centrifuged at 1000 × g for 5 min. The supernatant was removed by pipette, and the precipitate was washed twice with the solution containing 3 % sucrose (w/v) and 0.4 mol L−1 mannitol. Protoplasts floating on the surface of washing solution were collected and passed through 50-μm-pore-size nylon mesh filter. Microscope and hemocytometer were used to adjust the concentration of protoplasts to 5 × 106 mL−1 for detecting intracellular H+.
Protoplasts isolated from rice roots were incubated with the pH-sensitive fluorescent dye [2′, 7′-bis (carboxyethyl)-5(6)-carboxy] (BCECF) at 37 °C for 60 min under an atmosphere of 21 % O2–5 % CO2. Protoplasts were then washed with HEPES-buffered salt solution (pH 7.4) at 37 °C for 15 min to remove extracellular dye and retain complete desertification of cytosolic dye. Radiometric measurement of BCECF fluorescence was taken by a workstation (Intracellular Imaging Inc, Cincinnati, OH) consisting of a Nikon TSE 100 Ellipse inverted microscope with epifluorescence attachments. Wavelength of excited light was 500 nm, and wavelength of emitted light was 530 nm. The fluorescence signal was adjusted to zero background for cells without dying (Undem et al. 2012). The change of intracellular H+ in rice roots treated with acid rain at different pHs was evaluated on the basis of the BCECF fluorescence compared to the control.
Ammonium and nitrates concentration measurement
Fresh rice roots (1 g) was harvested and then cut into pieces. After being grounded in deionized water with a mortar and pestle, roots were transferred to Erlenmeyer flask with 20 mL deionized, and then oscillated for 3 min. After centrifugation (4000×g, 5 min), a 2 mL of supernatant was transferred to test tubes. The method of Berthelot (Scheiner 1976) was used to determine ammonium; the absorbance was measured at 550 nm by spectrophotometer (JINGHRI, Shanghai, China).
Nitrates content was measured according to the method described by Miranda et al. (2001), and fresh roots (0.5 g) were put into graduated test tube with 20 mL deionized water. Test tubes blocked with plugs were put into boiling water for 30 min and then cooled with flowing water. Extraction was filtered and diluted to 25 mL with deionized water. A 0.1 mL of extracting solution was transferred to test tubes with 0.4 mL 5 % (w/v) salicylic acid–H2SO4 solution. The mixture was shaken for 20 min at ambient temperature. After adding 9.5 mL 8 % NaOH, the mixture was shaken, cooling to room temperature. The absorbance was measured at 410 nm by a spectrophotometer (JINGHRI, Shanghai, China).
Total phosphorus concentration determination
Dried roots of each treatment were ground finely enough to pass a 1.0-mm screen. A total of 0.1 g powder was digested with 5 mL H2SO4 (98 %, v/v) and 3 mL hydrogen peroxide (30 %, v/v). After cooling, the digested sample was diluted to 100 ml with distilled water. The phosphorus concentration in the solution was measured according to the molybdate blue method (Chang et al. 2009). The absorbance was measured at 700 nm by a spectrophotometer (JINGHRI, Shanghai, China).
Growth determination
After harvesting, each rice seedling was weighed to determine the total biomass and roots fresh weight. Then, roots were dried separately in a forced-air oven at 70 °C for about 48 h for determination of dry weight. Root cap ratio is the ratio of the dry weight of root to the dry weight of shoot.
Statistical analysis
Data were presented as mean ± standard deviation. The difference significance between different treatments was analyzed by the one-way analysis of variance (ANOVA) using SPSS 11.5 (P < 0.05).
Result and discussion
Effect of SAR on ATP content, plasma membrane H+-ATPase activity, and intracellular H+ in rice roots
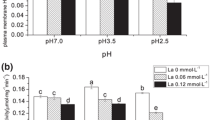
The plasma membrane H+-ATPase is a universal electrogenic H+ pump that provides the primary electrical gradient and proton motive force for absorbing nitrogen and phosphorus by hydrolysis of ATP as energy source to pump H+ across plasma membranes into the apoplast (Crawford and Glass 1998; Forde and Clarkson 1999; Arango et al. 2003; Santi et al. 2003; Vansuyt et al. 2003). We studied the effects of SAR (pH 5.0, 3.5, and 2.5) on ATP content (Fig. 1), plasma membrane H+-ATPase activity (Fig. 2), and intracellular H+ (Fig. 3) in rice roots. After a 5-day exposure, the ATP content in rice roots treated with SAR (pH 5.0, 3.5, and 2.5) was decreased by 15.7, 5.5, and 22.4 %, respectively, compared with that of the control (P < 0.05) (Fig. 1). As shown in Fig. 2, at pH 5.0 and 3.5, SAR increased the hydrolytic activity of plasma membrane H+-ATPase in rice roots by 89.9 and 17.6 % compared with that of the control, whereas SAR (pH 2.5) decreased the hydrolytic activity of plasma membrane H+-ATPase. The effects of SAR on intracellular H+ in rice roots are shown in Fig. 3. At pH 5.0 and 3.5, SAR increased intracellular H+ in rice roots by 45.2 % and 30.1 % compared with that of the control after a 5-day exposure. However, at pH 2.5, SAR decreased intracellular H + compared with that of the control. The increase in intracellular H+ caused by SAR (pH 5.0 or 3.5) was because of excessive accumulation of extracellular H+ (Ramos et al. 1976). Thus, the increase in intracellular H+ caused an increase in activity of plasma membrane H+-ATPase. Meanwhile, the decrease in ATP content in rice roots treated with SAR (pH 5.0 or 3.5) could have resulted from the hydrolysis of ATP catalyzed by plasma membrane H+-ATPase. Holyoak et al. (1996) also confirmed that the reductions in cellular ATP resulted from consumption of more ATP in Saccharomyces cerevisiae during growth period under acid between pH 5.7 and 3.8. When rice seedlings were exposed to SAR (pH 2.5), the activity of plasma membrane H+-ATPase was inhibited (Fig. 2). In addition, ATP content and intracellular H+ in rice roots were all decreased under SAR (pH 2.5). That is maybe the ATP synthesis was inhibited and cellular homeostasis was ruined in rice roots under SAR (pH 2.5). In the previous study (Liang et al. 2015), we found that high acidity of SAR (pH 2.5) decreased intracellular H+ in rice leaves because of the increase in leakage of plasma membrane under acid rain stress. The decrease in ATP content mainly resulted from the inhibition on the synthesis of ATP caused by high acidity of SAR.
Effect of simulated acid rain on intracellular H+ in rice roots. The intracellular H+ in rice root was expressed by relative BCECF fluorescence intensity which was measured by adjusting the fluorescence signal in cells without dying to zero. Significant difference at p < 0.05 during stress period is shown with different lowercase letters and, during recovery period, is shown with uppercase letters
After a 5-day recovery, ATP content (Fig. 1) and intracellular H+ (Fig. 3) in rice roots treated with SAR (pH 5.0) were at almost similar level with control, showing that the increase in intracellular H+ and the decrease in ATP content caused by SAR (pH 5.0) during the exposure period can be restored after a 5-day recovery. However, plasma membrane H+-ATPase activity (Fig. 2) was still higher than that of the control. Being different from that treated with SAR (pH 5.0), the intracellular H+ in rice roots treated with SAR (pH 3.5) was still higher than that of the control. That mainly resulted from more H+ entry into cells (the acidity of SAR was increased) and less pump-out (plasma membrane H+-ATPase activity was lower than that of pH 5.0 treatment group). However, the intracellular H+ in rice roots treated with SAR (pH 3.5) was lower than that measured during the exposure period, indicating that the intracellular H+ in rice roots under SAR (pH 3.5) can be decreased by the increase in plasma membrane H+-ATPase. For the group treated with SAR (pH 2.5), plasma membrane H+-ATPase activity in rice seedlings was still lower than that of the control after a 5-day recovery, indicating that SAR (pH 2.5) caused irrecoverable inhibition on the activity of plasma membrane H+-ATPase. The intracellular H+ in rice root treated with SAR (pH 2.5) was still lower than that of the control, but higher than that of the exposure period (Fig. 3). The changes in the intracellular H+ were related to osmotic adjustment and membrane permeability (Guo et al. 2009; Liang et al. 2015). In addition, ATP content in rice roots treated with SAR (pH 2.5) was still lower than that of the control, and lower than that measured during the exposure period, indicating that the ATP synthesis was inhibited by high acidity (pH 2.5), and the inhibition exceeded the self-regulating capacity of rice seedlings.
Effect of SAR on expression of plasma membrane H+-ATPase at transcriptional levels in rice roots
The relative transcriptional levels of genes (OSA1–OSA10) of plasma membrane H+-ATPase in rice roots treated with SAR during exposure and recovery periods are shown in Figs. 4, 5. After a 5-day exposure, at pH 5.0, SAR increased transcriptional levels of genes OSA1, OSA3, OSA4, OSA5, OSA6, OSA7, OSA8, OSA9, and OSA10, but the transcriptional level of OSA2 in roots was not significantly changed. At pH 3.5, SAR increased the transcriptional levels of genes OSA1, OSA4, OSA5, OSA6, OSA8, OSA9, and OSA10, and the transcriptional levels of OSA2, OSA3, and OSA7 in roots were not significantly changed. Under normal circumstances, genes such as subfamilies I (OSA1, 2, 3) and II (OSA5, 7) are expressed at higher level, whereas genes such as subfamilies III (OSA9), IV (OSA4, 6, 10), and V (OSA8) display their expression at lower or undetectable levels (Sperandio et al. 2011). Our results showed that transcriptional levels of genes of III, IV, and V subfamilies were higher under SAR (pH 5.0 or 3.5), to increase plasma membrane H+-ATPase activity. The increase in the transcriptional level of genes I (OSA1, 3), II (OSA5, 7), III (OSA9), IV (OSA4, 6, 10), and V (OSA8) in rice roots treated with SAR (pH 5.0) was higher than those in rice roots treated with SAR (pH 3.5), in line with the increased degree of the activity of plasma membrane H+-ATPase, indicating that SAR (pH 5.0 or 3.5) induced the transcriptional levels of all genes (except OSA2) in rice roots increased, and then increased the activity of plasma membrane H+-ATPase. The transcriptional level of OSA2 did not change under SAR (pH 5.0 or 3.5), so we inferred that OSA2 belongs to housekeeping gene. As Nicot et al. (2005) reported, housekeeping genes should not fluctuate during stress treatments. When rice roots were exposed to SAR (pH 2.5), transcriptional levels of 10 genes (OSA1–OSA10) were all lower than those of the control (Fig. 4b), and the activity of plasma membrane H+-ATPase was inhibited, indicating that SAR (pH 2.5) inhibited the activity of plasma membrane H+-ATPase by decreasing transcriptional levels of OSA1–OSA10.
a Agarose gel electrophoresis of the RT-PCR product for OSA1 and Actin during the stress period. The results of agarose gel electrophoresis [OSA1 (one of 10 genes of plasma membrane H+-ATPase) and Actin] were to verify the product of RT-PCR. b Relative expression of plasma membrane H+-ATPase at transcriptional level during stress period
a Agarose gel electrophoresis of the RT-PCR product for OSA1 and Actin during the recovery period. The results of agarose gel electrophoresis [OSA1 (one of 10 genes of plasma membrane H+-ATPase) and Actin] were to verify the product of RT-PCR. b Relative expression of plasma membrane H+-ATPase at transcriptional level during recovery period
After a 5-day recovery (Fig. 5b), the transcriptional levels of genes OSA1, OSA4, OSA5, OSA6, OSA7, OSA8, OSA9, and OSA10 in rice roots treated with SAR (pH 5.0) were still higher than those of the control, whereas transcriptional levels of OSA2 and OSA3 were at almost similar level with control. The up-regulated transcriptional levels of genes continued to increase the activity of plasma membrane H+-ATPase, contributing to the adaptation of rice roots to acid rain stress. For the group treated with SAR (pH 3.5), transcriptional levels of genes OSA1, OSA4, OSA5, OSA6, OSA8, OSA9, and OSA10 in rice roots were still higher than those of the control, whereas transcriptional levels of OSA2, OSA3, and OSA7 were at almost similar with control (Fig. 5b). When rice seedlings were treated with SAR (pH 2.5), transcriptional levels of 10 genes in rice roots were all still lower than those of the control, indicating that the inhibition of transcriptional levels of genes caused by strong acid rain (pH 2.5) cannot be recovered.
Effects of SAR on ammonium, nitrates, and phosphorus absorption in rice roots
Nitrogen and phosphorus are essential nutrients for plant growth and development (Tischer et al. 2015). Ammonium and nitrates are the two major nitrogen sources taken up by plant roots (Forde and Clarkson 1999). The transmembrane transport of ammonium is an active transport process, depending on ammonium transporter (Amtmann and Blatt 2009) on the cell membrane and proton motive force provided by plasma membrane H+-ATPase (Zhu et al. 2012). Our results showed that at pH 5.0 or 3.5 SAR promoted the absorption of ammonium in rice roots (Fig. 6). That mainly resulted from the increase in H+ gradient under SAR (pH 5.0 or 3.5). In addition, the absorption of ammonium can lead to rhizosphere acidification (Alvarez-Pizarro et al. 2011), and external acidification can promote the absorption of ammonium. However, for the group treated with SAR (pH 5.0), ammonium content in rice roots was decreased by 33.5 % compared with that of the control (Fig. 6), implying that the decrease in the activity of plasma membrane H+-ATPase and ATP content (Figs. 1, 2) decreased the absorption of ammonium. And it could also be because SAR (pH 2.5) caused the damage to integrity of plasma membrane, and led to the outflow of substances (ammonium) inside (Liang et al. 2015). The absorption of both nitrates and phosphorus is active absorption against electrochemical potential by plasma membrane H+-ATPase providing proton motive force (Miller and Smith 1996). The absorption of nitrates is via 2 H+:1 NO3 − symports, and the absorption of phosphorus is via an anion/H+ co-transport process (Glass et al. 2002; Zeng et al. 2012). In our experiments, at pH 5.0, SAR had no obvious effect on the absorption of nitrates and phosphorus in rice roots (Figs. 7, 8). At the same treatment, the activity of plasma membrane H+-ATPase and the transcriptional levels of all genes (except OSA2) were increased (Figs. 2, 4b), being beneficial for the hydrolysis of ATP to provide energy to maintain the absorption of nitrates and phosphorus. However, at pH 3.5, SAR decreased the concentration of nitrates and phosphorus by 26.3 % and 32.0 % compared with those of the control (Figs. 7, 8). That could have resulted from the decrease in proton gradient by the increase in the intracellular H+ (Fig. 3). The low external pH influences the electrical potential difference between the plasma membrane and the surrounding environment (Reid and Hayes 2003). The absorption of nitrates and phosphorus under SAR (pH 5.0 or 3.5) was different from ammonium because the absorption of nitrates and phosphorus needs to consume H+, whereas the absorption of ammonium produces H+ (Zhu et al. 2009). At pH 2.5, SAR decreased the concentration of nitrates and phosphorus in rice roots by 46.2 % and 63.0 % compared with those of the control (Figs. 7, 8), and that was due to the decrease in H+-ATPase activity (Fig. 2) and the decrease in ATP content (Fig. 1). In addition, at pH 2.5, SAR caused damage to the integrity of plasma membrane and led to the outflow of substances (nitrates and phosphorus) inside.
After a 5-day recovery, the ammonium concentration in rice roots treated with SAR (5.0 or 3.5) was still higher than that of the control (Fig. 6). At the same treatment, the intracellular H+ (Fig. 3) and ATP content (Fig. 1) were at almost similar level with control. We inferred that the excess ammonium stored in rice root during the exposure period could not be transported. The ammonium content in rice roots treated with SAR (pH 2.5) was lower than that of the control, and even worse than that measured during the exposure period (Fig. 6), resulting from the decrease in ATP content (Fig. 1), H+-ATPase activity (Fig. 2), and the increase in leakage of plasma membrane. The nitrates and phosphorus contents in rice roots treated with SAR (pH 5.0) had no obvious difference from that of the control (Figs. 7, 8). At the same treatment, the ATP content and intracellular H+ in rice roots were at almost similar level with control, and the activity of plasma membrane H+-ATPase in rice roots was increased. The phenomena showed that the increase in the activity of plasma membrane H+-ATPase was beneficial to maintaining the normal absorption of nitrates and phosphorus in rice roots under such SAR stress (pH 5.0). However, the content of nitrates and phosphorus in rice root treated with SAR (pH 3.5) was still lower than those of the control, but higher than those measured during the exposure period (Figs. 7, 8). Combined with the increase in plasma membrane H+-ATPase activity and the decrease in ATP content in rice root treated with pH 3.5 SAR, we found that the increase in the activity of plasma membrane H+-ATPase accelerated the hydrolysis of ATP during the recovery period. The nitrates and phosphorus contents in rice roots treated with SAR (pH 2.5) were still lower than that of the control, and also lower than that measured during the exposure period (Figs. 7, 8), indicating that the inhibition induced by SAR (pH 2.5) on nitrates and phosphorus absorption is unrecoverable. That could have resulted from the decrease in ATP content (Fig. 1) and plasma membrane H+-ATPase activity (Fig. 2) and an increase in the leakage of plasma membrane (Liang et al. 2015).
Effects of SAR on plant root cap ratio and biomass of rice seedlings
Growth can be used as an indicator to reflect indirectly if nutrient absorption in plants is limited under stress conditions. In our experiments, at pH 5.0, SAR did not significantly affect the root cap ratio and total biomass in rice seedlings after a 5-day exposure (Table 1). At the same treatment, an increase in the activity of plasma membrane H+-ATPase can maintain normal absorption of nitrates and phosphorus (Figs. 7, 8) and increased ammonium content (Fig. 6) in rice roots to ensure the normal growth of rice seedlings. Usually, sole ammonium nutrition at high concentration is toxic for many plants, inhibiting growth of plants (Guo et al. 2002; Wang and Li 2003). However, rice has a high tolerance to ammonium because excessive ammonium can be degraded or diffused in cell (Britto et al. 2001). However, at pH 3.5, SAR caused an increase in root cap ratio by 15.3 % and a decreased in the biomass of roots by 32.2 %, compared with those of the control (Table 1). The increase in root cap ratio indicated that the inhibition on rice leaves caused by SAR (pH 3.5) was heavier than that on rice roots. The decreased biomass was resulted from the decreased nitrates and phosphorus absorption in rice roots (Figs. 7, 8). In addition, at pH 2.5 SAR caused the reduction in the ratio of root to shoot and biomass of rice seedlings (Table 1). That was because low external pH (2.5) caused the absorption of ammonium, nitrates, and phosphorus decreased (Figs. 6, 7, 8) and the outflow of nutrient substances.
After a 5-day recovery, the root cap ratio and biomass in rice roots treated with SAR (pH 5.0) had no significant difference from those of the control (P < 0.05). Treated with SAR (pH 3.5), the root cap ratio was higher than that of the control, and the biomass of rice seedlings was restored at a certain degree (Table 1). This meant the biomass of rice leaves was better than that of roots. The root cap ratio and biomass of rice seedlings treated with SAR (pH 2.5) were still lower than those of the control, and those measured during the exposure period (Table 1). This meant the inhibition caused by SAR (pH 2.5) to plant growth cannot be restored because deficiency in nitrogen and phosphors nutrient was heavier by the decrease in plasma membrane H+-ATPase activity, indicating that the damage caused by SAR (pH 2.5) to growth exceeded the regulating capacity of plasma membrane H+-ATPase activity on absorption of nitrogen and phosphorus in rice roots.
Correlation analysis between nutritional content, growth, and H-ATPase activity with SAR
The correlation coefficients are shown in Table 2. During the exposure period, the positive correlation was significant between ammonium content, nitrates content, phosphorus content, and H+-ATPase activity (p < 0.05). And we also found that the correlation between intracellular H+, ATP content, and root cap ratio was significantly positive (p < 0.05). During the recovery period, the correlation was significantly positive between ammonium content, nitrates content, phosphorus content, root cap ratio, and H+-ATPase activity. In addition, we also found positive correlation between ammonium content, nitrates content, phosphorus content, biomass, and ATP content (p < 0.05).
Conclusion
Our study confirmed that: (1) at pH 5.0 or 3.5, SAR increased in the activity of plasma membrane H+-ATPase by inducing the high transcriptional levels of OSA1, 4, 5, 6, 7, 8, 9, and 10 in rice roots and also increased intracellular H+ and decreased ATP content in rice roots. However, significantly different change in range of plasma membrane H+-ATPase, intracellular H+, and the decrease in ATP content resulted in different H+ gradient and nutritional content in rice roots under SAR (pH 5.0 or 3.5). At pH 5.0, SAR was beneficial to providing energy to absorb ammonium and maintain normal absorption of nitrates and phosphorus. In addition, SAR (pH 3.5) decreased H+ gradient and limit the absorption of nitrates and phosphorus, being harmful to the growth of rice. (2) High acidity of SAR (pH 2.5) inhibited plasma membrane H+-ATPase activity by decreasing the expression of plasma membrane H+-ATPase at transcriptional levels (OSA1–OSA10), inhibited the absorption of nitrates and phosphorus, and limited the growth of rice seedlings. (3) During the recovery period, the increase in the activity of plasma membrane H+-ATPase and its increased expression at transcriptional levels (OSA1, 4, 5, 6, 8, 9, and 10) in rice roots treated with SAR (pH 3.5) was beneficial for the adaptation of rice seedlings to acid rain stress by alleviating the inhibition on nitrogen and phosphorus absorption. However, regulating effects on nitrogen and phosphorus absorption by plasma membrane H+-ATPase in rice seedlings depended on the acidity of SAR. These results will help us recognize the adverse environmental impact of acid rain; understand the adaptation of plant to acid rain stress from changes in genetic expression, biochemical, and growth; and develop measures to alleviate or eliminate this damage.
References
Alvarez-Pizarro JC, Gomes-Filho E, Prisco JT, Grossi-de-Sa MF, Neto OBO (2011) NH4 +-stimulated low-K+ uptake is associated with the induction of H+ extrusion by the plasma membrane H+-ATPase in sorghum roots under K+ deficiency. J Plant Physiol 168:1617–1626
Amtmann A, Blatt MR (2009) Regulation of macronutrient transport. New Phytol 181:35–52
Andonova LB (2007) Acid rain in a wider Europe: the post-communist transition and the future European acid rain policies. In: Visgillio G (ed) Acid in the environment: lessons learned and future prospects. Springer, US, pp 151–173
Arango M, Gevaudant F, Oufattole M, Boutry M (2003) The plasma membrane proton pump ATPase: the significance of gene subfamilies. Planta 216:355–365
Baginski E, Foa P, Zak B (1967) Determination of phosphate: study of labile organic phosphate interference. Clin Chim Acta 15:155–158
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Britto DT, Siddiqi MY, Glass ADM, Kronzucker HJ (2001) Futile transmembrane NH4 + cycling: a cellular hypothesis to explain ammonium toxicity in plants. Proc Natl Acad Sci USA 98:4255–4258
Chang C, Hu Y, Sun S, Zhu Y, Ma G, Xu G (2009) Proton pump OSA8 is linked to phosphorus uptake and translocation in rice. J Exp Bot 60:557–565
Crawford NM, Glass ADM (1998) Molecular and physiological aspects of nitrate uptake in plants. Trends Plant Sci 3:389–395
DeHayes DH, Schaberg PG, Hawley GJ, Strimbeck GR (1999) Acid rain impacts on calcium nutrition and forest health alteration of membrane-associated calcium leads to membrane destabilization and foliar injury in red spruce. Bioscience 49:789–800
Du Y, Wei M, Reddy KR, Liu Z, Jin F (2014) Effect of acid rain pH on leaching behavior of cement stabilized lead-contaminated soil. J Hazard Mater 271:131–140
Farid M, Ali S, Ishaque W, Shakoor MB, Niazi NK, Bibi I, Dawood M, Gill RA, Abbas F (2015) Exogenous application of ethylenediamminetetraacetic acid enhanced phytoremediation of cadmium by brassica napus l. Int J Environ Sci Technol 12(12):3981–3992
Forde BG, Clarkson DT (1999) Nitrate and ammonium nutrition of plants: physiological and molecular perspectives. Adv. Bot. Res. 30:1–90
Glass AD, Britto DT, Kaiser BN, Kinghorn JR, Kronzucker HJ, Kumar A, Okamoto M, Rawat S, Siddiqi M, Unkles SE (2002) The regulation of nitrate and ammonium transport systems in plants. J Exp Bot 53:855–864
Guo S, Brück H, Sattelmacher B (2002) Effects of supplied nitrogen form on growth and water uptake of French bean(Phaseolus vulgaris L.) plants. Plant Soil 239:267–275
Guo R, Shi L, Yang Y (2009) Germination, growth, osmotic adjustment and ionic balance of wheat in response to saline and alkaline stresses. Soil Sci Plant Nutr 55:667–679
Hodge A, Berta G, Doussan C, Merchan F, Crespi M (2009) Plant root growth, architecture and function. Plant Soil 321:153–187
Holyoak C, Stratford M, McMullin Z, Cole M, Crimmins K, Brown A, Coote P (1996) Activity of the plasma membrane H+-ATPase and optimal glycolytic flux are required for rapid adaptation and growth of Saccharomyces cerevisiae in the presence of the weak-acid preservative sorbic acid. Appl Environ Microbiol 62:3158–3164
Klobus G, Buczek J (1995) The role of plasma membrane oxidoreductase activity in proton transport. J Plant Physiol 146:103–107
Larsson C, Widell S, Kjellbom P (1987) Preparation of high-purity plasma membranes. Methods Enzymol 148:558–568
Liang CJ, Wang W (2013) Antioxidant response of soybean seedlings to joint stress of lanthanum and acid rain. Environ Sci Pollut Res 20:8182–8191
Liang CJ, van Dijk JP, Scholtens IM, Staats M, Prins TW, Voorhuijzen MM, da Silva AM, Arisi ACM, den Dunnen JT, Kok EJ (2014) Detecting authorized and unauthorized genetically modified organisms containing vip3A by real-time PCR and next-generation sequencing. Anal Bioanal Chem 406:2603–2611
Liang CJ, Ge YQ, Su L, Bu JJ (2015) Response of plasma membrane H+-ATPase in rice (Oryza sativa) seedlings to simulated acid rain. Environ Sci Pollut Res 22:535–545
Ling DJ, Huang QC, Ouyang Y (2010) Impacts of simulated acid rain on soil enzyme activities in a latosol. Ecotox. Environ. Safe. 73:1914–1918
Liu H, Jiang Y, Luo Y, Jiang W (2006) A simple and rapid determination of ATP, ADP and AMP concentrations in pericarp tissue of litchi fruit by high performance liquid chromatography. Food Technol. Biotechnol. 44:531–534
Menz FC, Seip HM (2004) Acid rain in Europe and the United States: an update. Environ Sci Policy 7:253–265
Miller AJ, Smith SJ (1996) Nitrate transport and compartmentation in cereal root cells. J Exp Bot 47:843–854
Miranda KM, Espey MG, Wink DA (2001) A rapid, simple spectrophotometric method for simultaneous detection of nitrate and nitrite. Nitric Oxide-Biol. Chem. 5:62–71
Nicot N, Hausman JF, Hoffmann L, Evers D (2005) Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress.J. Exp. Bot. 56:2907–2914
Palmgren MG (2001) Plant plasma membrane H+-ATPases: powerhouses for nutrient uptake. Annu. Rev. Plant Physiol. Plant 52:817–845
Pereira SIA, Barbosa L, Castro PML (2015) Rhizobacteria isolated from a metal-polluted area enhance plant growth in zinc and cadmium-contaminated soil. Int J Environ Sci Technol 12(7):2127–2142
Ramos S, Schuldiner S, Kaback HR (1976) The electrochemical gradient of protons and its relationship to active transport in Escherichia coli membrane vesicles. Proc Natl Acad Sci 73:1892–1896
Reid R, Hayes J (2003) Mechanisms and control of nutrient uptake in plants. In: Jeon KW (ed) International review of cytology—a survey of cell biology. Elsevier Academic Press Inc, San Diego, pp 73–114
Sanchez DG, Rios GFL, Garcia MAH (2010) Acid rain and forest ecosystems. Rev Chapingo Ser Cienc For Am 16:187–206
Santi S, Locci G, Monte R, Pinton R, Varanini Z (2003) Induction of nitrate uptake in maize roots: expression of a putative high-affinity nitrate transporter and plasma membrane H+-ATPase isoforms. J Exp Bot 54:1851–1864
Scheiner D (1976) Determination of ammonia and Kjeldahl nitrogen by indophenol method. Water Res 10:31–36
Sperandio MVL, Santos LA, Bucher CA, Fernandes MS, de Souza SR (2011) Isoforms of plasma membrane H+-ATPase in rice root and shoot are differentially induced by starvation and resupply of NO3 − or NH4 +. Plant Sci 180:251–258
Sun Z, Wang L, Zhou Q, Huang X (2013) Effects and mechanisms of the combined pollution of lanthanum and acid rain on the root phenotype of soybean seedlings. Chemosphere 93:344–352
Sun Z, Wang L, Wang Q, Zhou Q, Huang X (2014) Interactive effects of cadmium and acid rain on photosynthetic light reaction in soybean seedlings. Environ Toxicol Chem 33:2013–2019
Tarhanen S, Metsarinne S, Holopainen T, Oksanen J (1999) Membrane permeability response of lichen Bryoria fuscescens to wet deposited heavy metals and acid rain. Environ Pollut 104:121–129
Tischer A, Werisch M, Dobbelin F, Camenzind T, Rillig MC, Potthast K, Hamer U (2015) Above- and belowground linkages of a nitrogen and phosphorus co-limited tropical mountain pasture system-responses to nutrient enrichment. Plant Soil 391:333–352
Undem C, Rios EJ, Maylor J, Shimoda LA (2012) Endothelin-1 augments Na+/H+ exchange activity in murine pulmonary arterial smooth muscle cells via rho kinase. Plos One 7:e463032012
Vansuyt G, Souche G, Straczek A, Briat JF, Jaillard B (2003) Flux of protons released by wild type and ferritin over-expressor tobacco plants: effect of phosphorus and iron nutrition. Plant Physiol Biochem 41:27–33
Wakeel A, Hanstein S, Pitann B, Schubert S (2010) Hydrolytic and pumping activity of H+-ATPase from leaves of sugar beet (Betavulgaris L.) as affected by salt stress. J Plant Physiol 167:725–731
Wang Z, Li S (2003) Effects of N forms and rates on vegetable growth and nitrate accumulation. Pedosphere 13:309–316
Wen K, Liang C, Wang L, Hu G, Zhou Q (2011) Combined effects of lanthanumion and acid rain on growth, photosynthesis and chloroplast ultrastructure in soybean seedlings. Chemosphere 84:601–608
Yang NC, Ho WM, Chen YH, Hu ML (2002) A convenient one-step extraction of cellular ATP using boiling water for the luciferin-luciferase assay of ATP. Anal Biochem 306:323–327
Zeng H, Liu G, Kinoshita T, Zhang R, Zhu Y, Shen Q, Xu G (2012) Stimulation of phosphorus uptake by ammonium nutrition involves plasma membrane H+-ATPase in rice roots. Plant Soil 357:205–214
Zhang Y, Wu L, Wang X (1996) Effects of acid rain on leaf injury and physiological characteristics of crops. Agro-Environ Prot. 15:197–208
Zhang JE, Yu J, Ouyang Y, Xu H (2014) Impact of simulated acid rain on trace metals and aluminum leaching in Latosol from Guangdong province. China. Soil. Sediment. Contam. 23:725–735
Zhu Y, Di T, Xu G, Chen X, Zeng H, Yan F, Shen Q (2009) Adaptation of plasma membrane H+-ATPase of rice roots to low pH as related to ammonium nutrition. Plant, Cell Environ 32:1428–1440
Zhu Y, Zeng H, Shen Q, Ishikawa T, Subbarao GV (2012) Interplay among NH4 + uptake, rhizosphere pH and plasma membrane H+-ATPase determine the release of BNIs in sorghum roots—possible mechanisms and underlying hypothesis. Plant Soil 358:125–135
Acknowledgments
The authors are grateful for the financial support from the Natural Science Foundation of Jiangsu Province (No.BK20161131) and the National Natural Science Foundation of China (31000245, 31370517).
Author information
Authors and Affiliations
Corresponding author
Additional information
Editorial responsibility: M. Abbaspour
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, B., Bu, J. & Liang, C. Regulation of nitrogen and phosphorus absorption by plasma membrane H+-ATPase in rice roots under simulated acid rain. Int. J. Environ. Sci. Technol. 14, 101–112 (2017). https://doi.org/10.1007/s13762-016-1125-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13762-016-1125-x