Abstract
Accurate retrieval of liver CT images can help a specialist to decide on the type of lesion and treatment planning. However, the complex texture of the abnormality and its nonlinear characteristic reduces the recognition rate of a retrieval system. In this paper, we propose how to represent an abnormal region of a liver by individual attributes of a multi-phase CT image. The indexing of a medical image database is represented by a correlation graph distance, which considers nonlinear behavior of the feature space as well. The results showed that the average recall was improved by 7.5% using the proposed feature vector. Concerning a complex scheme for lesion representation and the manifold indexing technique, the recall of the system was increased by twice. The proposed indexing and feature representation prove the potential of our method in content-based medical image retrieval systems.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Liver abnormalities are classified into benign lesions, including focal nodular hyperplasia (FNH), hemangiomas (HEM) and hepatocellular adenomas (HCA), and malignant lesions such as hepatocellular carcinoma (HCC), and metastases. They exist as single/multiple lesions in men/woman. Their sizes can be small, or they may occupy large portions of the liver. A biopsy is a traditional technique for diagnosis with a risk of bleeding that has no benefit for benign lesions compared to radiologic diagnosis.
Interpretation of medical images is a less/non-invasive approach to obtain more information about the anatomy and function of a patient’s body. It results in early detection of abnormal tissues and can assist a physician in designating the type of the malignancy as well. Hepatic cancer with a rising rate of death is a good example which exploits the benefits of imaging technology [1]. Concerning liver lesions, multi-phase CT images provide extensive characteristics of the disease and help a specialist in improved decision making. However, the hard and time-consuming task of the manual image analysis is a major challenge that is released by machine learning algorithms. Moreover, there are subtle changes in images of various lesion types.
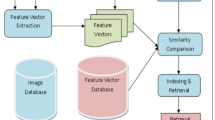
The increasing growth of medical images has opened a new field of content-based medical image retrieval (CBMIR) in disease diagnosis [2, 3]. Retrieving images with similar visual contents to an incoming patient’s data can assist a doctor in discriminating the class of the lesion and deciding on the treatment planning. There are two stages in a CBMIR system: feature representation and indexing [4, 5]. In the first stage, a large number of low/high-level characteristics are obtained from input images to represent the disease accurately. Typical low-level features are color and texture, and high-level descriptors are Gray-Level Co-occurrence Matrix (GLCM), variations of Local Binary Pattern (LBP), and Bag of Visual Words (BoVW) [6]. Feature indexing methods include Euclidean-based techniques [7], dictionary-based sparse representation, and hashing functions. An efficient CBMIR system relies on a proper selection of the features and an appropriate discriminative metric. Traditional content-based image retrieval (CBIR) systems are not effective in the retrieving medical images due to the semantic gap between low-level characteristics and high-level concepts, and the nonlinear nature of the corresponding features.
2 Previous researches
Regarding feature extraction techniques, the Gray-Level Co-occurrence Matrix (GLCM), Local Binary Pattern (LBP), and Bag of Visual Words (BOVW) are commonly utilized. The GLCM is a conventional feature with several variations, and the BOVW is a more complicated characteristic which is more used in recent researches. Malviya et al. [8] used the GLCM features to retrieve pathological CT images of the lung. Roy et al. employed spatiotemporal features to improve retrieval of liver CT images. They mainly used the intensity and GLCM matrix in different parts of a tumor in a multi-phase data [9]. Satish et al. [10] used a Local Binary Pattern (LBP) descriptor and enhanced it by the Scale Invariant Feature Transform (SIFT). Then, they formulated the retrieval problem in a Bayesian framework.
Concerning the nonlinear character of image features, some researchers employed a manifold approach to learn features and classify images [11,12,13,14]. Therefore, they considered the complexity of the data. They ranked objects according to the conventional feature vectors and used the graph theory to construct the relationship between the entities in a dataset. They used globally ranking methods to remove local biases. Ma et al. [14] combined visual and semantic similarities to reduce limitations of pairwise measures. The visual metric was conventional measures such as Euclidean and correlation distances, and the semantic metric used a classifier to calculate the probability of assigning a query image to a specific class. Then, the authors built a graph based on the two measures and traversed it using the shortest path algorithm to rank the images of the database. Wang et al. [15] extended the concept of linear sparse-coding using third-order tensors of multi-phase data to consider nonlinearity of the features.
Pedronette et al. [12] used the manifold learning approach in the CBIR systems. They used the correlation metric to set up a correlation graph distance. They used the strongly connected component (SCC) algorithm in an iterative approach refining the similarity of the input images [12]. They considered the non-symmetric distance between typical objects to improve the visual metrics. They also used the reciprocal K-nearest neighbor to improve the correlation graph [16]. Later, Pedronette and Torres [3] improved the ranking step by a diffusion process to reduce the complexity of the algorithm. Their recent method utilized the intrinsic manifold structure of the data more effectively. However, the processes required much time to run.
Some researchers have used hashing functions to relieve a CBIR algorithm from the expensive search step and to develop a scalable technique. Scalability is an important issue in designing a retrieval algorithm due to the increasing number of available medical images. Onjeti et al. employed hash functions to develop a large-scale image retrieval algorithm [4, 17].
The deep neural network is a recent trend in image processing that exploits features inherent in the whole input data. It has several variations, including auto-encoders, deep belief networks, convolutional neural networks (CNNs), and generative adversarial networks. Qayyum et al. [18] employed CNNs to classify healthy medical images with various modalities.
In this paper, we propose an unsupervised manifold learning algorithm to retrieve multi-phase CT images of hepatic tumors. Our main novelties are (1) using the correlation graph distance as a nonlinear approach measuring the similarity of pathological images, (2) modification of the SCC algorithm to fix the number of the clusters and (3) appropriate selection of the features for each phase to improve the overall results. As far as we know, this is the first research that employs a graph-based technique to measure the distance of pathological liver images. Concerning multi-phase CT data, researchers have employed identical features for all phases. However, based on our experience, a specific set of features work better for a phase compared to other phases. We compared our method with conventional features and distances, and our results outperformed other methods.
This paper is organized as follows. In Sect. 2, we describe the preliminaries of our technique and explain our method. Section 3 presents the results and discussions. In Sect. 4, we conclude the paper and give our plans.
3 The proposed method
The main steps of the proposed method are feature extraction and graph construction. The mask of a tumor is used to define its region-of-interest (ROI) to reduce the processing time and the required memory. For feature extraction, we followed two scenarios: (1) using statistical textures for the whole lesion’s region, (2) partitioning the lesion’s volume into three parts and extracting individual characteristics from each zone. The partitioning is based on the fact that a lesion shows different characteristics based on its growing stage. It means that the internal parts of a lesion have different features compared to the newly affected tissues. In each partition, shape, spatial, and temporal features are extracted. Then, we construct a graph in which database images constitute its nodes, and the edges define the similarity between images. The graph is a manifold representation of image similarities and denotes inherent nonlinearity of the relations.
3.1 Feature selection: the first scenario
The GLCM matrix is a common texture descriptor that represents the properties of the gray level and texture of an image. We used six attributes of the matrix: entropy, contrast, correlation, homogeneity, intensity, and energy, which are described in Eqs. (1)–(10):
In Eqs. (1–10), g (i, j) denotes the frequency of the gray-level pairs (i, j), and N is the total number of gray levels in the image.
Based on our experience, various features have their potentials in discriminating lesion categories. In Fig. 1, typical 2D and 3D plots of the lesions in different feature spaces are shown. As shown in Fig. 1a, homogeneity and entropy are good characteristics to label HCC (class-3) lesions, while type-II lesions are better discriminated by the correlation (Fig. 1b). We plotted lesion categories using several 2D and 3D features and found that using dissimilar features for different imaging phases is more appropriate for separating lesion types. Therefore, we selected conventional metrics of CBIR systems as cost functions to pick up the best characteristics of liver abnormalities. Based on our experience, the feature vector in Eq. 11 gives the highest class separation.
In (11), Cntr, Crr, Int, Hmg, and Ent refer to the contrast, correlation, intensity, homogeneity, and entropy, respectively.
3.2 Feature selection: the second scenario
Concerning the second setup, we divide the volume of a tumor into three partitions shown in Fig. 2: the inner (P1), intermediate (P2), and borderline (P3) partitions [9]. In each section, three categories of attributes are obtained: (1) density, (2) temporal density, (3) texture, and (4) temporal texture. Each attribute is calculated for individual image phases: non-contrast (NC), arterial (ART), and portal vein (PV). The partitions are obtained by morphological erosion with a size of 6 and 12 pixels corresponding to P2 and P3.
The density feature \( (F_{\text{D}} ) \) is the ratio of the tumor’s average intensity to the average intensity of the healthy region (e.g., \( \frac{{d_{{{\text{P}}1}}^{\text{NC}} }}{{d_{\text{liver}}^{\text{NC}} }} \)). It reflects the contrast enhancement of the lesion in each of the partitions P1 to P3. It is individually calculated for each phase (NC, ART, and PV) in the three partitions. Therefore, the density feature is a 9 × 1 vector described by Eq. (12).
The temporal density feature \( (F_{\text{TD}} ) \) is a 6 × 1 vector that shows the relative enhancement of the lesion’s intensity between different phases in each partition (Eq. 13).
The texture feature is the conventional GLCM descriptors consisting of the energy \( (g_{1} ) \), entropy \( (g_{2} ) \), homogeneity \( (g_{3} ) \), intensity \( (g_{4} ) \), contrast \( (g_{5} ) \), and correlation \( (g_{6} ) \) measures. It is calculated for the ART and PV phases in each of the three partitions. Therefore, it consists of 36 elements.
The temporal texture feature \( (F_{\text{TT}} ) \) depicts the variations of the GLCM descriptors concerning the acquisition phase. It is a heterogeneous feature vector that is composed of 36 elements that are described in Eqs. (14)–(17).
In Eqs. (17), median, min, and max are the median, minimum, and maximum operators and \( g_{{K,{\text{Part}} .}}^{\text{Phase}} \) is the average of the kth texture descriptor that is calculated for the three phases. Finally, we develop the spatiotemporal feature vector consisting of 87 elements.
3.3 The correlation graph distance
The construction of a correlation graph has three stages: (1) calculating the global correlation between database images, (2) designating strongly connected components (SCCs), and (3) construction of a “distance correlation graph.”
First, the correlation between an input image and the database images of the database is calculated, and they are ranked and put in a list based on their similarities. Images with a similarity lower than a threshold are removed from the list. Therefore, a graph is constructed between the remaining images using the correlation metric. The correlation graph formed in the previous step is modified by identifying the fully connected components and reinforcing the connection between graph nodes.
The distance obtained by the correlation graph is superior to conventional measures such as the Euclidean and local correlation metrics. The discrimination power of the graph proves itself where there is nonlinearity in the feature space. In Fig. 3, the known two-moon data are shown, and they are classified by (a) the K-means algorithm using the Euclidean distance (Fig. 3a), and (b) the correlation graph measure (Fig. 3b). The K-means algorithm labels the data based on their distance, which results in misclassification, as is shown in Fig. 3a. However, the correlation graph considers the structure of the intrinsic geometry of the data manifold. Details of the correlation graph distance are given below.
3.3.1 The correlation graph
Given a database \( C = \left\{ {img_{1} ,img_{2} , \ldots ,img_{n} } \right\} \) consisting of \( n \) images, we compare a query image \( img_{q} \) to these images using the Euclidean distance and then sort them in the ascending order \( \tau_{q} = (img_{1} ,img_{2} , \ldots ,img_{{n_{s} }} ) \). The correlation graph \( G = \left( {V, E} \right) \) includes a set of vertices V resembling available images and the links between them (E). Next, we calculate the correlation between the query image \( (img_{q} ) \) and an element of \( \tau_{q} \)\( (img_{j} ) \). To calculate the Pearson’s correlation coefficient between the two images, we use their K-nearest neighbor \( N_{k} \left( q \right) \). Then, the correlation between \( img_{q} \) and \( img_{j } \) is calculated using Eq. 18.
In (16), \( img_{i} \in N_{k} \left( {q,\,j} \right) \), \( X_{i} = \rho \left( {q,i} \right) , Y_{i} = \rho \left( {j,\,i} \right) \), \( \bar{X} \), and \( \bar{Y} \) are the corresponding arithmetic means, \( \rho \left( {q,i} \right) \) is the Euclidean distance between the two images \( q \) and \( i \), \( N_{k} \left( {q,j} \right) \) is the accumulated set of \( N_{k} \left( q \right) \) and \( N_{k} \left( j \right) \), and \( k_{u} \) is the cardinality of \( N_{k} \left( {q,\,j} \right) \). The value of the correlation coefficient lies in the range [− 1, + 1]. If the calculated correlation is greater than a threshold \( t_{\text{c}} \), there will be a link between \( img_{q} \) and \( img_{j} \).
3.3.2 Strongly connected components
To refine the correlation graph of the previous step, we obtain fully connected components using the Tarjan algorithm [19]. The general output of the algorithm is defined as a set of strongly connected components \( s = \left\{ {s_{1} ,s_{2} , \ldots ,s_{m} } \right\} \), and the improvement is performed repetitively. We increase the value of the correlation threshold \( (t_{\text{c}} ) \) in each iteration and stop the algorithm when the threshold reaches a value of 1.
3.3.3 Correlation graph distance
The constructed correlation graph and the components obtained at the previous stage are used to compute a similarity matrix. The similarity between the two images \( img_{i} \), \( img_{j} \) is initially set to \( w_{ij} \), and it is changed iteratively by calculating the affinity of non-similar images. More details of the algorithm are given in [9].
4 Result and discussions
4.1 Dataset and evaluation metrics
In this study, we used 411 CT liver data consisting of three phases: non-contrast (before injection of the contrast material), arterial (15–40 s after the injection), and portal vein (70–80 s after the injection of contrast agent). There were 137 lesions including 38 Cyst, 22 FNH, 28 HCC, 28 HEM, and 21 METS. The images belonged to Sir Run Run Shaw Hospital, Hangzhou, China [15, 20].
The proposed algorithm was implemented in the MATLAB 2015 environment on a personal computer running Windows 8, with a \( {\text{core}}^{\text{TM}} {\text{i}}5 \) quad-core processor and 4 GB of dynamic memory. Retrieval of a single image takes about 16 s on the above platform.
The evaluation metrics were recall and precision defined in Eqs. (19) and (20).
Consider distinguishing HCC lesions, TP (True Positive) is the number of HCC images that are correctly labeled as HCC, FN (False Negative) is the number of HCC data that are assigned to other classes mistakenly, FP (False Positive) is the number of other tumor types that are tagged as HCC [21]. The recall index is the ratio of the fetched HCC lesions to the total number of HCC data. The precision metric is the ratio of the fetched HCC lesions to the total number of retrieved images.
4.2 Quantitative results
As stated earlier, we used two scenarios for feature extraction. In the first scenario, the GLCM matrix was used to characterize a lesion. In the second scenario, the volume of a lesion was divided into three partitions, and several attributes were extracted in each section.
The results shown in Fig. 4 compare the retrieval of the lesions using single-phase data. Concerning the NC, ATR, and PV phases, we used “contrast–correlation–intensity,” “homogeneity–correlation–intensity,” and “contrast–correlation–entropy” features, respectively. Both the Euclidean measure and correlation graph distance were used for indexing.
As is shown in Fig. 4, using simple attributes of a single-phase image gives a low retrieval performance (both in terms of precision and recall). However, the correlation graph distance improves the outcome. Moreover, different acquisition phases are successful in recognition of different lesions. The recall and precision results confirm each other as well. The maximum of the recall is in the range [40, 45] for NC, ART, and PV phases.
In Fig. 5, we improved the describing attributes of the lesions and used an 87 × 1 feature vector. While the results of the Euclidean indexing are changed a little, the outcome of the correlation graph distance is nearly increased by 100%. The results prove that the essence of the lesion features is described by a manifold approach better than conventional metrics. Moreover, augmentation of imaging phases improves the results. Figure 5 also reveals that the ART phase is more descriptive compared to two other phases.
We compared our results with two other methods to evaluate the effectiveness of the proposed algorithm (Table 1). The first method was proposed by Roy et al. [9] that used 123 features using 4-phase CT data to recognize five types of hepatic lesions. The second method belonged to Wang et al. [15]. It used linear sparse-coding using third-order tensors of multi-phase data to consider nonlinearity of the features. The results in Table reveal that choosing a proper indexing technique can be as effective as a complex feature vector.
Concerning the challenges of the proposed method, it requires a multi-phase data to decide on the type of the lesion. However, the acquisition of several CT images from a single patient is hazardous. Therefore, it is required to suggest new features to replace the currently large feature vector of multi-phase data. Recently, we have researched to introduce new elements that describe a tumor better and improve discrimination between different tumor types as well. We also need to collect a larger dataset and further verify the effectiveness of our method. Our research proved the nonlinear nature of tumor features.
4.3 Sensitivity analysis
In this section, the sensitivity of the proposed method to the parameters of the algorithm is investigated, and the optimal values of the parameters are calculated. The inspected parameters in both scenarios are the number of the nearest neighbors (k), correlation threshold (tc), and the number of similar images to the search image \( (n_{\text{s}} ) \).In Fig. 6, the sensitivity analysis is shown. Based on the analysis, we set \( n_{\text{s}} = 28 \), \( k = 11 \), and \( t_{\text{c}} = 0.15 \).
5 Conclusions
In the paper, we proposed two approaches for liver lesion retrieval. In the first approach, individual features were selected for each of the imaging phases to improve the separability of the lesion classes. In the second approach, a lesion was represented by an 87 × 1 elements vector to describe various regions of an abnormal region. To measure the distance of an input image to other images of a database, we used the correlation graph distance that represents the nonlinear structure of the feature vector. We showed that using a manifold scheme to calculate the distance between images improves the discrimination power of a CBMIR system. It was proved that a more complex feature vector also improves the results as well. The overall recall of our results was averagely improved by 7.5% using the proposed feature vector.
We plan to employ sophisticated feature vectors and use more recent manifold techniques of natural image retrieval systems to increase the accuracy of a CBMIR system.
References
Golchin E, Maghooli K (2014) Overview of manifold learning and its application in medical data set. Int J Biomed Eng Sci (IJBES) 1(2):23–33
Li Z et al (2018) Large-scale retrieval for medical image analytics: a comprehensive review. Med Image Anal 43:66–84
Pedronette DCG, Torres RS (2017) Unsupervised rank diffusion for content-based image retrieval. Neurocomputing 260:478–489
Heidari H, Chalechale A, Mohammadabadi AA (2013) Parallel implementation of color based image retrieval using CUDA on the GPU. Int J Inf Technol Comput Sci (IJITCS) 6(1):33
Zin NAM, Yusof R, Lashari SA, Mustapha A, Senan N, Ibrahim R (2018) Content-based image retrieval in medical domain: a review. In: journal of physics: conference series, vol. 1019, no. 1, p. 012044. IOP Publishing
Datta R et al (2008) Image retrieval: ideas, influences, and trends of the new age. ACM Comput Surv (Csur) 40(2):5
Smeulders AWM et al (2000) Content-based image retrieval at the end of the early years. IEEE Trans Pattern Anal Mach Intell 12:1349–1380
Malviya N, Choudhary N, Jain K (2017) Content based medical image retrieval and clustering based segmentation to diagnose lung cancer. Adv Comput Sci Technol 10(6):1577–1594
Roy S et al (2014) Three-dimensional spatiotemporal features for fast content-based retrieval of focal liver lesions. IEEE Trans Biomed Eng 61(11):2768–2778
Satish B, Supreethi KP (2017) Content based medical image retrieval using relevance feedback Bayesian network. In: 2017 International conference on electrical, electronics, communication, computer, and optimization techniques (ICEECCOT). IEEE
Ghodsi A (2006) Dimensionality reduction a short tutorial. Department of Statistics and Actuarial Science, University of Waterloo, Ontario, Canada 37:38
Pedronette DCG, Torres RS (2016) A correlation graph approach for unsupervised manifold learning in image retrieval tasks. Neurocomputing 208:66–79
Webber W, Moffat A, Zobel J (2010) A similarity measure for indefinite rankings. ACM Trans Inf Syst (TOIS) 28(4):20
Ma L, Liu X, Gao Y, Zhao Y, Zhao X, Zhou C (2017) A new method of content based medical image retrieval and its applications to CT imaging sign retrieval. J Biomed Inf 66:148–158
Wang J, Li J, Han X-H, Lin L, Hu H, Xu Y, Chen Q, Iwamoto Y, Chen Y-W (2019) Tensor-based sparse representations of multi-phase medical images for classification of focal liver lesions. Pattern Recognit Lett. https://doi.org/10.1016/j.patrec.2019.01.001
Pedronette DCG, Gonçalves FMF, Guilherme IR (2018) Unsupervised manifold learning through reciprocal kNN graph and connected components for image retrieval tasks. Pattern Recognit 75:161–174
Conjeti S (2018) Learning to hash for large-scale medical image retrieval. Ph.D. Dissertation, Technische Universität München
Qayyum A et al (2017) Medical image retrieval using deep convolutional neural network. Neurocomputing 266:8–20
Tarjan R (1972) Depth-first search and linear graph algorithms. SIAM J Comput 1(2):146–160
Xu Y, Lin L, Hu H, Wang D, Liu Y, Wang J, Han X-H, Chen Y-W (2018) Texture-specific bag of visual words model and spatial cone matching based method for the retrieval of focal liver lesions using multiphase contrast-enhanced CT images. Int J Comput Assist Radiol Surg 13(1):151–164
Sokolova M, Lapalme G (2009) A systematic analysis of performance measures for classification tasks. Inf Process Manag 45(4):427–437
Acknowledgements
The authors would like to thank Prof. Yen-Wei Chen, Ritsumeikan University, Kansai, for the use of their images in this study.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Mirasadi, M.S., Foruzan, A.H. Content-based medical image retrieval of CT images of liver lesions using manifold learning. Int J Multimed Info Retr 8, 233–240 (2019). https://doi.org/10.1007/s13735-019-00179-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13735-019-00179-6