Abstract
Purpose
Connexin 43 (Cx43) is a widely expressed gap junction protein. It can also regulate various gap-junction independent processes, including cellular proliferation. The latter regulatory functions have been attributed to its carboxy-terminal domain, CT-Cx43. CT-Cx43 has been found to be expressed independent of full-length Cx43 in various cell types. Its nuclear localization has additionally raised the possibility that it may regulate the expression of particular genes, including miRNAs, known play a role in the regulation of cellular proliferation. Here, we set out to uncover the molecular mechanism(s) underlying CT-Cx43 mediated gene (de-)regulation in human breast cancer.
Methods
Western blotting and quantitative real time PCR were carried to assess the expression of CT-Cx43 and miR-125b in a panel of 60 primary human breast cancer tissues and its paired normal adjacent tissues. In addition, CT-Cx43 was exogenously expressed in the breast cancer-derived cell line MCF-7 and its effect on the expression of miR-125b and its downstream target p53 were evaluated, as well as its effect on cellular proliferation and death using MTT and LDH assays, respectively.
Results
We found that CT-Cx43, but not full-length Cx43, was down-regulated in low grade human breast cancers. In addition, we found that the tumor suppressor protein p53 exhibited a decreased expression in the CT-Cx43 down-regulated samples. Interestingly, we found that miR-125b, a negative regulator of p53, exhibited an inverse expression relationship with CT-Cx43 in the breast cancer samples tested. This inverse relationship was confirmed by exogenous expression of CT-Cx43 in MCF-7 cells. In addition, we found that CT-Cx43 up-regulation and subsequent miR-125b down-regulation resulted in a decreased proliferation of MCF-7 cells.
Conclusions
Our data suggest a mechanism by which CT-Cx43 may regulate cell proliferation. Targeting of CT-Cx43 and/or miR-125b may be instrumental for therapeutic intervention in human breast cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Intercellular communication, mediated by gap-junctions, allows the exchange of small ions and messengers between cells and, as such, plays an important role in tissue homeostasis [1, 2]. Gap junctions are composed of connexins and a large family of these proteins is known to be expressed in different cell types and tissues [3]. Connexin 43 (Cx43) is the most widely expressed gap junction protein and, besides mediating gap junctional communication, it is known to regulate various processes, including cellular proliferation [4]. Specifically, Cx43 is known to act as tumor suppressor [5] and its down-regulation has been associated with a number of pathological conditions, including cancer [6, 7]. Many of the gap-junction independent functions of Cx43 have been attributed to its carboxy-terminal domain, CT-Cx43 [8]. Interestingly, the expression of CT-Cx43 has been found to be independent of that of full-length Cx43 in various cell types [9] and its nuclear localization has raised the possibility that CT-Cx43 may directly regulate gene expression [10].
p53 is one of the most widely studied tumor suppressors and, as such, it is known to regulate various cellular processes [11], including proliferation [12–14]. Under normal conditions, the activity of p53 is maintained at a basal level [15], but this activity may increase in response to various stress conditions [16], such as different types of DNA damage [17] and oncogenic insults [18]. Deregulation of the p53 pathway is considered to be a key event in tumorigenesis, and in most human cancers p53 is either down-regulated or mutated [19–21]. In addition, a group of small RNAs, i.e., microRNAs (miRNAs), has been shown to be able to regulate the expression of genes implicated in various normal and pathological conditions, including cellular proliferation and cancer [22–27]. Specifically, the expression of miR-125b, a conserved homolog of the Caenorhabditis elegans miRNA lin-4, has been found to be altered in various cancers [28]. Whereas miR-125b is up-regulated in some cancers, in other cancers such as breast cancer miR-125b has been found to be down-regulated. Therefore, it has occasionally been labeled as a tumor suppressor.
In the present study, we provide evidence for a potential interplay between CT-Cx43, miR-125b and p53 in primary human breast cancers and for its significance in controlling the in vitro proliferation of MCF-7 breast cancer-derived cells.
2 Materials and methods
2.1 Patient specimens
Human breast cancer tissues and paired normal adjacent tissues were obtained from patients who underwent radical resection at the Department of General Surgery, Sheri-Kashmir Institute of Medical Sciences (SKIMS), Srinagar, J&K, India, from 2012 to 2014. In all cases, the clinical diagnosis was confirmed through histological examination. All samples were collected as per protocol approved by the Research Ethical committee, SKIMS. In addition, written informed consent was obtained from each patient. In each case, samples from the primary tumor and the corresponding normal breast tissue were collected for comparison. All samples were immediately preserved in RNA later® (Qiagen) and stored at −80 °C until use.
2.2 Primer designing and plasmid construction
The carboxy-terminal domain of connexin 43 (CT-Cx43) was cloned in a pCDNA 3.1(−) vector (Invitrogen) using its EcoR1 and BamH1 restriction sites. CT-Cx43 was amplified from human genomic DNA using forward primer 5`-GCCGAATTCAGCTGGTGGTGTCCTTGGTGTCTCTC-3`), having an EcoRI restriction site, and reverse primer 5’-GCAGGATCCTTAAATCTCCAGGTCATCAGGCCGAGG-3′, having a BamH1 restriction site. To construct a plasmid expressing miR-125b, we amplified a 300 bp DNA fragment encompassing miR-125b from human genomic DNA and cloned it into pCDNA 3.1(−) using forward primer 5`-GCGAATTCGTATGTGCGTGATTGTATATGCGC-3`, having an EcoRI restriction site, and reverse primer 5`-GCGGATCCTGCCACTCTCTGGTCACCTG-3`, having a BamHI restriction site. All clones were confirmed by sequencing (Scigenom Ltd., Kerala, India).
2.3 Cell culture and transfection
The human breast cancer-derived cell line MCF-7 was obtained from the National Centre for Cell Science (NCCS, Pune) and cultured in DMEM (Dulbellco’s modified essential medium) containing 10 % fetal bovine serum (FBS, Sigma-Aldrich). Transient transfections were carried out in MCF-7 cells using the Effectene reagent in accordance with the manufacturer’s protocol (Qiagen).
2.4 RNA isolation and miRNA quantification
Total RNA from the primary human breast tissue samples and from the MCF-7 cell line were isolated using TRIzol® reagent (Invitrogen) in accordance with the manufacturer’s instructions. After DNase treatment, RNA was subjected to reverse transcription using a standard protocol. The first cDNA strand was synthesized using Superscript III (Invitrogen) and a two-step qRT-PCR method was carried out using hairpin-looped RT primers specific for miR-125b and miR-16. miR-16 was used as an endogenous control. The RT-primers were designed as per Chen et al. [29]. The miR-125b RT primer sequence used was 5`-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTCACAA-3` and the miR-16 RT primer sequence used was 5`-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCGCCAA-3`. First strand cDNA was subjected to qRT-PCR using a Real Time PCR 7500 machine (Applied Biosystems) in conjunction with a maxima Sybrgreen® mix (Thermo Scientific) in accordance with the manufacturer’s protocol. The forward primer used for miR-125b amplification was 5`-TCCCTGAGACCCTAAC-3` and for miR-16 amplification 5`-GCGGCGGTAGCAGCACGTAAATA-3`. The common reverse primer used was 5`-GTGCAGGGTCCGAGGT-3`. All the reactions were run in triplicate.
2.5 Protein extraction
From the primary human breast tissues, protein was extracted after disintegration of the tissues in 0.5 % trypsin-EDTA solution (Sigma), followed by incubation at 37 °C for 5 min. Cell lysis was carried out using a lysis buffer (20 mM Tris–HCl pH 8.0, 137 mM NaCl, 10 % Glycerol, 1 % Triton-X-100, and 2 mM EDTA). A protease inhibitor cocktail, PMSF and phosphatase inhibitors (1 mM PMSF, 5-10 mM NaF) (Sigma-Aldrich) were freshly added to the lysis buffer. Finally, supernatants were collected and protein concentrations were determined using the Bradfords assay.
2.6 Western blotting
Protein extracts, preheated at 100 °C for 5 min in reducing SDS sample buffer containing 50 mM Tris–HCl (pH 6.8), 2 % SDS, 10 % glycerol, 0.1 % bromophenol blue, 100 mM β-mercaptoethanol, were run on 12 % SDS-polyacrylamide gels. After electrophoresis, separated proteins were transferred to PVDF membranes (Millipore) using a semi-dry transfer method in accordance with the manufacturer’s instructions (Hoefer). For immune-detection, the PVDF membranes were processed using standard protocols in conjunction with rabbit anti-Cx43 (1:2000) (Genescript Inc.) and rabbit anti-p53 (1:1000) (SantaCruz Biotechnologies) antibodies. As a loading control, mouse anti-β-actin (1:7000) (Sigma-Aldrich) was included. For protein detection secondary fluorescent anti-mouse IRDye 680 (1:20,000) or anti-rabbit IRDye 800 (1:10,000) antibodies were used in conjunction with an Odyssey infrared detection system (LI-COR).
2.7 Quantification of protein bands
The protein bands on the Western blots were quantified according to the protocol provided by LI-COR. Briefly, a standard was prepared by adding different concentrations of fluorescently labelled Licor secondary antibodies to the PVDF membranes. The fluorescent spots on the membranes were identified using a shape tool and the fluorescence emitted was measured. Similarly, the fluorescence emitted by the Western blot bands (Cx43, CT-Cx43 and p53) was measured and the amount of protein was estimated using the standard. Each protein band was normalized to ß-actin.
2.8 Cell proliferation assay
To perform cell proliferation analyses, ~104 cells were seeded in 12-well plates including the necessary controls and incubated overnight at 37 °C in a humidified 5 % CO2 atmosphere. The next day, the cells were transfected with the respective plasmids. All transfections were carried out at least in triplicate and the assays were independently repeated three times. 48 h post transfection, the culture medium was removed completely followed by the addition of 300 μl MTT (MTT Cell Growth Assay; Merck) solution (5 mg/ml in PBS). After 4 h of incubation at 37 °C, MTT was carefully removed and 300 μl MTT solvent was added (to dissolve formazan crystals) at room temperature for 30 min in the dark. Next, the optical density (OD) of the plate was read at 560 nm on an ELISA Plate Reader (BioTek instruments inc.) within 10 min.
2.9 Lactate dehydrogenase leakage assay
To monitor cell damage and injury, the amount of lactate dehydrogenase (LDH) released in the culture medium was measured. Briefly, optimum numbers of cells were seeded in 24-well plates and all the necessary controls were included. The plates were incubated overnight at 37 °C in a CO2 incubator to allow the cells to adhere and proliferate. The next day, the cells were transfected with the respective plasmids. All transfections were carried out at least in triplicate and the assays were independently repeated three times. After 48 h, the plates were removed from the incubator and 100 μl medium from each well was transferred to 96-well plates. Next, 100 μl LDH substrate, prepared according to the manufacturer’s instructions (LDH activity assay kit, Sigma-Aldrich), was added to each well. After a 20 min incubation period at room temperature, the enzyme reaction was arrested by adding 50 μl stop solution and the LDH activity was determined by measuring the absorbance at 490 nm.
2.10 Statistical analysis
In this study, representative experiments from three independent experiments are shown. Results for each experiment are given as a mean of triplicates ± SE. Statistically significant differences between sample groups were determined using the Student’s t-test. A p-value of <0.05 was considered significant.
3 Results
3.1 CT-Cx43 is down-regulated in primary human breast cancers
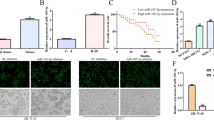
In addition to the full-length Cx43, the carboxy-terminal domain of Cx43 (CT-Cx43) is known to be independently expressed in various cells and tissues. In order to assess Cx43 and CT-Cx43 expression in primary human breast cancers, we performed Western blotting using a rabbit anti-Cx43 antibody on pathologically confirmed human breast cancer tissue samples and their adjacent normal control tissue samples. Out of the 60 samples tested, 23 samples showed a decreased expression of CT-Cx43 in the tumor tissues (T) compared to adjacent normal tissues (N), without an obvious change in the expression of full-length Cx43 (Fig. 1a). After quantification (see materials and methods) CT-Cx43 showed a ~4 fold decrease in expression in the 23 tumor tissues compared to the adjacent normal tissues, whereas full-length Cx43 showed no change (Fig. 1b). Interestingly, clinico-pathological evaluation of the 23 samples with a decreased CT-Cx43 expression indicated that all these samples were of low tumor grade (grade I, II; Table 1).
CT-Cx43 is down-regulated in low grade human breast cancers.A) Representative Western blots showing full-length (FL)-Cx43 and CT-Cx43 expression in the low grade human breast cancer tissues (T) and adjacent normal tissues (N) using an anti-Cx43 antibody that detects FL-Cx43 as well as CT-Cx43. β-actin was used as loading control. IDC: Infiltrating ductal carcinoma. SC: Squamous cell carcinoma. MU-C: Mucinous carcinoma. DCIS: Ductal carcinoma in situ. B) Quantitative analysis of immunoblots of 23 breast cancer tissues and their adjacent normal tissues (*p < 0.05). Protein bands were normalized to β- actin bands
3.2 CT-Cx43 expression is inversely related with miR-125b expression in primary human breast cancers
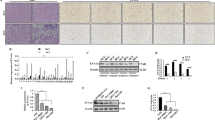
To reveal the influence of CT-Cx43 on the expression of miRNAs, we transfected MCF-7 cells with a CT-Cx43 expression construct and assessed miRNA expression patterns using end-point PCR. MCF-7 cells were chosen because they express wild-type p53 (see below) and are known to have retained several features characteristic of mammary epithelium. Our data (see supplementary Fig. 4) indicate that, next to other miRNAs, miR-125b was significantly up-regulated in CT-Cx43 transfected MCF-7 cells. This result prompted us to explore a putative correlation between CT-Cx43 and miR-125b expression in primary human breast cancers. For this purpose, the primary breast cancer samples included above were tested for the expression of miR-125b. To achieve this, total RNA was isolated from the tumor and corresponding normal tissues and subjected to qRT-PCR to quantify the miR-125b expression levels in these tissues. As shown in Fig. 2a, miR-125b expression was found to be ~4fold up-regulated in the tumor (T) tissues compared to the normal (N) tissues. This up-regulation correlated well with the previously observed down-regulation of CT-Cx43 in the same breast tumor samples. In order to further ascertain the role of CT-Cx43 in regulating the expression of miR-125b, we exogenously over-expressed CT-Cx43 in MCF-7 cells. The over-expression of CT-Cx43 was confirmed by Western blotting (Fig. 2b, compare the CT-Cx43 protein band in lane III with those in lanes I and II). Interestingly, qRT-PCR of total RNA extracted from the CT-Cx43 over-expressing MCF-7 cells revealed a ~10 fold decrease in the expression of miR-125b as compared to the un-transfected cells (Fig. 2c). These latter results confirm an inverse relationship between CT-Cx43 and miR-125b expression.
CT-Cx43 and miR-125b expression are inversely related in human breast cancers. A) qRT-PCR of miR-125b in human breast cancer tissues (T) and adjacent normal tissues (N). Values are expressed as fold expression of miR-125b in cancer tissue (T) relative to normal tissue (N) (*p < 0.05). B) Western blot showing over-expression of CT-Cx43 in transiently transfected MCF-7 cells (lane III) compared to un-transfected cells (lane I) and vehcile (plasmid only) transfected cells (lane II). β-actin was used as a loading control. C) qRT-PCR of miR-125b in CT-Cx43 transfected and control MCF-7 cells (n = 3). miR-125b expression was normalized to miR-16 as an internal control. Data are expressed as mean ± SEM
3.3 CT-Cx43 regulates p53 expression in breast cancer cells
Since, miR-125b is known to target p53 mRNA and, by doing so, to down-regulate its expression, we hypothesized that down-regulation of miR-125b by CT-Cx43 may influence the expression of p53. To confirm this, we first investigated the expression of p53 in the 23 primary human breast cancer tissues with a low CT-Cx43 expression (see above) and their adjacent normal tissues. For this purpose Western blotting, using a rabbit anti-p53 antibody, was performed on proteins extracted from the tumor and normal tissues. As shown in Fig. 3a, the expression of full-length p53 was markedly decreased in the tumor (T) tissues compared to the adjacent normal (N) tissues. As a loading control β-actin was used. Further quantification of the protein bands indicated that the expression of p53 was ~3fold less in the tumor tissues compared to the normal tissues (Fig. 3b). A comparison between the expression levels of Cx43, CT-Cx43, p53 and miR-125b and the pathological grades of the 23 breast tumor samples tested is provided in Table 1.
CT-Cx43 positively regulates p53 expression in human breast cancers.A) Western blots showing the expression of p53 in primary human breast cancer tissues (T) compared to adjacent normal tissues (N). The same set of breast cancer tissues was used for CT-Cx43 and p53 expression analyses. β-actin was used as loading control. B) Quantitative analysis of cancer tissues (T) and adjacent normal tissues (N) (*p < 0.05). The fluorescence of p53 bands was normalized to that of β-actin. C) Western blot showing over-expression of CT-Cx43 in MCF-7 cells (middle panel, compare un-transfected and vechile control cells, lane I & II, with CT-Cx43 transfected cells, lane III) and its effect on p53 expression. β-actin was used as loading control (bottom panel). D) Bar graph representing densitometeric analyses of FL-Cx43, CT-Cx43 and p53 protein bands (n = 3) in un-transfected, vechile (plasmid only) transfected and CT-Cx43 transfected MCF-7 cells. Data are expressed as mean ± SEM
To confirm the link between CT-Cx43 and p53 expression regulation, we exogenously over-expressed CT-Cx43 in MCF-cells. The effect of this over-expression on the expression of p53 was investigated by Western blotting. As shown in Fig. 3c, CT-Cx43 over-expression (middle panel, lane III) resulted in an increased expression of p53 (top panel, lane III) compared to un-transfected (lane I) and plasmid-only (lane II) transfected MCF-7 cells. As a loading control β-actin was used. Further quantification of the protein bands indicated that full-length p53 showed a ~2.5 fold increase in expression in the CT-Cx43 transfected cells compared to the un-transfected and vehicle control transfected cells (Fig. 3d). Full-length Cx43 showed no significant changes in expression between the un-transfected, vehicle control and CT-Cx43 transfected cells. Together, the above results indicate that CT-Cx43 regulates the expression of p53 and that this regulation may be mediated by miR-125b.
3.4 Down-regulation of miR-125b by CT-Cx43 results in decreased cell proliferation
In order to ascertain the functional implications of CT-Cx43 and miR-125b expression regulation, we performed MTT cell proliferation assays. For this purpose, 1 × 104 MCF-7 cells were seeded in 12-well plates and after 24 h, the cells were either separately transfected with CT-Cx43 or miR-125b expression constructs, or co-transfected with these constructs. After 48 h, the MTT assay was performed as described in the materials and methods section. As shown in Fig. 4a, exogenous over-expression of CT-Cx43 in the MCF-7 cells resulted in a significant decrease of metabolically active cells compared to the control (i.e., reagents plus cells only). Moreover, exogenous over-expression of miR-125b resulted in an increased number of metabolically active cells compared to the control cells. Co-transfection of miR-125b and CT-Cx43 revealed that miR-125b over-expression counteracted the anti-proliferative effect of CT-Cx43 on MCF-7 cells. From the above results we conclude that miR-125b and CT-Cx43 have an inverse relationship in terms of expression and physiological function in MCF-7 cells.
CT-Cx43 inhibits the proliferative activity of miR-125b on MCF-7 cells. MCF-7 cells were transfected with CT-Cx43 or miR-125b, or co-transfected with CT-Cx43 and miR-125b. 48 h post transfection, cell viability and cell death assays were performed. A) MTT cell viability assay showing that CT-Cx43 decreases proliferation of MCF-7 cells compared to control (reagent plus empty vector only) MCF-7 cells (*p < 0.05), whereas miR-125b increases the proliferation of MCF-7 cells. CT-Cx43 seems to counteract the proliferative activity of miR-125b after co-transfection (CT-Cx43 + miR-125b). B) LDH cell death assay showing that, in comparison to the negative control (cont.: reagents + cells only), transfection with CT-Cx43 or miR-125b did not induce any significant change in the death of MCF-7 cells. As a positive control (+ve cont.) for cell death, MCF-7 cells were treated with tritonX-100 (2 % v/v). The data shown represent three independent experiments performed in triplicate. Data are expressed as mean ± SEM
Since the MTT assay is based on the presence of viable cells, we wondered whether the over-expression of CT-Cx43 decreases cell proliferation or, alternatively, increases cell death. To address this issue, we performed a lactate dehydrogenase (LDH) leakage assay. For this purpose, 1 × 104 MCF-7 cells were seeded in 12-well plates and after 24 h the cells were either separately transfected with the CT-Cx43 or miR-125b expression constructs, or co-transected with both these constructs. After 48 h, the LDH leakage assay was performed as described in the materials and methods section. As shown in Fig. 4b, exogenous over-expression of CT-Cx43 did not result in a significant change in the release of LDH compared to the control cells (i.e., cells plus reagent only). Similarly, exogenous over-expression of miR-125b alone or miR-125b and CT-Cx43 combined did not result in a significant change in the release of LDH compared to the control cells. From these results we conclude that over-expression of CT-Cx43 does not increase cell death but, instead, decrease cell proliferation.
4 Discussion
Here, we provide evidence for a potential role of the carboxy-terminal domain of Cx43 (CT-Cx43) in controlling cellular proliferation. Earlier studies have shown the independent expression of CT-Cx43 in various cells and tissues [8]. Insight into the molecular mechanism underlying this expression was recently provided by Hussain et al. [30]. Additionally, it has been shown by many groups that, in addition to full length Cx43, CT-Cx43 is equally responsible for controlling various aspects of cellular growth and proliferation [10]. These observations prompted us to explore the expression status of Cx43 and CT-Cx43 in primary human breast cancer samples. Out of 60 samples tested, 30 showed down-regulation of both full-length Cx43 and CT-Cx43. In addition, 23 samples showed down-regulation of CT-Cx43 compared to adjacent normal tissue samples with no apparent change in full length Cx43 expression. Interestingly, all these 23 samples were well-differentiated and, based on clinico-pathological characteristics, classified as being at the primary stages of breast cancer development (grade I and II). Additionally, we found that exogenous over-expression of CT-Cx43 in MCF-7 breast cancer-derived cells affected the expression of various miRNAs, in particular miR-125b. These observations prompted us to explore the effect of CT-Cx43 down-regulation on the expression of miR-125b in the 23 primary breast cancer tissues and their adjacent normal tissues. We found that miR-125b was significantly up-regulated in the breast cancer tissues compared to the normal tissues. Although these results are in agreement with an oncogenic role of miR-125b [31], other studies have indicated that miR-125b may acts as a tumor suppressor in breast cancer [32]. A possible explanation for this presumed contradiction may be the cellular heterogeneity of the tumors examined [33] and/or alterations in their molecular make-up during disease progression (i.e., increasing grade). Moreover, recent reports have shown that in breast cancer stem cells [34] and in chemo-resistant breast cancers [35] the expression of miR-125b is significantly up-regulated. To further ascertain whether a link exists between CT-Cx43 and miR-125b expression, we exogenously (co-) expressed CT-Cx43 and miR-125b inMCF-7 cells. We found that over-expression of CT-Cx43 resulted in a significant decrease in the expression of miR-125b. Although the mechanism underlying this effect remains to be elucidated, it has been found that CT-Cx43 can affect the expression of various genes, either directly or indirectly [36, 37]. Interestingly, the observed localization of CT-Cx43 in the cell nucleus lends credence to the notion that CT-Cx43 may directly affect gene regulation [10]. To uncover the significance of CT-Cx43 and miR-125b expression on breast cancer development, we set out to investigate the effect of miR-125b on the expression of one of its known targets, p53. First, we assessed the expression of p53 in primary breast cancer samples, and found that p53 was down-regulated in samples that showed a lower expression of CT-Cx43 compared to its adjacent normal tissues. p53 is known for its tumor suppressive properties and mutations in p53 have been reported in various cancers, including breast cancer [38, 39]. Our primary breast cancer data were confirmed through over-expression of CT-Cx43 in MCF-7 cells, with a differential down-regulation of the miR-125b level and a concomitant increase in the p53 level. The physiological significance of the CT-Cx43/miR-125b regulatory network was established using MTT proliferation and LDH leakage assays. From the combined results we conclude that CT-Cx43 has a proliferation inhibitory function, while miR-125b has a proliferation stimulatory function. Interestingly, we found that the stimulatory function of miR-125b can be counteracted by CT-Cx43. It has been well-established that CT-Cx43 can regulate cellular growth and proliferation independent from full length Cx43, without the formation of gap-junctions [8]. Hence, our data provide an interesting lead to understanding the molecular network underlying breast epithelial cell proliferation and its derailment during carcinogenesis.
References
G. Mese, G. Richard, T.W. White, Gap junctions: basic structure and function. J. Investig. Dermatol. 127, 2516–2524 (2007)
H.A. Dbouk, R.M. Mroue, M.E. El-Sabban, R.S. Talhouk, Connexins: a myriad of functions extending beyond assembly of gap junction channels, Cell. Commun. Signals 7, 4 (2009)
K. Willecke, J. Eiberger, J. Degen, D. Eckardt, A. Romualdi, M. Guldenagel, U. Deutsch, G. Sohl, Structural and functional diversity of connexin genes in the mouse and human genome. Biol. Chem. 383, 725–737 (2002)
G. Pointis, J. Gilleron, D. Carette, D. Segretain, Physiological and physiopathological aspects of connexins and communicating gap junctions in spermatogenesis, philosophical transactions of the royal society of london. Series B, Biol. Sci. 365, 1607–1620 (2010)
S. Sirnes, J. Bruun, M. Kolberg, A. Kjenseth, G.E. Lind, A. Svindland, A. Brech, A. Nesbakken, R.A. Lothe, E. Leithe, E. Rivedal, Connexin43 acts as a colorectal cancer tumor suppressor and predicts disease outcome. Int. J. Cancer 131, 570–581 (2012)
M. Oyamada, Y. Oyamada, T. Takamatsu, Regulation of connexin expression. Biochim. Bophys. Acta 1719, 6–23 (2005)
R. Ismail, R. Rashid, K. Andrabi, F.Q. Parray, S. Besina, M.A. Shah, M. Ul Hussain, Pathological implications of Cx43 down-regulation in human colon cancer. Asian Pac. J. Cancer Prev. 15, 2987–2991 (2014)
C. Moorby, M. Patel, Dual functions for connexins: Cx43 regulates growth independently of gap junction formation. Exp. Cell Res. 271, 238–248 (2001)
J.E. Trosko, R.J. Ruch, Gap junctions as targets for cancer chemoprevention and chemotherapy. Curr. Drug Targets 3, 465–482 (2002)
X. Dang, B.W. Doble, E. Kardami, The carboxy-tail of connexin-43 localizes to the nucleus and inhibits cell growth. Mol. Cell. Biochem. 242, 35–38 (2003)
W.D. Foulkes, p53–master and commander. New Eng. J. Med. 357, 2539–2541 (2007)
E. Wawryk-Gawda, P. Chylinska-Wrzos, M. Lis-Sochocka, K. Chlapek, K. Bulak, M. Jedrych, B. Jodlowska-Jedrych, P53 protein in proliferation, repair and apoptosis of cells. Protoplasma 251, 525–533 (2014)
L.A. Voutsadakis, The chemosensitivity of testicular germ cell tumors. Cell. Oncol. 37, 79–94 (2014)
V.S. Bielicka, P. Domagala, D. Bielicki, K. Safranow, W. Domagala, Thymidylate synthase expression and P21WAF1/P53 phenotype of colon cancers identify patients who may benefit from 3-flurouracil based therapy. Cell. Oncol. 37, 17–28 (2014)
N. Almog, V. Rotter, Involvement of p53 in cell differentiation and development. Biochim. Biophys. Acta 1333, F1–27 (1997)
A.J. Giaccia, M.B. Kastan, The complexity of p53 modulation: emerging patterns from divergent signals. Genes Dev. 12, 2973–2983 (1998)
K. Sakaguchi, J.E. Herrera, S. Saito, T. Miki, M. Bustin, A. Vassilev, C.W. Anderson, E. Appella, DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 12, 2831–2841 (1998)
S.W. Lowe, Activation of p53 by oncogenes. Endocr. Relat. Cancer 6, 45–48 (1999)
L. Zheng, J.Q. Ren, H. Li, Z.L. Kong, H.G. Zhu, Downregulation of wild-type p53 protein by HER-2/neu mediated PI3K pathway activation in human breast cancer cells: its effect on cell proliferation and implication for therapy. Cell Res. 14, 497–506 (2004)
N. Rivlin, R. Brosh, M. Oren, V. Rotter, Mutations in the p53 tumor suppressor gene: important milestones at the various steps of tumorigenesis. Genes Cancer 2, 466–474 (2011)
P.A. Muller, K.H. Vousden, p53 mutations in cancer. Nat. Cell Biol. 15, 2–8 (2013)
M. Ul Hussain, Micro-RNAs (miRNAs): genomic organisation, biogenesis and mode of action. Cell Tissue Res. 349, 405–413 (2012)
R. Maqbool, M. Ul Hussain, MicroRNAs and human diseases: diagnostic and therapeutic potential. Cell Tissue Res. 358, 1–15 (2014)
R. Nagadia, P. Pandit, W.B. Coman, J.C. White, C. Punyadeera, MicroRNAs in head and neck cancer revisited. Cell. Oncol. 36, 1–7 (2013)
Y. Wang, M. Li, W. Zang, Y. Ma, N. Wang, P. Li, G. Zhao, MiR-429 upregulation induces apoptosis and suppresses invasion by targeting Bcl-2 and SP-1 in esophageal carcinoma. Cell. Oncol. 36, 385--394 (2013)
L. Rask, E. Balslev, R. Sokilde, E. Hogdall, H. Flyger, J. Eriksen, T. Litman, Differential expression of miR-139, miR-486 and miR-21 in breast cancer patients sub classified according to lymph node status. Cell. Oncol. 37, 215–227 (2014)
C. Salazar, R. Nagadia, P. Pandit, J.C. White, N. Banerjee, N. Dimitrova, W.B. Coman, C. Pundyadeera, A novel saliva based microRNA biomarker panel to detect head and neck cancers. Cell. Oncol. 37, 331–338 (2014)
J. Banzhaf-Strathmann, D. Edbauer, Good guy or bad guy: the opposing roles of microRNA 125b in cancer. Cell Commun. Signal 12, 30 (2014)
C. Chen, D.A. Ridzon, A.J. Broomer, Z. Zhou, D.H. Lee, J.T. Nguyen, M. Barbisin, N.L. Xu, V.R. Mahuvakar, M.R. Andersen, K.Q. Lao, K.J. Livak, K.J. Guegler, Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 33, e179 (2005)
M. Ul-Hussain, S. Olk, B. Schoenebeck, B. Wasielewski, C. Meier, N. Prochnow, C. May, S. Galozzi, K. Marcus, G. Zoidl, R. Dermietzel, Internal ribosomal entry site (IRES) activity generates endogenous carboxyl-terminal domains of Cx43 and is responsive to hypoxic conditions. J. Biol. Chem. 289, 20979–20990 (2014)
N. Wu, X. Lin, X. Zhao, L. Zheng, L. Xiao, J. Liu, L. Ge, S. Cao, MiR-125b acts as an oncogene in glioblastoma cells and inhibits cell apoptosis through p53 and p38MAPK-independent pathways. Br. J. Cancer 109, 2853–2863 (2013)
A. Feliciano, J. Castellvi, A. Artero-Castro, J.A. Leal, C. Romagosa, J. Hernandez-Losa, V. Peg, A. Fabra, F. Vidal, H. Kondoh, Y.C.S. Ramon, M.E. Lleonart, miR-125b acts as a tumor suppressor in breast tumorigenesis via its novel direct targets ENPEP, CK2-alpha, CCNJ, and MEGF9. PLoS One 8, e76247 (2013)
A. Geurts van Kessel, The cancer genome: from structure to function. Cell. Oncol. 37, 155–165 (2014)
H.J. Wang, Y.Q. Guo, G. Tan, L. Dong, L. Cheng, K.J. Li, Z.Y. Wang, H.F. Luo, miR-125b regulates side population in breast cancer and confers a chemoresistant phenotype, J.Cell. Biochem. 114, 2248–2257 (2013)
H. Wang, G. Tan, L. Dong, L. Cheng, K. Li, Z. Wang, H. Luo, Circulating MiR-125b as a marker predicting chemoresistance in breast cancer, PloS One.7, e34210 (2012)
D.A. Iacobas, E. Scemes, D.C. Spray, Gene expression alterations in connexin null mice extend beyond the gap junction. Neurochem. Int. 45, 243–250 (2004)
E. Kardami, X. Dang, D.A. Iacobas, B.E. Nickel, M. Jeyaraman, W. Srisakuldee, J. Makazan, S. Tanguy, D.C. Spray, The role of connexins in controlling cell growth and gene expression. Prog. Biophys. Mol. Biol. 94, 245–264 (2007)
M. Hollstein, D. Sidransky, B. Vogelstein, C.C. Harris, p53 mutations in human cancers. Science 253, 49–53 (1991)
U.M. Moll, G. Riou, A.J. Levine, Two distinct mechanisms alter p53 in breast cancer: mutation and nuclear exclusion. Proc. Natl. Acad. Sci. U. S. A. 89, 7262–7266 (1992)
Acknowledgments
The junior research fellowships to RM (UGC-JRF 19-06/2011 Ci EU-IV), SN (ICMR JRF-2012/HRD-29) and project fellowships to RI (BT/PR14336/MED/30/493/2010) and RR (SR/WOS-A/LS-661/2012) are highly acknowledged. The present work was supported by project grant (BT/PR11917/MED/30/181/2009) from the Department of Biotechnology (DBT), New Delhi, to MUH.
Conflict of interest
All authors declare no conflict of interest
Author information
Authors and Affiliations
Corresponding author
Additional information
Raihana Maqbool and Rabiya Rashid contributed equally to the work.
Electronic supplementary material
Supplementary Fig. 1
Total RNA was isolated from the normal and breast cancer samples as described in material and methods. To check the integrity of the RNA, 5 μl of each sample was loaded on formaldehyde agarose gel. After electrophoresis, the agarose gel was incubated for 5–10 min in MOPS buffer containing ethidium bromide. The gel was visualized in gel-documentation apparatus. As shown in suppl. Fig. 1, intact and prominent bands, corresponding to 28S and18S rRNA can be detected. Moreover, RNA bands corresponding to small RNAs can also be detected. Thus, the presence of intact RNA bands established the integrity of the RNA obtained from the tissue samples. (GIF 181 kb)
Supplementary Fig. 2
Total protein was extracted from the tumor and adjacent normal breast samples as described in material and methods. 30 μg of protein was loaded on SDS-PAGE and electrophoresis was performed. After PAGE, Western blot was performed using anti-Cx43 antibody as described previously. As shown in suppl. Fig. 2, anti-Cx43 detected FL-Cx43 and CT-Cx43 in the normal samples (N). However, the tumor samples (T) showed decreased expression of FL-Cx43 as well as CT-Cx43. (GIF 169 kb)
Supplementary Fig. 3
To investigate whether CT-Cx43 has the potential to bind the miR-125b promoter, we expressed CT-Cx43 as GST fusion protein in bacterial system. The GST-CT-Cx43 was purified from the bacterial lysate using Gluthatione-agarose beads. Additionally, 200 bps of miR-125b promoter (upstream of transcriptional site containing the Tata box; Gene ID: 406,911) was PCR amplified using the forward primers 5`-TTTGAAAAGACACTAAATCCT-3` and the reverse primer 5’ACAATGTTTTCTTTCTTGAGA-3`. Prior to PCR, the primers were 32p labeled using T4 polynucleotide kinase and gamma phosphate (32P) labeled ATP. The 200 bp DNA probe and purified GST-CT-CX43 were incubated together and gel-shift assay was performed. As shown in suppl. Fig. 3, lane II, addition of CT-Cx43 did not show any binding to the miR-125 DNA. Moreover, addition of anti-CT-Cx43 did not show and supershift of miR-125b probe (lane III). Thus, our initial data indicate that CT-Cx43 does not bind the proximal promoter element of miR-125b. Further upstream of 200 bp needs to further screened for any binding of CT-Cx43. (GIF 62.9 kb)
Supplementary Fig. 4
In order study the effect of CT-Cx43 on the expression of various growth related miRNAs, we transiently transfected MCF-7 cells with either CT-Cx43 plasmid or with empty plasmid. After 36 h transfection, RNA was isolated and using stem-loop primers strategy (for details consult material methods of the main manuscript) many miRNAs were PCR amplified (here denoted as “a, b, c, d, e, f, g, h, I, j), including the miR-125b. As shown in suppl Fig. 4, various miRNAs showed differential regulation between empty plasmid and CT-Cx43 transfected MCF-7 cells (compare “a & a`; e & e`; f & f`). Interestingly, miR-125b showed highly significant up-regulation in CT-Cx43 transfected MCF-7 cells. (GIF 291 kb)
Supplementary Fig. 5
In order to establish the possibility of nuclear localization of CT-Cx43 in breast samples, we isolated the nuclear and cytosolic fraction from the normal and tumor breast cancer samples using nuclear extraction kit (Sigma adrich). 30 μgs of each fraction was run on 12 % SDS-PAG and Western blot was performed using anti-Cx43 antibody as described in material and methods. As shown in Suppl. Fig. 5, anti-Cx43 antibody detected FL-Cx43 in the cytosolic fraction (Cyt) fraction of normal (N) as well as in the tumor (T) samples. Moreover, CT-Cx43 was detected in the cytosolic (Cyt.) and nuclear fractions (Nuc.) of the normal samples (N). However, very less/or no signal of CT-Cx43 was detected from the nuclear fraction (Nuc.) of tumor samples (T). Hence, it may be concluded that CT-Cx43 has the potential to localize in cell nucleus when it is expressed in the cells. (GIF 122 kb)
Rights and permissions
About this article
Cite this article
Maqbool, R., Rashid, R., Ismail, R. et al. The carboxy-terminal domain of connexin 43 (CT-Cx43) modulates the expression of p53 by altering miR-125b expression in low-grade human breast cancers . Cell Oncol. 38, 443–451 (2015). https://doi.org/10.1007/s13402-015-0240-x
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13402-015-0240-x