Abstract
The goal of Big Data analysis is delineating hidden patterns from data and leverage them into strategies and plans to support informed decision making in a diversity of situations. Big Data are characterized by large volume, high velocity, wide variety, and high value, which may represent difficulties in storage and processing. Research on Big Data repositories has contributed promising results that primarily address how to efficiently mine a variety of large volume of structured and unstructured data. However, innovative insights can emerge while leveraging the value characteristic of Big Data. In other words, any given data can be big if analytics can draw a big value from it. In this paper, we demonstrate the potential of five machine learning algorithms to leverage the value of medium size microscopic blood smear images to classify patients with chronic lymphocytic leukemia (CLL). The maximum majority voting method is used to fuse the predications made by the five classifier models. To validate this work, 11 CLL patients are refereed by flow cytometry equipment and the results are compared to the proposed classifier model. The proposed method proceeds through a sequence of steps while working with the lymphocyte images: it segments the lymphocyte images, extracts/selects features, classifies the selected features using five classifiers, and calculates the majority class for the test image. The proposed composite classifier model has an accuracy of 87.0%, true-positive rate of 84.95%, and 10.96% false-positive rate and can correctly identify 9 out of 11 patients as positive for CLL.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Big Data is the term used to describe datasets having the “4V” characterization: volume, variety, velocity, and value. Such datasets present problems with storage, analysis, and visualization (Rajaraman and Ullman 2012). Initial research of Big Data analytics mainly had focused on the data volume, examples include: clinical Big Data analysis (Dai et al. 2012), public databases (Wang et al. 2011), biometrics (Kohlwey et al. 2011), genome and protein analysis (Wang 2014), and biomedical image analysis (Wang et al. 2012a).
Health care systems, in general, suffer unsustainable costs and lack data on utilization (Kaplan and Porter 2011). Therefore, there is a pressing need to find solutions that can reduce unnecessary costs. Cost control measures in healthcare can benefit from using data resources. The problem in health care systems is not the lack of data; it is the lack of information that can be utilized to support critical decision making (Musen et al. 2014).
Big Data by itself usually confers little direct advantage; however, intelligent tuned analytics can reveal many actionable insights that may prove useful in a clinical environment. The basic motivation behind this research is to leverage big value from medium size medical datasets to significantly enhance the medical services provided to patients. This goal is achieved by demonstrating a case study of classification of a wide spread cancer among Canadian adults, namely chronic lymphocytic leukemia (CLL).
CLL is a blood cancer which develops in the soft spongy center of long bones known as bone marrow. It is characterized by the proliferation of abnormal lymphocytes in the bone marrow, which do not respond to cell growth inhibitors (Bain 2008). CLL has a widespread prevalence among adults in Canada (Canadian Cancer Society 2016; Canadian Cancer Statistics 2016; Healey et al. 2015). Moreover, the CLL cells morphology and size variations may be similar to normal lymphocytes in the early stages (Seftel et al. 2009) and it may require a pathologist to identify the lymphocyte as CLL. For these reasons, the clinical course and the phenotypic presentation of CLL are highly diverse, and there are limited treatment options (Grever et al. 2007). Thus, the current clinical best practices emphasize delaying the treatment until a patient demonstrates either symptomatic or progressive disease, which do not necessarily correlate with the optimal treatment outcomes or long-term survival (Grever et al. 2007). In fact, there is a tendency for diagnostic pathology to rely on automated systems (McPherson and Pincus 2011) to aid in diagnosis. No single genetic mutation or abnormality responsible for CLL development has been identified. Rather, the disease is characterized by a variety of chromosomal abnormalities (Tam et al. 2008).
According to the clinical practice guideline of Alberta Health Care Services “LYHE-007 Version 2”, CLL is the most common adult leukemia in the western world, accounting for approximately 7% of non-Hodgkin lymphomas and the average age at diagnosis of CLL is 67 years; and it is rarely seen in children (Herring et al. 2016).
Big Data analytics principles can be applied across different healthcare domains to implement data-driven healthcare solutions that enable better-quality clinical decisions, patient outcomes, survival rates, and reduce the cost of healthcare services (Chen 2016; Mathews et al. 2013; Rothwell et al. 2012; Vollset et al. 2013).
Big Data analytics methods can leverage complex imaging and genotype data analysis, processing data streams from wearables and medical devices to increase cancer survival rates, which can be illustrated in the following:
-
1.
Accelerating genomic sequencing applications that producing better knowledge on different diseases (Chen et al. 2014; Wang 2014).
-
2.
Promoting informed decision making by creating an integrated patient-specific treatment program (Kohlwey et al. 2011; Wang et al. 2011).
-
3.
Designing machine learning algorithms that predict different patient outcomes, e.g., blood and tissue screening, hospitalization period, survival rate, etc. (Kaplan and Porter 2011; Musen et al. 2014).
In this paper, we focus on incorporating different types in machine learning algorithm in analyzing a medium size blood image data to build a patient screening tool for CLL to support informed decision by a hematopathologist.
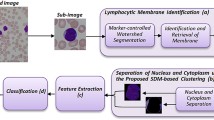
Figure 1 shows the processing steps of the proposed method for CLL cell classification. The proposed method uses machine learning algorithms (MLA) such as the Support Vector Machine (SVM) and the Artificial Neural Network (ANN) to segment, i.e., extract, the lymphocyte parts from the complicated blood smear image. The image segmentation is followed by selecting measures that can identify CLL cells from normal cells with good accuracy. The features are then used to train five different classifier models to label CLL and normal cells for further patient treatment plans. The proposed method can aggregate the results of any number of classifier models in an attempt to increase the accuracy of the aggregated model. To solve the tie problem, an odd number of classifier models can be utilized.
Different classifier models have different structure, learning algorithm and separation capabilities (Sobajic et al. 2010) and, therefore, different classifiers can make different errors when classifying the same features; however, the classifiers aggregate result can provide a better accuracy for the features under test. An ensemble of classifiers, i.e., multiple classifiers system, is used to compose a composite classifier model (Alpaydin 2007) with higher classification accuracy than any individual classifier mode.
A novel Deep Semi-Nonnegative Matrix Factorization (NMF) model was proposed in Trigeorgis et al. (2014) to learn hidden representations in large datasets, e.g., images. The proposed model allowed for an interpretation of pattern clustering according to unknown features of a given dataset. The purpose of this model was to learn low-dimensional transformation of the data that were better for clustering. A novel approach based on NMF was proposed to reduce big datasets dimensionality for better clustering (Allab et al. 2017). This approach utilized the mutual reinforcement between data reduction and clustering tasks to better approximate the dimension reduction solution.
Active learning (Burbidge et al. 2007) (AL) signifies a family of approaches which selectively label training samples to build classifiers with maximum prediction accuracy. Compared to passive learning, which labels samples in a random manner, a new method based on AL and optimal subset selection (ALOSS) was proposed in Fu et al. (2013) to consider sample correlations for selecting the most important instance subset for labeling. The authors proved that the ALOSS method was better than other sample selection methods based on AL.
Feature selection (FS) is an important step in data mining of large-scale high-dimensional data. A novel algorithm known as the Semi-Supervised Representatives Feature Selection algorithm based on information theory (SRFS) (Wang et al. 2017) was proposed to select the most significant features from high-dimensional features datasets. This algorithm was independent of any algorithm used for classification learning, and can identify and remove the less relevant features. The results of this algorithm on several benchmark datasets surpassed the state of the art supervised and semi-supervised algorithms. Discriminative Sparse Flexible Manifold Embedding (SparseFME) method with novel graph was proposed (Zhang et al. 2017b) to enhance the representation and label prediction of FME by improving the reliability and robustness of distance metric. Thus, more accurate identification of hard labels can be obtained. In this study, a novel graph weight construction method was proposed to integrate class information and consider a certain kind of similarity/dissimilarity of samples so that the true neighborhoods can be discovered.
A unified framework for improved structure estimation and feature selection was proposed in Zhang et al. (2017a) to learn data structure and feature selection. A higher order description of the neighborhood structures was presented in the data using hypergraph learning to select the most significant higher order features. A single objective function was designed to capture and regularize the hypergraph weight estimation and feature selection processes. The objective function captured the global discriminative structures in 9 benchmark datasets.
This paper is structured as follows:
Section 2 contains background information on the CLL cancer, white blood cell (WBC) microscopic image segmentation, feature extraction/selection, WBC classification, and multiple classifier system. Section 3 presents the details about the dataset and methodologies used in this paper. Section 4 presents detailed discussion of the results of the proposed method, overall performance, and validation. Section 5 is there to draw the final conclusions, limitations, and future works.
2 Literature review
2.1 Chronic lymphocytic leukemia
Microscopic examination of blood smear images is the main source of information that indicates changes in the development of specific diseases. Blood smear images consist of leukocyte cells, red blood cells, platelets, and background.
CLL is a cancer of lymphocytes, which are blood cells involved in the body’s immune system. CLL/small lymphocytic lymphoma (SLL) is classified by the World Health Organization (WHO) as a low-grade (slow-growing) non-Hodgkin lymphoma and is synonymous with SLL (Oliai 2013). The CLL cells are mainly found in the lymph nodes (glands), as in most other lymphomas. CLL is a disease which mainly affects older people and which has limited treatment options. As more CLL cells accumulate, they can release chemicals which cause tiredness, weight loss, and sweating. If they accumulate in the bone marrow, they can also stop the bone marrow from working properly.
CLL is usually diagnosed by microscopic examination of blood smear films. It is suspected when the blood count shows a large number of lymphocytes (Oliai 2013). The microscopic examination of the CLL cells shows that CLL is small cells with condensed chromatin (which is found in the central nucleus of the cell) and very little cytoplasm. The diagnosis is confirmed by a technique called ‘immunophenotyping’, which involves the detection of the characteristic proteins (or ‘antigens’) on the surface of the lymphocytes (Craig and Foon 2008).
2.2 Microscopic image segmentation
Accurate segmentation of lymphocyte nucleus and cytoplasm from a microscopic blood smear image is a mandatory step to aid in the automatic detection and diagnosis of CLL. Segmentation is the process of correctly and accurately extracting different parts of an image. Leukocytes have different, wide variations of cell morphology, and size which make them difficult to be segmented accurately.
Multispectral WBC segmentation using an SVM (Guo et al. 2007) showed robust, effective, and insensitive results to blood smear staining and illumination condition. However, the results showed low nucleus segmentation accuracy. This is due to the variation of the nucleus color. A method based on feature scale-space filtering and watershed clustering obtained 98.9% maximum cell accuracy for WBC segmentation (Jiang et al. 2006). However, it suffers from the over-segmentation due to plateau of the watershed lines. A framework for WBCs segmentation (Sadeghian et al. 2009) showed accuracy of 92% for nucleus and 78% for cytoplasm using active contours and a Zack thresholding algorithm. This low accuracy resulted from the utilization of the nucleus segmentation results into the Zack thresholding algorithm which further increased the error in the cytoplasm segmentation. A method based on a combination of automatic contrast stretching, arithmetic image operation, minimum filter, and global threshold techniques was proposed as a localization and segmentation method for WBCs nucleus (Madhloom et al. 2010). This proposed method was simple; however, the results showed that the proposed method managed to obtain a wide range of accuracy (85–98%). A stepwise merging rules and a gradient vector flow snake method were proposed to automatically segment WBCs (Ko et al. 2011). This method reduced the over-segmentation problem by 10.31% and the under-segmentation by 1.32%, but the algorithm was computationally expensive.
A method for localization and segmentation of lymphoblast cells using peripheral blood images was proposed in the study (Madhloom et al. 2012); it gave an accuracy of 90–95% in restoring the lymphoblast pixels from the original image; and this was due to the color inconsistency. Color image segmentation using SVM and fuzzy C-means was proposed in Wang et al. (2012b). This method had the advantage of segmenting any type of images accurately and fast. However, it suffered the problems of over and under-segmentation.
A Self-Organizing Map (SOM) neural network along with wavelets was used to segment WBCs (Jaffar et al. 2010). The results showed that if the SOM training was performed on the wavelet-transformed image, it reduced the SOM training time and made more compact segments. This method had the following advantages: it yielded more homogeneous regions than those of other methods for color images, it reduced the spurious blobs, and it removed noisy spots. However, the method was computationally expensive. A pulse-coupled neural network (PCNN) with multichannel (MPCNN) linking and feeding fields was proposed for color image segmentation (Zhuang et al. 2012). Pulse-based radial basis function units were introduced into the model neurons of PCNN to determine the fast links among neurons with respect to their spectral feature vectors and spatial proximity.
2.3 Feature extraction and selection
The complications of interpreting an accurate diagnostic decision in pathology are limited by the lack of definitive measurable features for detecting and characterizing diseases, and their corresponding histological and/or cytological features. The peripheral blood smear of patients is routinely investigated for abnormalities; however, the delicate visible differences exhibited by some disorders can lead to a significant number of false negatives during microscopic examination of the peripheral blood smears.
Measuring object properties has been a subject of study since the early 1970s and is considered to be the conclusion of considerable development (Fukunaga 1990). Feature extraction is the process of converting a given segmented object, i.e., mask (cell, nucleus, and cytoplasm masks), into a set of measurements. There are many features that can be measured for a given object in an image (Fukunaga 1990).
The feature selection is defined as choosing a subset of the extracted features that have minimum redundancy and maximum relevance to the object of interest. The generic purpose pursued is the improvement of the classifier output, either in terms of learning speed, generalization capacity, or simplicity of the representation.
The goal of the feature selection algorithm is to reduce the dimensionality of the classifiers input data by selecting the most distinctive features, which maximize the correct classification rate (CCR). There are basically two types of Features Selection Algorithm (FSA). The first type is the filter type (Freeman et al. 2015), in which statistical analysis such as mutual information is used to rank the features according to the information represented by the features. The performance of a single feature classifier can be used to select features according to their individual predictive power. The predictive power of the feature can be measured in terms of error rate; however, ranking criteria based on the CCR cannot distinguish between the top-ranking variables where there are a large number of features that separate the data perfectly. The FSA based on filter type requires the use of the features probability density function (PDF) which is not easily computed.
The second type of the FSA is the wrapper type, which assess subsets of features according to their usefulness to a given classifier (Hu et al. 2015). The wrapper type offers a simple and powerful way to address the problem of features selection, regardless of the chosen classifier algorithm. It is based on using the classifier performance to assess the relative usefulness of subsets of features. The wrapper type requires a methodology to search the space of all possible features subsets, which is computationally expensive.
2.4 CLL cell classification
2.4.1 SVM classifier
The SVM model is a representation of the features dataset as points in the feature space, mapped in a way that the features of the CLL and normal cases are separated by a clear gap (Margin) which is as wide as possible. The unknown lymphocyte features are then mapped into the same feature space and are predicted based on which side of the gap they fall on. The features of the training dataset of the lymphocyte images are overlapped, which makes it difficult for an SVM classifier to linearly separate the two classes; however, an SVM classifier can efficiently perform a non-linear classification using the kernel trick, which maps the features into a higher dimensional feature space. In this paper, the Soft Margin method (Xu et al. 2013) is used to determine the parameters of the hyper-plane that separate into two classes.
The Soft Margin problem is solved by the Lagrange multiplier method (Lawson and Hanson 1974). The Soft Margin parameters are the slack variable ξ that represents the degree of misclassification and C factor that represents the penalty cost of misclassification. Increasing C places more weight on the slack variable ξ, which means that the optimization attempts to make a stricter separation between classes. Equivalently, reducing C towards 0 makes the misclassification less important. The Quad Programming (QP) (Gould and Toint 2004) method and the Gaussian RBF are used to solve the optimization problem for the SVM training as recommended by Hsu et al. (2003).
2.4.2 ANN classifier
The ANN topology plays an important role in the classification process and the optimal topology depends upon the problem in hand. In this paper, the ANN topology is a Multi-Layer Perceptron (MLP) network with 20 neurons in the input layer, 43 neurons in one hidden layer, and 2 neurons in the output layer to represent the classes. The training of the ANN is conducted to select the number of neurons in the hidden layer based on the cross-validation error. The training is stopped when the cross-validation error started to increase to avoid network over-fitting.
All network weights were initialized with random values in the range [−1, +1]. The ANN is trained with the conjugate gradient descent (CGD) back-propagation algorithm (Yegnanarayana 2006). The back-propagation training method is simple even for complex models having hundreds or thousands of features. The cross-validation error is calculated for every training set of the features for ten times and the average cross-validation error is reported to choose the best ANN topology.
2.4.3 Decision tree
Decision tree (Loh 2011) is a popular technique used in classification problems as they are accurate, relatively simple to implement, produce a model that is easy to interpret and understand, and have built-in dimension reduction. A decision tree is a structure that is either a leaf, indicating a class, or a decision node that specifies some test to be carried out on a feature (or a combination of features), with a branch and sub-tree for each possible outcome of the test. The decision at each node of the tree is made to reveal the structure in the data. The traditional version of a decision tree algorithm creates tests at each node that involve a single feature. As the test at each node is very simple, it is easy for the domain expert to interpret the tree.
2.4.4 K-Nearest Neighborhood (K-NN)
The K-Nearest Neighbor (KNN) algorithm belongs to the category of instance-based learning, in which the learning occurs only when the data items are to be classified (Ripley 2002). The classification algorithm typically classifies the data items as belonging to the nearest class that is represented by a set of measured features. The KNN algorithm assigns to an unlabeled item the most frequently occurring class label among the k most similar data items. The similar data items are obtained using different distance metrics between the feature vectors such as Euclidean distance and city-block distance metrics. The k-nearest neighbor can also be applied using weights, where the neighbors which are closer to the query item have larger weights.
2.4.5 Adaptive Boosting (AdaBoost)
Adaptive Boosting is founded on the notion of using a set of weak classifier models and pooling the classification results of such models to produce a stronger composite classifier model. In the sequence of weak models used, each classifier focuses its discriminatory power on the training samples misclassified by the previous weak classifier. The main reference for the AdaBoost algorithm is the original paper by Freund and Schapire (1995). AdaBoost maintains a probability distribution over all the training samples. This distribution is modified iteratively with each selection of a weak classifier. Initially, the probability distribution is uniform over the training samples. The AdaBoost algorithm can utilize up to (T) weak classifiers which can be as simple as individual attributes or individual features that provide some discrimination between the objects of interest.
2.4.6 Multiple classifier system (MCS)
An approach in classification which has gained much acceptance in the community of data mining and data fusion is the concept of ensembles or committees of classifiers, which involve combining multiple models of classifiers to form a composite, stronger one. The idea behind this is very simple, in which the training dataset is used to train several different models, each of which is used to assign a class label to a previously unseen instance. These class labels are then combined suitably to generate a single class label for the instance. This has been found to improve the accuracy of the resulting model (Alpaydin 2007); however, the process is computationally expensive and it is hard to understand how the decision was obtained and this is depending on the fusion technique used which can be one of the following techniques: majority voting, maximum, minimum, average, sum, and decision.
2.5 Using MCS to classify the WBCs
Reliable detection of pathological blood samples is of major importance in clinical laboratories (Houwen 2001). Even though current automated cell counters used in hospitals are based largely on laser-light scatter principles, a quarter of the blood samples require microscopic review by experts. However, few algorithms allowed for automatic cell classification using image processing. A method based on Bayes classifier and neural networks using four granulo-metric nuclei features without cytoplasm was used to automatically classify WBCs (Shivhare and Shrivastava 2012). An algorithm to optimize the pattern recognition of different WBC types in flow cytometry was introduced in study (Adjouadi et al. 2005). In this algorithm, an SVM classifier was used to cluster parametric data in a multidimensional space. An automated approach to WBCs classification was introduced in (Ramoser 2008). This approach used a pairwise SVM classifier to label cytoplasm and nucleus features. A method based on a two-phase methodology to analyze the morphology of abnormal leukocytes images was used to classify acute leukemia subtypes using image processing and data mining techniques (Reta et al. 2010).
A method based on fuzzy image segmentation and an SVM classifier was proposed for automated leukemia detection and classification (Abdul Nasir et al. 2012). This method managed to classify the lymphocytic cell nucleus as either lymphocyte or lymphoblast. In this method, a fuzzy-based two-stage color segmentation method was used to segment the WBCs. The result showed that an accuracy of 93% was achieved.
3 Methodology
3.1 Lymphocyte images
Giemsa stained peripheral blood smear slides are used to acquire 6345 images using the commercial CellaVision™DM96 system (CellaVision Company 2016) with a 100× oil-immersed objective (Calgary Laboratory Services 2016). The system searches for the WBC and takes an image with the cell at the center of the image. The images resolution is 363 × 360 pixels. The images acquired from the CellaVision™DM96 are categorized as follows: 1010 images manually categorized into CLL and normal images, and 5335 images are acquired from 11 positively identified CLL cases using a commercial flow cytometry device (Calgary Laboratory Services 2016). A randomly selected dataset (out of all the images available for this study) of 140 images are segmented manually to calculate the accuracy of the segmentation algorithms. Table 1 shows the number of images used to design and validate the segmentation algorithms as well as the training and validation images used by the classifier model used in the proposed method.
3.2 Lymphocyte cell segmentation and feature selection
The purpose of the lymphocyte (CLL and normal) cell segmentation is to extract the lymphocyte nucleus and cytoplasm from other different parts in a microscopic blood smear image. Blood smear images consist of WBC “nucleus and cytoplasm”, red blood cells (RBCs), platelets, and background.
3.2.1 Lymphocyte cell segmentation using machine learning algorithm (MLA)
In this section, we describe a method to segment the lymphocyte cell using MLA, i.e., SVM and ANN classifiers.
The problem of lymphocyte nucleus segmentation is considered as a classification problem. The goal of the segmentation is to classify every pixel as a nucleus pixel or background pixel. It is important to select the training dataset for the nucleus area to robustly identify the nucleus pixels from other pixels. The background is defined as any non-nucleus pixels, i.e., cytoplasm, RBCs, platelets, and image background. The algorithm starts with a training phase which includes 12 images for normal and CLL cells.
The training dataset is extracted as follows: thresholding the original image using Otsu’s method (Otsu 1975) to select the nucleus pixels and removing the pixels that represent the color values of the cytoplasm trapped inside the nucleus region. Then, collecting the pixels that belong to the nucleus only and labeling it as ‘NUCLEUS’.
The positive class ‘NUCLEUS’ and the negative class ‘BACKGROUND’ are grouped into a feature matrix: (3xn) data points and (1xn) label vector, where (n) is the number of observations (pixels). The features matrix is composed of the color component (Red, Green, and Blue). The same classification problem applies to the cell segmentation. The subtle difference is that the training dataset contains the cytoplasm color pixels plus the pixels belonging to the nucleus area. The training process is repeated until the best performance of minimum error is achieved for both classifiers, then the best classifier is used for lymphocyte segmentation. This method is described in more detail in our previous work (Mohammed et al. 2013).
3.2.2 Segmentation accuracy measurement
The closed contour area overlapping is a measurement metric used to evaluate the segmentation accuracy (Clinton et al. 2008). The overlapping area between the segmented mask and the ground truth mask represents the segmentation accuracy. The higher the overlapping area, the higher is the accuracy of the produced mask. Let ASec represents the intersection area between any ground truth and the corresponding output mask, AG represents the area of the ground truth image, and AM represents the area of the segmented mask.
We can compute the overlapping area as shown (Clinton et al. 2008):
The overlapping area can be modified to measure the over-segmentation and under-segmentation error as follows:
3.2.3 Feature extraction and feature selection algorithms
The output of the segmentation algorithm for every image is a set of three masks, which are cell mask, nucleus mask and cytoplasm mask as shown in Fig. 2. These masks are used to extract descriptive and distinctive features that can differentiate between the CLL and normal lymphocyte cells. The masks illustrated in Fig. 2 show the three output masks resulting from the proposed segmentation process. The measured features from the three masks are based on the geometrical shape and the statistical measurements of the masks, which are used to extract scale, translation, and rotation invariant features.
A dataset of 129 CLL and 82 normal training lymphocyte images is used to train and measure the accuracy of the segmentation algorithms. There is no golden rule for the number of the training images used in a supervised classification (Ripley 2002). However, the training images must reflect the common characteristics of both types of cells (CLL and normal).
4 Results and discussion
4.1 Segmentation of the lymphocyte cell (CLL and normal)
4.1.1 Performance comparison
Figure 3 shows the segmentation accuracy comparison chart for the two algorithms used to segment a lymphocyte test images. Figure 3 illustrates the nucleus under-segmentation error resulting from using the proposed segmentation method in which the SVM-based method exhibits the smallest under-segmentation error because the separation hyper-plane generated from the SVM training phase tends to maximize the margin between the cell pixels and the background pixels.
The ANN-based segmentation algorithm comes second, as the network topology uses only 10 neurons in its single hidden layer to speed up the segmentation process; however, this leads to increase in over-segmentation and under-segmentation error slightly more than that of the SVM.
The execution time of the segmentation algorithms is recorded, which represents the time required by the segmentation method to extract the cell from the complicated background and further divides the cell into nucleus and cytoplasm masks. The execution time is measured using Intel® quad core CPU i5 2.53 GHz, 4 GB DDR3 RAM PC Windows® 7 64-bit using MATLAB® 2011b (The Language of Technical Computing 2016). Figure 4 shows the execution time for the segmentation methods for the 140 lymphocyte images, in which the SVM algorithm demonstrates the minimum processing time because the SVM segmentation method requires only finding the side of the hyper-plane at which every pixel resides. The ANN requires the substitution of the pixel color values into the network model to find its category, which may increase the processing time. According to the aforementioned performance, we select the SVM-based segmentation method to segment the testing images.
4.1.2 Segmentation performance comparison with published work
Study (Guo et al. 2007) used an SVM for the nucleus segmentation and showed accuracy up to 94% while the proposed method yields up to 98.43%. Study (Madhloom et al. 2010) showed up to 95% for cytoplasm segmentation accuracy while the proposed method yields up to 99.85%. The proposed segmentation method results show that the application of the SVM and the ANN classifiers with a k-means algorithm has high accuracy in lymphocyte cell segmentation.
The study of Madhloom et al. (2012) achieved an accuracy of 90–95% in restoring the lymphoblast pixels from the original image. The proposed method in this paper shows an enhancement in cell segmentation. Figure 3 shows the results of the nucleus segmentation using closed contour area overlapping metric with the under-segmentation error for every image. Image number 37 (blue circle) has a high under-segmentation error as the ANN algorithm has misclassified it.
4.2 Feature selection
The algorithm used for features selection in this paper is the wrapper method based on Sequential Forward Selection (SFS) and Sequential Backward Selection (SBS) (Ververidis and Kotropoulos 2008). A software component (Feature Selection Software Component 2016) is used to decide on the subset features with highest separation power between the classes using SFS and SBS with Naïve Bayes classifier. The resulting features are described in Table 2.
4.3 Lymphocyte cell classification
In this section, we present the steps of the classifier models learning process to find and tweak the model parameters.
Table 3 shows the classifier performance parameters used to evaluate and compare the classifier models used by the proposed method.
4.3.1 The SVM classifier
The training algorithm of the SVM classifier model searches for the radial basis function (RBF) sigma and the box constraint values which maximize the CCR and minimize the misclassified samples. The results of searching for the local minima (error) in the space for the possible values of sigma and the box constraint are illustrated in Table 4. The search is repeated ten times one for each fold of the cross-validation process. The search process uses the optimization technique of multidimensional unconstrained non-linear minimization (Nelder–Mead) (Lagarias et al. 1998).
Table 4 shows that there is more than one local minima that are very close to each other and represent possible candidates for the optimum parameters. To get the best results, the concept of reinforcement learning is utilized in which the candidate local minima at (0.19, 0.21, 0.23, 0.26, and 0.27) are used to train an SVM model with the corresponding sigma and box constraint values, and some values outside the boundary of the chosen parameters are used in the reinforcement learning process to check the goodness of the local minima parameters. The chosen sigma and box constraints values are 1.4796 and 5.1185, respectively.
A dataset (testing dataset) of 799 pre-classified lymphocyte images (662 images of CLL cells and 137 images of normal cells) is used in the reinforcement learning process at which the bias parameter of the model is determined, which represents the closeness of the decision plane to a specific cell type (CLL or normal). If the decision plane is close to one specific cell type, it would be biased to this cell type. A good decision plane would be equally apart from the two types of cells. The reinforcement learning process is repeated for 100 times and the best run setting is chosen, which is the iteration number 60, which yields the highest accuracy for the two-cell class (CLL and normal), as illustrated in Fig. 5.
The SVM classifier model has a sensitivity of 85.97%, specificity of 84.02%, false-positive rate of 15.98%, and total accuracy of 84.99%. The SVM model considers the features of the CLL cells to be the positive samples and the features of the normal lymphocyte cells to be the negative samples. The sensitivity represents the percentage of the true positive (TP) classified as CLL lymphocyte cells by the algorithm and the specificity represents the percentage of the true negative (TN) classified as normal lymphocyte cells by the algorithm. The false-positive rate represents the images classified as CLL while they are Normal.
4.3.2 ANN classifier
The training algorithm for the ANN searches for the weights and the bias for a given set of neurons per single hidden layer of the net using the conjugate gradient descent back-propagation algorithm. The training algorithm increases the number of neurons and finds the corresponding weights and bias for the net topology which aims to maximize the CCR and minimize the misclassified instances. For every net topology, the training data are divided randomly into 70% for training, 15% for validation and 15% for testing. The process for calculating the cross-validation process is repeated and the parameters—the weights and bias—for the minimum cross-validation error are chosen. Then, the net is used to classify a pre-classified 799 lymphocyte test dataset images in the concept of reinforcement learning and the accuracy is recorded.
The experiment is run for 100 iterations, in which the number of neurons is increased by one and the whole process is repeated. At the last step, the net topology that yields the minimum classification error is chosen. As shown in Fig. 6, the number of neurons that give the maximum accuracy for both classes is 43 neurons. This net topology is selected and used by the proposed method to classify a test lymphocyte. The ANN classifier has a sensitivity of 84.32%, specificity of 82.19%, false-positive rate of 17.81%, and total accuracy of 83.26%.
4.3.3 KNN classifier
The parameters for the KNN classifier are the number of the neighbors ‘k’ that is used to label a test lymphocyte image and the distance metric used to rank the neighbors. The used distance metric, in this paper, is the Euclidean distance. To determine the number of k, the training data are divided tenfold for test and training. Initially, k is set to 1 and the re-substitution and the cross-validation errors are averaged for 10 times. The algorithm repeats the process with increasing k from 1 to 100. The optimum value for k is 1, as choosing k = 2 has the effect of increasing the processing time without any gain in the classification accuracy. The KNN classifier has a sensitivity of 80.78%, specificity of 83.1%, false-positive rate of 16.9%, and total accuracy of 81.94%.
4.3.4 Decision tree classifier
The training process of the decision tree aims to get the optimum terminal leaf nodes count. This number is used to prune the tree to speed up the classification process. The leaf node count is determined as illustrated in Fig. 7 in which the re-substitution and cross-validation errors are used to select the tree terminal leaf nodes. The best choice is a 3-leaf node model, which is chosen because the re-substitution and cross-validation errors are almost the same, and this reduces the effect of over-fitting and maximizes the CCR for new unseen test lymphocyte images.
The decision tree classifier has a sensitivity of 72.19%, specificity of 87.67%, false-positive rate of 12.33%, and total accuracy of 79.93%.
4.3.5 AdaBoost classifier
The AdaBoost classifier depends on scaling the training dataset by the classification error; in which the weak classifier focuses more on misclassified instances, which is achieved by putting more weights on the misclassified samples and less on the truly identified ones. The weak classifier used by the AdaBoost algorithm is the decision tree explained earlier; therefore, the same number of terminal leafs is used by the AdaBoost decision tree algorithm. The other parameter needed by the AdaBoost is the number of weak learners (classifiers) which is determined by finding the number of trees that have the best CCR, as illustrated in Fig. 7, in which the classification results and the cross-validation errors are used to select the number of weak learners in which the error curve shows a decreasing value of the classification error, and the cross-validation error is closely following the classification error while increasing the number of trees. Choosing the number of learners is a critical step to ensure good classification results and not to bias the aggregation of the results. The chosen number of learners is 285 at which the classification error stops fluctuating and the cross-validation error is fairly small.
Figure 8 shows how to select the number of decision trees used by the AdaBoost classification algorithm which is used to classify the 799 test lymphocyte images. It shows that the AdaBoost classifier has a sensitivity of 81.16%, specificity of 75.8%, false-positive rate of 24.20%, and total accuracy of 78.48%.
4.3.6 Fusion of the classifier models results
The main idea behind classifiers fusion is to build a composite model with a better CCR from a group of relatively simple classifiers, which is based on the assumption that every classifier may make different mistakes in the classification process; however, if the classifiers made exactly the same mistakes, using any fusion method will not yield any better performance and may lead to classification biasing toward a specific cell class.. In this paper, the majority voting fusion method is used to fuse the classifier models output. Table 5 shows the classification performance attributes of every classifier model along with the fusion results using the majority voting method.
The majority voting fusion method offers fusing any number of classifier models; however, choosing the optimum combination of models can guarantee a faster and better classification performance.
In this paper, ten ensembles of three and five classifier composites are fused by the majority voting method and ranked by the smallest FPR and highest accuracy, and the best composite is used to classify the lymphocyte test images. Table 6 shows the classification performance attributes of the tested aggregates, in which the fusion of the KNN, the SVM and the decision classifier aggregate gives the best results.
If two or more aggregates have very close accuracy and FPR, the one with small number of classifier models is chosen as it results in shorter classification time and reduces the method complexity.
4.3.7 Validation
The validation process is conducted in two phases. The first phase is the reinforcement learning where 799 pre-classified lymphocyte images are used to calculate the classification performance parameters as illustrated in Table 6. The second phase is a comparison between flow cytometry results and the proposed classification method, in which a flow cytometry device is used to analyze blood samples from 11 patients (CLL cases) (Calgary Laboratory Services 2016).
Direct correlation between the number of the CLL cells identified by the flow cytometry and the number of CLL cells identified by the proposed method cannot be achieved, as it appears that the cells scanned by CellaVision™DM96 are not sampled the same way as the flow cytometry device. However, the flow cytometry data can be used to conduct a decision comparison in which other blood samples are acquired from the same 11 CLL cases and analyzed using the CellaVision™DM96. The proposed classification method is used to analyze the lymphocyte images and if the percentage of CLL cells found is ≥70% of the total lymphocyte cells, then there is a chance of CLL case, and if the percentage of CLL is ≤40%, then the images are sampled from a normal case, and in between 40 and 70% a suggestion for re-examination of the blood smear for that case is supported.
Table 7 shows the flow cytometry analysis results for the 11 CLL cases with suggestions for the analyzed slides/cases. The results show that the proposed method is capable of identifying the CLL cases in a way that matched with the flow cytometry results.
Table 8 shows a comparison between other studies (Sabino et al. 2004; Ushizima et al. 2005) and the proposed method in this paper for CLL classification. The accuracy of the proposed method is 87.0%, while the accuracy of the Leuko proposed in Sabino et al. (2004) is 72%. The proposed method uses less features (20) and classifiers (3) than the Leuko system (62 features and 5 classifiers Ushizima et al. 2005).
The accuracy of the proposed system is 3% less than that of the Leuko system with 5 SVMs classifier, and it was verified using more CLL images than the Leuko system. The classifier performance attributes are not available for studies (Sabino et al. 2004; Ushizima et al. 2005). Moreover, the cross-validation method used in these studies is the ‘Hold-out’ cross-validation, which tends to give optimistic results, whereas tenfold cross-validation method is used to verify the proposed method.
4.4 Proposed method output
The proposed method main objective is to provide the hematopathologist with the percentage of CLL and the normal lymphocyte cells present in the test images along with the diagnostic suggestion for the case under examination. The proposed method functional blocks are illustrated in Fig. 1, which represent a complete system for image analysis CLL. The user can run a training phase to learn how to distinguish between the CLL normal cell. The training phase includes extracting the lymphocyte cell from the complicated blood smear images and further divides it into nucleus and cytoplasm parts. This is proceeded by extracting different features for each part and then train several machine learning algorithms to identify the CLL and normal lymphocyte cells. The training phase can be done offline with any number of lymphocyte images.
The system is then ready to process and classify unseen lymphocyte images and present the output to the use. The output is compiled into a report that contains a graph representing a comparison between the chosen classifier models and the fused composite classifier model. The report contains a suggestion for the hematopathologist in which a CLL case is suggested when the proposed method recognizes CLL cells ≥70% of the processed cells, or suggests a normal case if the recognized CLL cells ≤40%, otherwise a re-examination suggestion of the case otherwise. Figure 9 shows a sample report of a CLL case produced by the output phase of the proposed method.
5 Conclusion
Many studies have addressed efficient methods to process and analyze the variety of massive data volume arriving at very high velocity to draw solid conclusions to support informed decisions. However, there is little effort dedicated to leverage beneficial values from small to medium sizes of data; this is mostly because—hypothetically—more interesting patterns can be found analyzing big volumes of data rather than small to medium size data.
In this paper, we steer the attention to the analysis of a moderate size blood smear image dataset and focus on integrating many machine learning algorithms to accurately detect CLL cases. This in turn shows that a cheap imaging modality (microscopic image analysis) can be a good candidate to build a clinical screening system for CLL which shows the power of analytics using medium size data to draw high value with actionable insight.
The scope of this paper is the analysis of lymphocyte microscopic blood images to detect CLL cells. The proposed method consists of lymphocyte segmentation, feature extraction/selection, and multiple classifier models, and the method output is presented in a report format. The design and performance of the proposed method have been validated using 6345 microscopic blood images. The proposed method can segment the lymphocyte cells with high accuracy, and label the cell as CLL or normal. A report is presented to the hematopathologist containing the percentage of the obtained CLL and normal lymphocyte cells in a graphical representation along with a suggested decision.
In this paper, a method for lymphocyte color cell segmentation using the SVM algorithm and ANN is discussed. For segmentation accuracy measurement, 140 images are used and 12 of these images are used for training of the SVM and the ANN algorithms. The SVM algorithm has a superior segmentation performance, in which it obtains 97.0% ± 0.5 average accuracy for nucleus segmentation and 97.62% ± 0.77 for cell segmentation. The cytoplasm region can be extracted by 92.08% ± 9.24 average accuracy with simple mask subtraction.
MCS is a technique used to enhance the results of the classification process, in which an ensemble of classifiers is trained using part or all of the training dataset in a parallel, serial, or hybrid fashion and used to classify a test image, and then the results of the ensemble are fused using a fusion method which can be a majority voting, minimum, maximum, and product. The composite classifier model is composed of the SVM, the KNN, and the decision tree classifiers, which have the best classification performance attributes: 89.04% specificity, 84.95% sensitivity, 10.96% FPR, and 87% overall accuracy.
The proposed method can be used as a cheap and fast pre-screening tool to delineate the CLL cells in a blood smear slide, which will significantly reduce the reliance on the flow cytometry devices as they are costly, and not appropriate as screening tools.
The proposed method contributes in reducing the cost for CLL refereeing by complicated expensive equipment, e.g., flow cytometry, by providing a high accuracy cheap screening tool. This is true as the average cost of a flow cytometry device per patient is approximately 105$, whereas the cost for the CellaVision™DM96 is approximately 5$ per patient (Calgary Laboratory Services 2016).
The proposed method can be used as a generic tool to screen other blood cancer/disease by carefully tuning the extracted and selected features that can differentiate the target cell from the background and then an MCS can be trained to identify the target cell.
6 Limitations and future works
The lymphocyte images used in this study are acquired from the commercial hematopathology equipment CellaVision™DM96 which impose some image quality attributes such as image resolution, objective settings, and specimen illumination and, thus, the results of this research are dependent on these settings. The CellaVision™DM96 acquires images with the cell at the center of the image, which facilitates the localization of the cell in the segmented mask; however, if this is not true for any other equipment settings, a search technique for the cells must be used.
The classification methods used in this paper are based on the non-parametric supervised classifier models and, thus, the results are limited to the training dataset, training algorithms, the reinforcement learning process, and the optimization method used by every classifier model.
Future work of this research focuses on implementing and deploying a clinical screening system for CLL and other common blood cancer with clinical controlled trial experiments involving many different positive and negative patients.
References
Abdul Nasir A, Mashor M, Hassan R (2012) Leukaemia screening based on fuzzy ARTMAP and simplified fuzzy ARTMAP neural networks. In: 2012 IEEE EMBS conference on biomedical engineering and sciences (IECBES), IEEE, pp 11–16
Adjouadi M, Zong N, Ayala M (2005) Multidimensional pattern recognition and classification of white blood cells using support vector machines. Part Part Syst Charact 22:107–118
Allab K, Labiod L, Nadif M (2017) A semi-NMF-PCA unified framework for data clustering. IEEE Trans Knowl Data Eng 29:2–16
Alpaydin E (2007) Combining pattern classifiers: methods and algorithms (kuncheva, li; 2004) [book review]. IEEE Trans Neural Netw 18:964
Bain BJ (2008) A beginner’s guide to blood cells, 2nd edn. Wiley, San Francisco
Burbidge R, Rowland JJ, King RD (2007) Active learning for regression based on query by committee. In: International conference on intelligent data engineering and automated learning. Springer, pp 209–218
Calgary Laboratory Services (2016) https://www.calgarylabservices.com/. Accessed 30 Dec 2016
Canadian Cancer Society (2016) http://www.cancer.ca/. Accessed 30 Dec 2016
Canadian Cancer Statistics (2016) http://www.cancer.ca/~/media/cancer.ca/CW/cancer%20information/cancer%20101/Canadian%20cancer%20statistics/canadian-cancer-statistics-2013-EN.pdf. Accessed 30 Dec 2016
CellaVision Company (2016) http://www.cellavision.com. Accessed 08 Dec 2016
Chen T-T (2016) Predicting analysis times in randomized clinical trials with cancer immunotherapy. BMC Med Res Methodol 16:1
Chen W-P, Hung C-L, Tsai S-JJ, Lin Y-L (2014) Novel and efficient tag SNPs selection algorithms. Bio-Med Mater Eng 24:1383–1389
Clinton N, Holt A, Yan L, Gong P (2008) An accuracy assessment measure for object based image segmentation. Int Arch Photogramm Remote Sens Spat Inf Sci 37:1189–1194
Craig FE, Foon KA (2008) Flow cytometric immunophenotyping for hematologic neoplasms. Blood 111:3941–3967
Dai L, Gao X, Guo Y, Xiao J, Zhang Z (2012) Bioinformatics clouds for big data manipulation. Biol Direct 7:43
Feature Selection Software Component (2016) http://www.mathworks.com/matlabcentral/fileexchange/22970-feature-selection-using-matlab. Accessed 21 Dec 2016
Freeman C, Kulić D, Basir O (2015) An evaluation of classifier-specific filter measure performance for feature selection. Pattern Recognit 48:1812–1826
Freund Y, Schapire RE (1995) A desicion-theoretic generalization of on-line learning and an application to boosting. In: European conference on computational learning theory. Springer, pp 23–37
Fu Y, Zhu X, Elmagarmid AK (2013) Active learning with optimal instance subset selection. IEEE Trans Cybern 43:464–475
Fukunaga K (1990) Introduction to statistical pattern recognition, 1st edn. Academic, San Diego
Gould N, Toint PL (2004) Preprocessing for quadratic programming. Math Program 100:95–132
Grever MR et al (2007) Comprehensive assessment of genetic and molecular features predicting outcome in patients with chronic lymphocytic leukemia: results from the US Intergroup Phase III Trial E2997. J Clin Oncol 25:799–804
Guo N, Zeng L, Wu Q (2007) A method based on multispectral imaging technique for white blood cell segmentation. Comput Biol Med 37:70–76
Healey R, Patel JL, de Koning L, Naugler C (2015) Incidence of chronic lymphocytic leukemia and monoclonal B-cell lymphocytosis in Calgary, Alberta, Canada. Leuk Res 39:429–434
Herring W, Pearson I, Purser M, Nakhaipour HR, Haiderali A, Wolowacz S, Jayasundara K (2016) Cost effectiveness of ofatumumab plus chlorambucil in first-line chronic lymphocytic leukaemia in Canada. PharmacoEconomics 34:77–90
Houwen B (2001) The differential cell count. Lab Hematol 7:89–100
Hsu C-W, Chang C-C, Lin C-J (2003) A practical guide to support vector classification. Data Sci Assoc 1–16
Hu Z, Bao Y, Xiong T, Chiong R (2015) Hybrid filter–wrapper feature selection for short-term load forecasting. Eng Appl Artif Intell 40:17–27
Jaffar MA, Ishtiaq M, Ahmed B (2010) Fuzzy wavelet-based color image segmentation using self-organizing neural network. Intern J Innov Comput Inf Control (IJICIC) 6(11):4813–4824
Jiang K, Liao Q-M, Xiong Y (2006) A novel white blood cell segmentation scheme based on feature space clustering. Soft Comput 10:12–19
Kaplan RS, Porter ME (2011) How to solve the cost crisis in health care. Harv Bus Rev 89:46–52
Ko BC, Gim J-W, Nam J-Y (2011) Automatic white blood cell segmentation using stepwise merging rules and gradient vector flow snake. Micron 42:695–705
Kohlwey E, Sussman A, Trost J, Maurer A (2011) Leveraging the cloud for big data biometrics: meeting the performance requirements of the next generation biometric systems. In: 2011 IEEE World Congress on Services (SERVICES), IEEE, pp 597–601
Lagarias JC, Reeds JA, Wright MH, Wright PE (1998) Convergence properties of the Nelder–Mead simplex method in low dimensions. SIAM J Optim 9:112–147
Lawson CL, Hanson RJ (1974) Solving least squares problems, vol 161. SIAM, Philadelphia, PA, USA
Loh WY (2011) Classification and regression trees. Wiley Interdiscip Rev Data Min Knowl Discov 1:14–23
Lorena AC, de Carvalho AC (2005) Minimum spanning trees in hierarchical multiclass support vector machines generation. In: Ali M, Esposito F (eds) Innovations in applied artificial intelligence. Springer, pp 422–431
Madhloom H, Kareem S, Ariffin H, Zaidan A, Alanazi H, Zaidan B (2010) An automated white blood cell nucleus localization and segmentation using image arithmetic and automatic threshold. J Appl Sci 10:959–966
Madhloom HT, Kareem SA, Ariffin H (2012) An image processing application for the localization and segmentation of lymphoblast cell using peripheral blood images. J Med Syst 36:2149–2158
Mathews JD et al (2013) Cancer risk in 680 000 people exposed to computed tomography scans in childhood or adolescence: data linkage study of 11 million Australians. BMJ: Br Med J 346(10):1–18
McPherson RA, Pincus MR (2011) Henry’s clinical diagnosis and management by laboratory methods, 22nd edn. Elsevier Health Sciences, Philadelphia
Mohammed E, Mohamed M, Naugler C, Far B (2013) Application of support vector machine and k-means clustering algorithms for robust chronic lymphocytic leukemia color cell segmentation. In: Proceedings of the 15th IEEE international conference on e-Health Networking, Application and Services HEALTHCOM, Lisbon. IEEE, pp 622–626. doi:10.1109/HealthCom.2013.6720751
Musen MA, Middleton B, Greenes RA (2014) Clinical decision-support systems. In: Shortliffe EH, Cimino JJ (eds) Biomedical informatics. Springer, pp 643–674
Oliai C (2013) Small lymphocytic lymphoma. In: Brady LW, Yaeger TE (eds) Encyclopedia of radiation oncology. Springer, pp 798–798
Otsu N (1975) A threshold selection method from gray-level histograms. Automatica 11:23–27
Rajaraman A, Ullman JD (2012) Mining of massive datasets. Cambridge University Press, Cambridge, United Kingdom
Ramoser H (2008) Leukocyte segmentation and SVM classification in blood smear images. Mach Graph Vis Int J 17:187–200
Reta C, Robles LA, Gonzalez JA, Diaz R, Guichard JS (2010) Segmentation of bone marrow cell images for morphological classification of acute leukemia. In: FLAIRS Conference
Ripley B (2002) Statistical data mining. Springer, New York
Rothwell PM et al (2012) Short-term effects of daily aspirin on cancer incidence, mortality, and non-vascular death: analysis of the time course of risks and benefits in 51 randomised controlled trials. Lancet 379:1602–1612
Sabino DMU, Costa LDF, Rizzatti E, Zago M (2004) Toward leukocyte recognition using morphometry, texture and color. In: IEEE international symposium on biomedical imaging: nano to macro. IEEE, pp 121–124
Sadeghian F, Seman Z, Ramli AR, Kahar BA, Saripan M-I (2009) A framework for white blood cell segmentation in microscopic blood images using digital image processing. Biol Proced Online 11:196–206
Seftel M et al (2009) High incidence of chronic lymphocytic leukemia (CLL) diagnosed by immunophenotyping: a population-based Canadian cohort. Leuk Res 33:1463–1468
Shivhare S, Shrivastava R (2012) Morphological granulometric feature of nucleus in automatic bone marrow white blood cell classification. Int J Sci Res Publ 2:1–7
Sobajic O, Moussavi M, Far B (2010) Parameterized strategy pattern. In: Proceedings of the 17th conference on pattern languages of programs. ACM, p 9
Tam CS et al (2008) Chronic lymphocytic leukaemia CD20 expression is dependent on the genetic subtype: a study of quantitative flow cytometry and fluorescent in situ hybridization in 510 patients. Br J Haematol 141:36–40
The Language of Technical Computing (2016) http://www.mathworks.com/products/matlab/. Accessed 20 Dec 2016
Trigeorgis G, Bousmalis K, Zafeiriou S, Schuller B (2014) A deep semi-NMF model for learning hidden representations. In: ICML, pp 1692–1700
Ushizima DM, Lorena AC, De Carvalho A (2005) Support vector machines applied to white blood cell recognition. In: Fifth international conference on hybrid intelligent systems, 2005. HIS’05. IEEE, pp 6–11
Ververidis D, Kotropoulos C (2008) Fast and accurate sequential floating forward feature selection with the Bayes classifier applied to speech emotion recognition. Signal Process 88:2956–2970
Vollset SE et al (2013) Effects of folic acid supplementation on overall and site-specific cancer incidence during the randomised trials: meta-analyses of data on 50 000 individuals. Lancet 381:1029–1036
Wang K (2014) BioPig a Hadoop-based analytic toolkit for large scale sequence data. Bioinformatics 29(23):3014–3019
Wang W, Haerian K, Salmasian H, Harpaz R, Chase H, Friedman C (2011) A drug-adverse event extraction algorithm to support pharmacovigilance knowledge mining from PubMed citations. In: AMIA annual symposium proceedings, 2011. American Medical Informatics Association, p 1464
Wang L, Chen D, Ranjan R, Khan SU, KolOdziej J, Wang J (2012a) Parallel processing of massive EEG data with MapReduce. In: ICPADS, pp 164–171
Wang X-Y, Zhang X-J, Yang H-Y, Bu J (2012b) A pixel-based color image segmentation using support vector machine and fuzzy C-means. Neural Netw 33:148–159
Wang Y, Wang J, Liao H, Chen H (2017) An efficient semi-supervised representatives feature selection algorithm based on information theory. Pattern Recognit 61:511–523
Xu X, Tsang IW, Xu D (2013) Soft margin multiple kernel learning. IEEE Trans Neural Netw Learn Syst 24:749–761
Yegnanarayana B (2006) Artificial neural networks, 1st edn. PHI Learning Pvt. Ltd., India Institute of Technology, New Delhi, India
Zhang Z, Bai L, Liang Y, Hancock E (2017a) Joint hypergraph learning and sparse regression for feature selection. Pattern Recognit 63:291–309
Zhang Z, Zhang Y, Li F, Zhao M, Zhang L, Yan S (2017b) Discriminative sparse flexible manifold embedding with novel graph for robust visual representation and label propagation. Pattern Recognit 61:492–510
Zhuang H, Low K-S, Yau W-Y (2012) Multichannel pulse-coupled-neural-network-based color image segmentation for object detection. IEEE Trans Ind Electron 59:3299–3308
Acknowledgements
This work has been supported and funded by SmartLabs Ltd., Calgary, AB, Canada and MITACS Accelerate program under Grant IT01892/FR02553.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Mohammed, E.A., Mohamed, M.M.A., Naugler, C. et al. Toward leveraging big value from data: chronic lymphocytic leukemia cell classification. Netw Model Anal Health Inform Bioinforma 6, 6 (2017). https://doi.org/10.1007/s13721-017-0146-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13721-017-0146-9