Abstract
The present study for the first time describes biological and molecular characterization of Citrus tristeza virus (CTV) occurring in the Terai area of Uttarakhand State in Northern Himalaya region of India. Direct antigen coated-ELISA and reverse transcriptase-polymerase chain reaction (RT-PCR) detected the CTV infection in Acid lime cv. Pant lemon (Citrus aurantifolia) orchards of Pantnagar with an estimated disease incidence of 16.6–20.5 %. To know the biological and genetic properties, an isolate, CTV Pant 4 was characterized. Isolate Pant 4 could be graft transmitted to Kinnow, Nagpur and Darjeeling mandarins, Mosambi sweet orange, Kagzi lime, Sweet lime, Sour orange but not to Rough lemon. The sequence analyses of the 5′ORF1a (3038 nucleotides) of LPro domain and 3′end (2058 nt) covering ORF7–ORF10 regions of the CTV genome revealed that Pant 4 was closely related to the previously reported Indian CTV isolate, Kpg3 from Northeastern Himalaya region with 97 and 98 % sequence identity, respectively. Whereas, it differed from the previously reported CTV isolate B165 from Southern India with 79 and 92 % identity, respectively for 5′ORF1a and 3′ end regions. Recombination and SplitsTree decomposition analyses indicated that CTV isolate Pant 4 was a recombinant isolate originating from Kpg3 as a major and B165 as a minor donor.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Citrus tristeza virus (CTV), one of the most destructive pathogens infecting citrus causes huge crop losses by killing over 100 million citrus trees worldwide including about one million trees in India [1, 3, 18]. CTV, a phloem limited plant virus with long flexuous particle, 2,000× 11–12 nm in size belonging to the genus Closterovirus is transmitted predominantly by brown citrus aphid, Toxoptera citricida in semi-persistent manner, and spread over a distant areas by transportation of infected planting material [18]. CTV infects nearly all the citrus species, cultivars, and inter-generic hybrids and some citrus relatives, inducing symptoms like decline, stem pitting, seedling yellows, vein clearing and flecking [5, 6, 14, 23]. CTV isolates differ in their biological characteristics, types and intensity of symptoms induced in different citrus hosts and aphid transmissibility [12], however, molecular determinant for symptom development in citrus hosts are not yet clearly known.
The CTV genome contains single stranded RNA of about 19.3 kb with 12 open reading frames (ORFs) encoding at least 19 proteins [12, 21]. The ORFs 1a and 1b encode replication related proteins, whereas, the 3′ end ten ORFs (ORFs 2–ORF11) encodes proteins p33, p6, p65, p61, p27, p25, p18, p13, p20 and p23 [11, 22]. Analysis of 19 complete genomes CTV isolates available in GenBank data base revealed extensive variation in sequence among the CTV isolates and enabled them to be grouped into six genotypes, recognized as T36 (Florida), T30 (Florida), VT (Israel), B165 (India), HA16-5 (Hawaii) and RB genotypes (New Zealand) [6, 10, 16, 19].
In India, CTV occurs in all the citrus growing geographical zones: Northeast, Northwest, Central and South with an estimated disease incidence ranging from 10 to 90 % [1, 4]. Occurrence of genetic diversity in CTV has been reported from India [5, 9, 11, 20, 23]. Analysis of the viral sequence of several CTV isolates from citrus growing areas of Northeast, North, South and Central India has documented for extensive genetic diversity classifying the Indian CTV isolates into at least six to eight genogroups. [7].
In the Uttarakhand Terai area of Northern Himalaya region of India, Acid lime cv. Pant lemon (Citrus aurantifolia) is cultivated at commercial level. Previously, there was no report of CTV occurring in citrus orchards of this region. During our survey, the citrus orchards surveyed appeared to be very poor in growth and showing declining symptoms; those diseases resembled to virus or virus-like diseases. Thus, in the present study, an effort was made to characterize the virus associated to cause the disease in this plant through biological indexing and sequence analysis of viral genome, and then to estimate disease incidence. Present study demonstrated the association of CTV with the infected Acid lime in this region, and further molecular analysis determined putative demographic history of this virus and its origin through recombination events.
Materials and Methods
Survey and Sample Collection
Survey was conducted in the two Pant lemon (C. aurantifolia) orchards, orchard-1 and orchard-2 at Experimental Farm at G. B. Pant Univ. of Agric. and Technology (GBPUA&T), Pantnagar, Uttarakhand of Northern Himalaya region of India in the month of September 2010. Total number of 52 samples, 34 from orchard 1 and 18 from Orchard 2 with one to two twigs were collected from each of the randomly selected trees from both the orchards and brought to the laboratory for molecular assay. For detection of CTV and the percent plant infection direct antigen coated-ELISA (DAC-ELISA) was carried out as described earlier [4, 5] using CTV specific antisera obtained from Advanced Centre for Plant Virology, Indian Agricultural Research Institute, New Delhi.
Host Range
To determine biological properties of CTV in Terai region of Uttarakhand, host range of an isolate designated as CTV Pant 4 was selected randomly from Pant lemon orchard-2 was studied through bud wood graft transmission using eight citrus hosts (Table 1), viz., Kinnow mandarin (C. reticulata), Nagpur mandarin (C. reticulata), Darjeeling mandarin (C. reticulata), Kagzi lime (C. aurantifolia), Mosambi sweet orange (C. sinensis), Rough lemon (C. jambhiri), sweet lime (C. limettoides) and Sour orange (C. aurantium) following the method described earlier [4]. The successful graft unions were considered and maintained in an insect-proof greenhouse for observation of CTV induced symptoms.
RNA Isolation, cDNA Synthesis and Cloning of Viral Sequences
A set of nine pairs of specific and degenerate primers (Table 2); five of them targeting the sequence of 5′ORF1a region of LPro domain and four targeting for the 3′end region covering ORF7–ORF10 of CTV genome described earlier [6] were used for amplification of the viral nucleotide sequences. Total plant RNA was isolated and first strand cDNA was synthesized using the protocols developed previously [26]. The RT-PCR was performed using the protocol described previously [4] with different sets of primers adapting appropriate annealing temperature. PCR product was purified (Promega, Madison, USA), cloned in the T&A cloning vector (RBC, UK) following manufacturer’ recommendation and the positive clones were selected using standard method of colony PCR. Two clones for each PCR product were sequenced outsourcing in both the directions using the vector derived primers M13F and M13R in automatic sequencer (AB 13130 Genetic analyser, Chromous Biotech, Bangalore). Overlapped contig sequences were assembled in DNA STAR SeqMan software and consensus sequences were used for further molecular analysis.
Sequence Analysis
The sequence of CTV isolate Pant 4 and corresponding sequence of six CTV isolates, T36 (U16304), T30 (AF260651), VT (U56902), B165 (EU076703), HA16-5 (GQ454870) and NZRB-G90 (FJ525432) representing each of the six internationally recognized CTV genotypes, and one previously reported Indian isolate, Kpg3 (HM573451) [6, 10, 16] were taken for sequence analyses and comparisons. The sequences were aligned using ClustalX version 1.81 [27]. Sequence identity matrix was generated using computer based program GeneDoc, version 2.6.002. To study nucleotide mutation and relative rate of dN and dS and its difference among the CTV sequences, the Tajima’s neutrality test was carried out using MEGA4 [24, 25]. Maximum likelihood phylogenetic tree analyses were conducted using the program MEGA4 [25]. The potential recombination events were identified using recombination-detecting program RDP3 program v.3.34; implemented with seven algorithms, RDP, GENECONV, Bootscan, MaxChi, Chimaera, 3Seq and SiScan [15]. Recombination events were checked manually and confirmed by UPGMA (Unweighted pair group method with arithmetic mean) tree using RDP3.
Results
Detection of CTV and Estimation of Disease Incidence
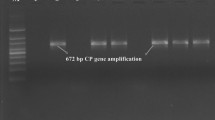
The poor growth of trees along with general decline was observed in the Pant lemon orchards, Orchard-1 and orchard-2 surveyed. Both the orchards were found to be infected by CTV when assayed with DAC-ELISA using CTV specific antisera. Three out of 18 samples in Orchard-1 (16.6 % disease incidence) and seven out of 34 samples in Orchard-2 (20.5 % disease incidence) were found to be CTV infected. The infected samples showed OD values of 0.74–1.57, whereas, healthy control showed OD value of 0.46. The virus infection in the ELISA positive samples were confirmed by RT-PCR for amplification of 404 nt sequence fragment of 5′ORF1a of CTV genome using primers, KLM 488 and KLM 491 described previously [5].
Host Range of CTV Isolates Pant 4
The CTV isolate Pant 4 was biologically indexed on eight citrus hosts using bud wood grafting (Table 1). Infection of virus was assayed, and different kinds of symptoms induced by the virus in different hosts were recorded after 6 months of graft inoculation. Isolate Pant 4 was found to be transmitted to all citrus hosts tested in the present study, except to Rough lemon. The virus infection in the inoculated hosts was detected by DAC-ELISA. Vein clearing and decline symptoms were observed in Kagzi lime and Sweet lime. In addition, vein-corking in Kagzi lime, and bushy appearance in Sweet lime was also observed. Infected Nagpur mandarin was found to be asymptomatic. Sweet orange and Sour orange showed chlorosis, and additionally sweet orange appeared stunted. Darjeeling mandarin showed no visible symptoms, although poor growth with decline symptoms was observed after 6 months of inoculation.
Sequence Comparison
A total length of 3038 nt (Acc No. JN974901) at nucleotide coordinate positioning from 924 to 3992 nt of 5′ORF1a (5′ORF1a) and 2078 nt (Acc. No. JN974902) positioned from 16110 to 18267 nt (as per the full genome sequence of isolate Kpg3) comprising ORF7 (672 nt), ORF8 (504 nt), ORF9 (360 nt) and ORF10 (549 nt) at 3′ end of CTV genome of isolate Pant 4 were sequenced and compared with other CTV isolates. Sequence analyses revealed that the viral sequences and organisation of ORFs of isolate Pant 4 were similar to other CTV isolates.
Pair wise nucleotide identities for the 5′ORF1a, 3′end and the individual ORFs among CTV isolates including isolate Pant 4 were analysed and compared. For the 5′ORF1a, isolate Pant 4 shared 97–98 % nt identity with previously reported Darjeeling CTV isolate Kpg3 and Israel isolate VT, 89 % with Florida mild isolate T30 and 72–79 % with other CTV isolates. For the 3′end sequence analysis, Pant 4 was found to be very close with Kpg3 by 97 % identity, but distant from other CTV isolates by 91–94 % identities. Analyses of individual ORFs showed that Pant 4 had 99–100 % nt identity with Kpg3 for ORF7, 9 and 10, 96 % identity with B165 for ORF10 and 99 % with Hawaii isolate HA16-5 for ORF9.
Sequence identity of deduced amino acid (aa) of ORF7 to ORF10 of Pant 4 were compared and patterns of aa sequence identity were found to be similar as observed in nucleotide sequence identities. Isolate Pant 4 showed 99–100 % aa identity for ORF7, 9 and 10 with Kpg3 and 95–98 % with B165 for ORF 7, 8 and 10, however differed from Kpg3 with 91 % identity and from B165 by 87 % for ORF8. Interestingly, isolate Pant 4 had 100 % aa identity with HA16-5 for ORF9.
Eight CTV isolates including two Indian isolates, Pant 4 and Kpg3, and six isolates representing internationally recognized genotypes, were analyzed to estimate genetic variation using bio-statistical parameters in MEGA4 (Table 3). The sequence diversity (π) was maximum (π = 0.190) in the 5′ORF1a region due to high mutational rate (θ = 0.161) compared to the 3′end region (π = 0.070–0.083) among CTV isolates including present isolate Pant 4 (Table 3). Of four ORFs in 3′end region analyzed, maximum sequence diversity (π = 0.083) was observed in ORF9 and minimum (π = 0.070) in ORF7. Positive Tajima’s D values were observed for 5′ORF1a (0.994) and ORF7 (0.043) that indicated low level, whereas, negative D values for ORFs 8–10 (−0.098 to −0.215) signified a high level of frequency of polymorphism (Table 3). The ratio of dN to dS for the 5′ORF1a region was higher (0.724) compared to other ORFs of CTV genome. The dN/dS value less than 1.0 implied that the CTV encoded proteins are not much variable within the populations thus not under intense purifying selection.
Phylogenetic Analysis
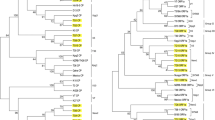
Phylogenetic analysis for the 5′ORF1a of Pant 4 and other CTV isolates generated overall six phylogroups; isolate Pant 4 grouped with Indian isolate Kpg3 and Israel isolate VT (Fig. 1a). Analysis for the 3′end sequences showed that Pant 4 grouped with isolate Kpg3 (Fig. 1b). In the analyses of individual ORFs, Pant 4 grouped together with Kpg3 and HA16-5 for ORF7 and ORF9, and only with Kpg3 for ORF10 (Figure not shown). Isolate Pant 4 was found to be a distinct in the phylogenetic tree for ORF8 sequence (Fig. 1c).
Phylogenetic relationships of the present CTV isolate Pant 4 with other isolates based on the nucleotide sequences of the 5′ORF1a region (a), assembled four ORFs (ORF7-10) at 3′end region (b) and ORF8 sequence (c) of the viral genomes, using maximum parsimony in the program MEGA4. The significance of the nodes was estimated with 1,000 bootstrap repetitions. The description of CTV isolates used for sequence analysis are of previously reported Indian isolate Kpg3 (HM 573451), Florida severe isolate T36 (U16304), Florida mild isolate T30 (AF260651), Israel severe isolate VT (U56902), New Zealand isolate NZRB-G90 (FJ525432), Indian isolate B165 (EU076703) and Hawaii isolate HA16-5 (GQ454870)
Neighbor-net decomposition (SplitsTree) analysis presented reticulate network among the CTV isolate (Figure not shown), indicating a complex evolutionary history among the CTV isolates. Analysis of the 5′ORF1a showed that isolates Pant 4, Kpg3 and VT were originated from common ancestors. The analysis of the 3′end also constructed reticulate network relationship among the CTV isolates indicating a close evolutionary relationship between Pant 4 and Kpg3 which might have been originated from distantly related ancestors.
Recombination Analysis
To detect recombination events, the 5′ORF1a and 3′end sequences of CTV isolates were examined using RDP3 program. No recombination event was identified in the 5′ORF1a, while one recombination event was detected at the 3′end at position of 655–1142 nt (ORF8), supported by maximum P value of 6.529 × 10−15 involving Kpg3 as major and B165 as minor donor. In UPGMA analysis, isolate Pant 4 grouped with major donor Kpg3 for non-recombinant consensus sequence (1143–654 nt) and with minor donor B165 for recombinant sequence (653–1142 nt), confirming recombination origin of isolate Pant 4 (Fig. 2).
Recombination analysis for assembled sequences of four ORFs (ORF 7-10) at 3′ end region of CTV genome of isolate Pant 4 using Chimaera algorithm that is implemented in recombination-detecting program RDP3. Position of sequence alignment and log P value were represented in X and Y axis, respectively. Sequence with recombination origin was indicated by deep shaded area
Discussion
Acid lime cv. Pant lemon is an important citrus species grown in Pantnagar area of Uttarakhand Terai region of Northern Himalaya region of India. The present study reported that the Pant lemon in this area is infected by CTV with an estimated disease incidence of 16–20 % using ELISA and RT-PCR. It is interesting to note that even under highly favourable environmental condition that prevailed in Uttarakhand Terai region of India, the isolate Pant 4 could not show typical visible symptoms on Pant lemon under field condition. Diagnosis of CTV based on visible symptoms was not possible as no CTV specific symptoms in Pant lemon orchard were observed. It was reported earlier that CTV infected citrus trees do not always produce external symptoms, as production of symptoms depends on hosts, virus strains and environmental conditions [2, 3, 13].
The symptomatology and experimental host range of isolate Pant 4 were found to be closely related with previously reported CTV isolate Kpg3 of the Darjeeling hills [5], suggesting isolate Pant 4 and Kpg3 are similar in pathogenicity. Sequence comparisons and phylogenetic tree analyses for 5′ORF1a and 3′end region revealed that isolate Pant 4 is closely related with Kpg3 and differed from previously reported another Indian isolate B165 of Bangalore region [19]. Isolate Pant 4 showed 99–100 % aa sequence identity with Kpg3 for ORF7, 9 and 10, indicating both the isolates are similar in pathogenecity, ORF7 encodes for major CP and RNAi suppressor, ORF9 for p13 protein with unknown function and ORF10 for RNAi suppressor [17]. ORF encodes p18 protein the function of which is unknown [17]; ORF8 of Pant 4 is different from Kpg3 isolate, indicating that p18 might have role in host specificity as Rough lemon is infected by the isolate Kpg3 in the Darjeeling hills but not by the isolate Pant 4 in glass conditions. A close relationship in amino acid sequence (99–100 % identity) of isolates Pant 4 and Kpg3 with Hawaii isolate HA16-5 for ORF7 and 9 was observed, that indicated both the Indian isolates might be related to Hawaii isolate in pathogenecity.
Homologous and non-homologous recombination is reported to be a fast means for evolution of CTV [6, 10, 15, 16, 19]. Present results indicated that Pant 4 is a recombinant CTV isolate showing ORF8 likely to be originated through recombination events by exchanging sequences of isolates Kpg3 and B165. This supports that RNA recombination has played a role in evolution of this CTV isolate. Earlier, isolates HA16-5, SY568, B165 and Kpg3 are reported to be recombinant/hybrid originated by multiple recombination events between CTV variants [6, 16, 19]. The SplitsTree analysis supported the phylogenetic relationship and evolutionary history of Pant 4 and Kpg3, showing that both the isolates originated from combination of common and distantly related ancestors. Previously, SplitsTree analyses demonstrated that phylogenetic relationships among CTV sequences are interconnected with a complex network [15].
The present study showed more sequence diversity in sequence of 5′ORF1a region and no recombination event was detected in this region. However, the biostatical analysis carried out in the present study suggested that 5′ORF1a region might have high mutation rate resulting in occurrence of genetic diversity among the CTV isolates. Previously, high rate of mutations, in addition to recombination and other biological and molecular phenomena, was reported to contribute for overall diversity and evolution of CTV isolates [8].
In conclusion, CTV occurs in Acid lime cv.Pant lemon orchards in Uttarakhand Terai area of Northern Himalaya region of India. The symtomatology, host range and sequence analysis suggested that CTV isolate Pant 4 is closely related to decline inducing Darjeeling CTV isolate Kpg3 but differs from South Indian severe isolate B165. The sequence, recombination and SplitsTree decomposition analyses indicated that CTV isolate Pant 4 is a putative recombinant isolate originating from Kpg3 as a major and B165 as a minor donor.
References
Ahlawat YS. Viruses, greening bacterium and viroids associated with citrus (Citrus species) decline in India. Indian J Agric Sci. 1997;67:51–7.
Ahlawat YS, Pant RP. Major virus and virus-like diseases of citrus in India: their diagnosis and management. Ann Rev Plant Pathol. 2003;2:447–74.
Bar-Joseph M, Dawson WO. Citrus tristeza virus. Encyclopedia Virol. 2008;1:520–5.
Biswas KK. Molecular diagnosis of Citrus tristeza virus in mandarin (Citrus reticulata) orchards of hills of West Bengal. Indian J Virol. 2008;19:26–31.
Biswas KK. Molecular characterization of Citrus tristeza virus isolates from the Northeastern Himalayan region of India. Arch Virol. 2010;155:959–63.
Biswas KK, Tarafdar A, Sharma SK. Complete genome of mandarin decline Citrus tristeza virus of Northeastern Himalayan hill region of India: comparative analyses determine recombinant. Arch Virol. 2012;157:579–83.
Biswas KK, Tarafdar A, Diwedi S, Lee RF. Distribution, genetic diversity and recombination analysisof Citrus tristeza virus of India. Virus Genes. 2012;45:139–48.
Cheng X, Wu X, Wang H, Sun Y, Qian Y, Luo L. High codon adaptation in Citrus tristeza virus to its citrus host. Virol J. 2012;9:113.
Ghosh DK, Aghave B, Roy A, Ahlawat YS. Molecular cloning, sequencing and phylogenetic analysis of coat protein gene of a biologically distinct CTV isolate occurring in central India. J Plant Biochem Biotechnol. 2009;18:105–8.
Harper SJ, Dawson TE, Pearson MN. Isolates of Citrus tristeza virus that overcome Poncirus trifoliata resistance comprise a novel strain. Arch Virol. 2010;155:471–80.
Hilf ME, Mavrodieva VA, Garnsey SM. Genetic marker analysis of a global collection of isolates of Citrus tristeza virus: characterization and distribution of CTV genotypes and association with symptoms. Phytopathology. 2005;95:909–17.
Karasev AV, Boyko VP, Gowda S, Nikolaeva OV, Hilf ME, Koonin EV, Niblett CL, Cline K, Gumpf DJ, Lee RF. Complete sequence of the Citrus tristeza virus RNA genome. Virology. 1995;208:511–20.
Korkmaz S, Cevik B, Onder S, Koc NK. Biological, serological and molecular characterization of Citrus tristeza virus isolates from different citrus cultivation regions of Turkey. Turk J Agric. 2008;32:369–79.
Lee RF, Bar-Joseph M. Tristeza. In: Compendium of citrus diseases, 2nd ed. St. Paul, MN, USA, APS Press; 2000. pp 61–3.
Martin S, Sambade A, Rubio L, Vives MC, Moya P, Guerri J, Elena SF, Moreno P. Contribution of recombination and selection to molecular evolution of Citrus tristeza virus. J Gen Virol. 2009;90:1527–38.
Melzer MJ, Borth WB, Sether DM, Ferreira S, Gonsalves D, Hu JS. Genetic diversity and evidence for recent modular recombination in Hawaiian Citrus tristeza virus. Virus Genes. 2010;40:111–8.
Moreno P, Ambros S, Albiach-Marti MR, Guerri J, Pena L. Citrus tristeza virus: a pathogen that changed the course of the citrus industry. Mol Plant Pathol. 2008;9(2):251–68.
Rocha-Pena MA, Lee RF, Lastra R, Niblett CL, Ochoa-Corona FM, Garnsey SM, Yokomi RK. Citrus tristeza virus and its aphid vector Toxoptera citricida: threats to citrus production in the Caribbean and Central and North America. Plant Dis. 1995;79:437–45.
Roy A, Brlansky RH. Genome analysis of an orange stem pitting Citrus tristeza virus isolate reveals a novel recombinant genotype. Virus Res. 2010;151:118–30.
Roy A, Manjunath KL, Brlansky RH. Assessment of sequence diversity in the 5-terminal region of Citrus tristeza virus from India. Virus Res. 2005;113:132–42.
Satyanayanana T, Gowda S, Boyko VP, Albiach-Marti MR, Mawassi M, Navas-Castillo J, Karasev AV, Dolja V, Hilf ME, Lewandowsky DJ, Moreno P, Bar-Joseph M, Garnsey SM, Dawson WO. An engineered closterovirus RNA replicon and analysis of heterologous terminal sequences for replication. Proc Natl Acad Sci U S A. 1999;96:7433–8.
Satyanayanana T, Gowda S, Mawassi M, Albiach-Marti MR, Ayllon MA, Robertson C, Garnsey SM, Dawson WO. Closterovirus encoded HSP70 homolog and p61 in addition to both coat proteins function in efficient virion assembly. Virology. 2000;278:253–65.
Sharma SK, Tarafdar A, Khatun D, Kumari S, Biswas KK. Intra-farm diversity and evidence of genetic recombination of Citrus tristeza virus isolates in Delhi region of India. J Plant Biochem Biotechnol. 2012;21:38–43.
Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–95.
Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–9.
Tarafdar A, Ghosh PD, Biswas KK. In planta distribution, accumulation, movement and persistence of Citrus tristeza virus in citrus host. Indian Phytopathol. 2012;65:184–8.
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The clustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;24:4876–82.
Acknowledgments
The authors are grateful to Head, Division of Plant Pathology; Director, IARI, New Delhi for providing financial and laboratory support; Prof. Anupam Varma for his guidance in this work. This study has been supported by Research Project (Code No. 24-33), Department of Biotechnology, Govt. of India. The first author is grateful to ICAR for financial assistance as JRF and thankful to Dr. A. K. Singh and Dr. (Mrs.) P. Sharma, IARI, for valuable suggestions. We are thankful to N. Trivedi, GBPAU&T, Pantnagar for his assistance during survey and sample collection.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Singh, J.K., Tarafdar, A., Sharma, S.K. et al. Evidence of Recombinant Citrus tristeza virus Isolate Occurring in Acid Lime cv. Pant Lemon Orchard in Uttarakhand Terai Region of Northern Himalaya in India. Indian J. Virol. 24, 35–41 (2013). https://doi.org/10.1007/s13337-012-0118-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13337-012-0118-8