Abstract
Aberrant expression of miR-137 has been reported in many kinds of cancers, but its mechanisms seem to be diversely. In the present study, we compared the expression level of miR-137 in 18 paired gastric cancer (GC) samples and surgical margin (SM) samples by RNA extraction and quantitative real-time PCR (QRT-PCR). Then, we investigated the effects of miR-137 on cell proliferation, cell cycle, and cell migration separately by cell growth counting assay, cell cycle analysis, and transwell assay. Candidate targets of miR-137 were selected by biological information analysis from the intersection of miRDB, Pictar, and TarScan. Finally, mRNA and protein expression level of Krűppel-like factor 12 (KLF12) and Myosin 1C (MYO1C) were tested by QRT-PCR and western blotting assay, followed by the Luciferase reporter assay to investigate the direct interaction between them and miR-137. The results showed that miR-137 was down-regulated in GC samples than in SM samples. The expression level of miR-137 was significantly higher in patients without the vascular embolus than those with vascular embolus. And the overall survival time of patients with high miR-137 expression was longer than those with low miR-137 expression. Over expression of miR-137 could inhibit the cell migration, proliferation, and promote cell cycle arrest in G0/G1 stage in BGC-823 and SGC-7901 cell lines. KLF12 and MYO1C might be the candidate target genes of miR-137 with direct interactions between them and miR-137. In conclusion, miR-137 plays tumor suppressor roles in gastric cancer cell lines by targeting KLF12 and MYO1C.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gastric cancer is one of the most mortality diseases in worldwide [1]. Scientists have found various related etiologies for cancerous progression. These etiologies often involved in abnormal expression and function of molecules, which could be helped to the diagnosis, prediction, and prognosis of gastric cancer. Among them, miRNA seems to play more and more important roles and will provide new insights of the target for gastric cancer.
miRNA is a kind of non-coding small RNA with 20–24 nt in length. miRNAs play roles in the post transcriptional level by pairing with 3′-UTR of target mRNAs in various biological processes. Some of them play as oncogenes, such as miR-21 [2–5], which is confirmed as serum biomarker in different cancers, and miR-17-92 [6, 7], which is frequently linked with amplification of myc gene region. Others play as tumor suppressors, such as miR-9 [8–10], which has often been reported to be methylated in different cancers, and let-7 family [11–13], which were reported down-regulated in different cancers.
miR-137 has been reported in many studies. Although some studies reported it plays as an oncogene in bladder cancer [14], most studies reported its tumor suppressor role in different kinds of cancers, such as lung cancer [15, 16], hepatocellular cancer [17], and gastric cancer [18, 19]. In line with related reports, our group also confirmed the methylated station of miR-137 during gastric cancer carcinogenesis [10]. However, the functions of miR-137 during gastric cancer progression have to be elucidated.
In the present study, we investigated the effects of miR-137 on cell proliferation, cell cycle, and cell migration in BGC-823 cell line and SGC-7901 cell line. We also identified that miR-137 plays as a tumor suppressor gene during gastric cancer progression through targeting MYO1C and KLF12.
Materials and methods
Tissue samples, cell lines, and cell transfection
Eighteen pairs of GC samples and SM samples were collected from Beijing cancer hospital (average age 55.8 years [range, 33–74]; 14 males and 4 females). This study was approved by the local medical ethics committee and informed consent was obtained from all patients. BGC-823 and SGC-7901 cell lines were obtained from Beijing institute for cancer research, and were cultured at 37 °C with 5 % CO2 by 90 % RPMI1640 with 10 % fetal bovine serum (both from HyClone, USA) and 1 % penicillin/streptomycin (Solarbio, China). All transfection assays were performed using FuGENE® HD Transfection Reagent (Promega, USA) according to the manufacturer’s instructions. Transfected cells were collected after 24 h, and all transfection assays were carried out two repeats.
RNA extraction and QRT-PCR
Total RNA of tissue samples and gastric cancer cell lines were extracted by RNA extraction kit (Beijing ComWin Biotech Co. Ltd., China) according to the manufacturer’s protocol. The expression of miRNA and relevant reference gene were tested by (QRT-PCR) using a SYBR green mix (Genecopoeia, China) on illumine Eco system. The primer of miR-137 and RNU6B were supplemented by kit. The expressions of target genes were performed using a SYBR green mix (TAKARA, Dalian, China) on illumine Eco system. The primers of KLF12, MYO1C, and GAPDH were as follow: KLF12-PF: TGGCAAAGCACAAATGGAC; KLF12-PR: CTAAATGGTGAAATTGAACAAGG; MYO1C-PF: ACCTGTACCTGGTGAAGGGC; MYO1C-PR: CTGAGGAGCCTGGTCAGATACT; GAPDH-PF: CTGGGCTACACTGAGCACCAG; and GAPDH-PR: CCAGCGTCAAAGGTGGAG.
Transwell assay
Both of the gastric cancer cell lines were transfected with miR-137 mimic and inhibitor, and about 4 × 104 cells were seeded within 200 μl RPMI1640 onto the upper layers of chamber. The lower layers were added about 500 μl mixed cultures with 90 % RPMI 1640 and 10 % FBS. Twenty-four hours after transferring and culturing in 37 °C with 5 % CO2, the transfected cell was fixed by 2 % formaldehyde for 20 min, followed with washing twice by 1× PBS, and was then stained with 1 % crystal violet for another 30 min. Cells on the upper layer of chambers were removed with cotton swabs, and cells on the opposite membranes of chamber were counted by microscope system. Each chamber was counted six fields.
Cell growth counting assay
About 1 × 104 of transfected miR-137 mimic and inhibitor cells were separately seeded in 400 μl mixed culture with 90 % RPMI 1640 and 10 % FBS per well and cultured in 37 °C with 5 % CO2. Each assay was tested by triplicates. Cells were then trypsinized and counted by microscope system.
Cell cycle assay
Twenty-four hours after transfection of miR-137 mimic and inhibitor, BGC-823 and SGC-7901 cell lines were trypsinized and washed twice by 1× PBS and were fixed with 70 % ethanol at 4 °C for 1 h. After centrifugation, cells were re-suspended and incubated in 1× PBS combined with 0.05 mg/ml propidium iodide (PI), 0.1 mg/ml RNaseA, and 0.02 % Triton X-100 for another 30 min. The cell cycle was then analyzed by flow cytometry (BD-C6, USA).
Western blot assay
After 24-h transfection of miR-137 mimic and inhibitor, cells were lysed with RIPA lysis buffer and protein concentration was determined. The proteins (20 μg) were electrophoresed by SDS-PAGE and transferred onto PVDF membranes. Membranes were blocked for 2 h at room temperature with 5 % non-fat milk. The membranes were incubated with primary antibodies (1:1000, anti-Myo1C, [EPR14771], ab194828, abcam; 1:200, KLF12, N-16, sc-84347, Santa Cruz; 1:1000, Anti GAPDH Rabbit Polyclonal Antibody, CW0101, CWBIO) overnight at 4 °C, followed by secondary antibody (1:3000, Goat Anti-Rabbit IgG, HPR conjugated) for another 1 h. Protein expression was assessed by LI-COR model 3600 (LI-COR, USA).
Plasmid construction and luciferase reporter assay
The region of 3′-UTR which contains the last seed sequence of wild type (WT) and mutant type (MUT) of MYO1C and KLF12 was synthesized chemically and cloned into pmirGLO Dual-Luciferase miRNA vector (XhoΙ/Sac Ι). All sequences were synthesized by GenePharma (ShanghaiGenePharmaCo., Ltd., China). The WT or MUT 3′-UTR vectors were co-transfected with miR-137 mimic, miR-137 inhibitor, or negative control into 293 T cell line using FuGENE® HD Transfection Reagent (Promega, USA). After 24 and 48 h transfection, luciferase activity was measured using the Dual-Glo Luciferase assay kit (Promega, USA) according to the manufacturer’s protocol. The normalized firefly luciferase activity was calculated as quotient of firefly/Renilla luciferase activity.
Statistical analysis
Statistical analysis was performed using SPSS 18.0 software. Paired t-test was performed to compare the difference of miR-137 expression between GC samples and SM samples. The Mann-Whitney U test and Student’s t-test were used to analyze the association between the miR-137 expression level and the clinicopathological features. Fisher’s exact test and Trend-test were used to analyze the association between high expression rate of miR-137 and the clinicopathological features. ROC curve was set up to separate high expression of miR-137 from low expression group. Kaplan-Meier analysis was used to compare overall survival of GC patients with different miR-137 expression level in univariate analysis. Student t-test was used to analyze the difference of cell proliferation curve, percentage of cell cycle stage, numbers of cell migration, expression level mRNA and protein, and results of luciferase reporter assay after transfection of mimic and inhibitor. All statistical tests were two-sided, and P < 0.05 was considered statistically significant.
Results
miR-137 is down-regulated in gastric cancer patients
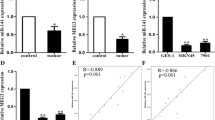
To analyze the expression of miR-137 in gastric cancer patients, we measured the expression of miR-137 in 18 pairs of GC samples and SM samples by QRT-PCR. As shown in Fig. 1, although there was no significant difference between GC group and SM group (Fig. 1, P = 0.052, paired t-test), the expression level of miR-137 was much lower in GC sample than in SM samples. According to those results, it indicated that miR-137 was down-regulated in GC patients, which might play as a tumor suppressor in gastric cancer. We further analyzed the clinicopathological features separately by GC samples and SM samples (Table 1 and Table 2). As the consequent, the clinicopathological features analyzed by GC samples showed that miR-137 was significantly higher expressed in no vascular embolus group compared with the vascular embolus group (Table 1. Median: 0.051 vs. 0.003, Mann-Whiney U test, P = 0.023). ROC curve was formulated by vascular embolus (Fig. 1, cut off value = 0.005, AUC = 0.841, P = 0.023). Kaplan-Meier analysis showed that the overall survival of patients with high miR-137 expression was longer than those with low miR-137 expression (Fig. 1, P = 0.386).
Expression levels and overall survival time analysis of miR-137 in gastric cancer samples. a QRT-PCR was used to test the difference of miR-137 expression level between 18 paired gastric cancer (GC) samples and surgical margin (SM) samples; b ROC curve was formulated by vascular embolus; c Kaplan-Meier analysis by miR-137 expression level
miR-137 inhibit gastric cancer cells migration, cell proliferation, and cell cycle
Based on the hypothesis of its tumor suppresser role, we separately over expressed and knocked down the expression of miR-137 in gastric cancer cell BGC-823 and SGC-7901 (Fig. 2). We then detected the migration capability changes between different treatments of both cell lines and found that over expression of miR-137 by miR-137 mimic could inhibit the migration of both kinds of cells (P < 0.001, Fig. 3 (A and B)), while knock down the expression of miR-137 by miR-137 inhibitor could promote the migration of both kinds of cell lines (P < 0.001, Fig. 3 (C and D)). The growth curves were also compared by cell counts for 5 days between above different treatments. Although there was no significant difference between miR-137 inhibitor NC and miR-137 inhibitor groups in both kinds of cell lines (Fig. 4c and Fig. 4d), cells in miR-137 mimic group grew more rapidly than in miR-137 mimic NC group (P < 0.05, Fig. 4a and Fig. 4b). Cell cycle assay showed that over expression of miR-137 could inhibit the vast majority of cells to stalled at G0/G1 stage in BGC823 cell lines (P = 0.006, Fig. 5 (B—g)), while knock down the expression of miR-137 could promote more cells to go into the S stage and ready for cell division in SGC7901 cell lines (P = 0.052, Fig. 5 (C—g)).
Overexpression and knock down of miR-137 in BGC823 and SGC7901 gastric cancer cell lines. miR-137 mimic and inhibitor were separately transfected into BGC823 and SGC7901 cell lines. QRT-PCR was used to confirm the changes of miR-137 expression level after 24 h of transfection. a, b Overexpression and knock down of miR-137 in BGC823 cell line; c, d Overexpression and knock down of miR-137 in SGC7901 cell line. Note: miR-137 mimic NC is miR-137 mimic control; miR-137 inhibitor NC is miR-137 inhibitor control
miR-137 inhibits the migration of BGC823 and SGC7901 gastric cancer cell lines. Transwell assays were performed when miR-137 was overexpressed or knocked down in BGC823 and SGC7901 cell lines. A (a and b) and C (a and b) Overexpression or knock down of miR-137 in SGC7901 cell line; B (a and b) and D (a and b) Overexpression or knock down of miR-137 in BGC823 cell line
KLF12 and MYO1C are potential targets of miR-137
We then analyzed the targets of miR-137 among three kinds of bioinformatics databases including miRDB, targetscan, and pictar. There were about 544, 1144, and 468 targets of miR-137 separately supported by miRDB, targetscan, and pictar (Fig. 6a). And we found 206 kinds of targets predicted by all three kinds of databases (Fig. 6a). And among them, KLF12 and MYO1C were predicted with relatively much higher scores than others, with five target sites for KLF12 and two target sites for MYO 1C. And based on the reports of their important roles in cancer, we selected them as the candidate target genes of miR-137(Fig. 6c). The QRT-PCR assay suggested that KLF12 and MYO1C showed negatively correlation with miR-137 in mRNA expression level (Fig. 7a and Fig. 7c, KLF12, P = 0.024 in BGC823 and P = 0.038 in SGC7901 by miR-137 mimic; Fig. 7e and Fig. 7h, MYO1C, P < 0.001 in BGC823 by miR-137 mimic and P = 0.007 in SGC7901 by miR-137 inhibitor). And western blot assay also suggested that KLF12 and MYO1C showed negatively correlation with miR-137 in protein expression level (Fig. 8). Luciferase reporter assay showed that the 3′-UTR of KLF12 and MYO1C could direct interact with miR-137 (Fig. 9a and Fig. 9c, KLF12, 24 and 48 h P < 0.001; Fig. 9e and Fig. 9g, MYO1C, 24 h P < 0.001, 48 h P = 0.005), and the mutant 3′-UTR of them could inhibit their interactions (Fig. 9b/D/F/H, P > 0.05).
Biological information analysis and reporter constructs of miR-137 target genes. a The candidate target genes of miR-137 were analyzed separately in miRDB, Pictar, and Targetscan, and the candidate target genes were selected from the intersection of three sets; b The vector for reporter constructs of 3′-UTR of target genes; c Diagram of 3′-UTR containing reporter constructs for KLF12 and MYO1C
KLF12 and MYO1C were negatively related to miR-137 in mRNA expression level by QRT-PCR assay. a, b The expression level of KLF12 when overexpression or knock down of miR-137 in BGC823 cell line; c, d The expression level of KLF12 when overexpression or knock down of miR-137 in SGC7901 cell line; e, f The expression level of MYO1C when overexpression or knock down of miR-137 in BGC823 cell line; g, h The expression level of MYO1C when overexpression or knock down of miR-137 in SGC7901 cell line
KLF12 and MYO1C were negatively related to miR-137 in protein expression level by western blot assay. A (a and c) The expression level of KLF12 when overexpression or knock down of miR-137 in BGC823 cell line; A (b and d) The expression level of KLF12 when overexpression or knock down of miR-137 in SGC7901 cell line; B (a and b) The expression level of MYO1C when overexpression or knock down of miR-137 in BGC823 and SGC7901 cell lines
Luciferase reporter assays. The relative luciferase activity was measured in 293 T cell line at the time of 24 or 48 h after co-transfection of the target gene-WT or target gene-MUT luciferase construct with either miR-137 or control (NC). a, b 24 h, KLF12-WT or KLF12-MUT; c, d 48 h, KLF12-WT or KLF12-MUT; e, f 24 h, MYO1C-WT or MYO1C-MUT; g, h 48 h, MYO1C-WT or MYO1C-MUT
Discussions
miR-137 has been reported in different cancers. However, because of the tissue specific characteristic of miRNAs, it is not surprised that the same miRNA plays different roles among various kinds of cancer types. Some of them suggested that miR-137 might play as an oncogene role, while others showed that it might play as a tumor suppressor role. Xiu et al. have reported that miR-137 was over expressed in bladder cancer and was associated with pTNM stage of clinical patients [14]. They also suggested that miR-137 could promote bladder cell proliferation, migration, and invasion by targeting PAQR3 [14]. However, Zhang et al. reported that miR-137 could inhibit NSCLC cell proliferation, migration, and invasion by targeting SLC22A18 and might be an independent prognostic factor of NSCLC [15]. Besides, another reports about NSCLC suggested that miR-137 might inhibit NSCLC cell proliferation by targeting KIT [16]. Liu et al. have reported that miR-137 could be regulated by FoxD3 and then targeted AKT2 to inhibit HCC progression [17]. Cheng et al. have reported that miR-137 might play a tumor suppressor role by targeting Cox-2 and then regulate the signaling pathway of PI3K/AKT in gastric cancer [20]. Similar phenomena that miR-137 played tumor suppressor role in gastric cancer were also been reported by Liu et al. [19], in which miR-137 inhibited EMT by targeting Twist1, and by Zheng et al. [18], in which miR-137 inhibited proliferation found by miRNA array. We have reported that miR-137 was silenced in GC samples compared with SM samples in gastric cancer [10], and it suggested that miR-137 might play as tumor suppressor role in gastric cancer. In the present study, we displayed miR-137 played the tumor suppressor role in two kinds of gastric cancer cell lines, including cell proliferation, cell migration, and cell cycle.
Although there is no significant difference between GC samples and SM samples in 18 paired patients, the trend of the expression level showed that miR-137 was down-regulated in GC samples and the significance between these two groups may be raised with the size of sample. Similarly, although we could only observed that miR-137 was significantly expressed in no vascular embolus group than in vascular embolus group (Table 1, Median, 0.051 vs. 0.003, Mann-Whiney U test, P = 0.023), the clinicopathological features showed that miR-137 was much higher expressed in no lymph node metastasis group, no distant metastasis group and early pTNM stages compared with lymph node metastasis group, distant metastasis group and late pTNM stages (Table 1). Furthermore, we set up the cut off value by vascular embolus (Fig. 1, cut off value = 0.005, AUC = 0.841, P = 0.023) and analyzed the overall survival time of gastric cancer patients, which suggested that the overall survival of patients with high miR-137 expression was longer than those with low miR-137 expression (Fig. 1, P = 0.386). However, limited to the present sample size, we could not receive more significant results.
We first transfected the miR-137 mimic and inhibitor into gastric cancer cell lines before we conducted the follow-up assays. Although the miR-137 mimic group significantly increased the expression of miR-137 in both kinds of gastric cancer lines, the miR-137 inhibitor group just down-regulated the expression of miR-137 without significant difference compared with inhibitor NC group. It was easy to understand the working mechanism of mature miRNAs. miRNA mimic imitated the same process of miRNA itself. However, we could not arrive at the truth of the real working mechanisms of miRNA inhibitor. Some of them could achieve the predictive result, while others could not found the difference compared with control groups, so that several companies suggested adding 3–5 times more than miRNA mimic to obtain the predictive results. As for the undesirable down regulation of miR-137 in two kinds of gastric cancer cell lines, it might be explained as many ways and one of them might be the insufficient dosage of materials. However, no matter how the mechanisms behind such phenomenon, the miR-137 inhibitor reduced the expression of miR-137 in both kinds of gastric cancer cell lines.
The migration capabilities of two kinds of gastric cancer cell lines were compared by transfecting with miR-137 mimic and inhibitor. And miR-137 really inhibited the migration capability of gastric cancer cell lines. As we could observe that number of the cell migration in miR-137 inhibitor group was significant lower than in miR-137 mimic group. Such phenomenon could be explained as we separately conducted the above transwell assays so that some differences existed in cell state between them. Besides, limited to present conditions, we did not further confirm this phenomenon by the transwell invasion assay and the animal model of metastasis assay.
We also compared the proliferation and cell cycle after the transfection with miR-137 mimic and inhibitor. Over expression of miR-137 could inhibit the proliferation of BGC823 and SGC7901, while knock down of miR-137 had no significant difference between miR-137 inhibitor and miR-137 inhibitor NC in both of the above cell lines. Compared with the results of cell proliferation, the only changes of cell cycles happened in BGC823 cell lines. More cells stayed in G0/G1 phrase and decreased in S phrase, which indicated that overexpression of miR-137 could promote the cell cycle arrest, and although there was no difference between the miR-137 inhibitor and miR-137 inhibitor NC, the trend suggested that knock down the expression of miR-137 could increase the rates of cells in G0/G1 phrase. It was interested that overexpression of miR-137 seemed to increase the rates of S stage cells in SGC7901, and knockdown the expression of miR-137 could reduce this trend. However, there was no significant difference in SGC7901 assay group. Besides, recently studies suggested that the S stage of the cell cycle should be rediscovered, because the replication of genetic material might happened not only in S phrase but also in G2/M phrase [21]. Thus, we could not make the conclusion by comparing them in only S stage without considering the G2/M phrase. However, the G0/G1 stage was still realized as cell cycle arrest check point. And the only conclusion of this assay might be overexpression of miR-137 could promote the cell cycle to arrest in G0/G1 phrase.
Based on the above results of miR-137 function in gastric cancer cell lines, we found that KLF12 and MYO1C might be another two target genes of miR-137. Firstly, we tested the expression level of KLF12 and MYO1C after transfection of miR-137 mimic and inhibitor. We found that the mRNA expression level of KLF12 was negatively related to miR-137 both in BGC823 cell line and SGC7901 cell line, while the mRNA expression level of MYO1C was negatively related with miR-137 in SGC7901 cell line. MYO1C was lower expressed in BGC823 cell lines with miR-137 overexpression, but not versa. However, in the protein level, both target proteins were negatively related to the expression of miR-137 in BGC823 cell line. While the negative relationship in SGC7901 cell line could only be observed when knockdown the miR-137 in BGC823 cell line for KLF12 or when overexpression of miR-137 in SGC7901 cell line for MYO1C. Because of the mRNA level and protein level were different working stages of miRNA machinery in different cell type, and we did not obtain them together, so that we might seized one moment of the long stage, which might be not be unanimously in different expression levels and in different cell types. But the trend of negative relationship always indicated that they were the candidates target gene of miR-137.
KLF12 have been reported in numerous kinds of cancers. It belongs to the KLFs’ family, which comprises a highly conserved family of zinc finger transcription factors that are involved in a series of cellular processes, ranging from proliferation and apoptosis to differentiation, migration, and pluripotency. Giefing et al. showed that the amplification region of 13q22.1–22.2 in 13/29 cases harbored KLF12 genes in salivary gland tumor [22]. And Xu et al. confirmed that miR-382 could inhibit the growth and chemosensitivity in osteosarcorna by targeting KLF12 and HIPK3 [23]. Hünten et al. used the means of next generation sequencing (NGS) and found that KLF12 might be the target of P53. All above reports were indirect proof of the oncogene role of KLF12. Meanwhile, Nakamura et al. supplied the direct proof of the oncogene role of KLF12, that knockdown the expression of KLF12 induced significant growth arrest, while over expression of KLF12 could promote the invasive potential in HGC27 gastric cancer cell lines [24]. In the present study, our results showed that the expression of KLF12 was negatively related to miR-137, which suggested it might play as an oncogene in human gastric cancer.
MYO1C was reported to be more related to hearing [25, 26]. But we found only two papers about MYO1C has the relationships with human cancers. One predicted it as tumor suppressor gene, because region of MYO1C neighbors with TP53, and other members of P53 family plays as a tumor suppressor role [27]. Another paper reported that MYO1C had three isoforms (isoform A, B, and C), and isoform A is specific high expressed in prostate cancer, which may be a potential maker for detection prostate cancer [28]. It was interesting that our data about MYO1C was also based on the isoform A. As our result, it was showed that the expression of MYO1C was negatively related to miR-137, which suggested it might play as an oncogene in human gastric cancer.
In conclusion, we found that miR-137 was down-regulated in clinical specimens of gastric cancer. It was significantly high expressed in patients without vascular embolus than with vascular embolus ones. And the overall survival time of miR-137 high expression patients was longer than miR-137 low expression ones. Overexpression of miR-137 could inhibit the capabilities of cell migration, cell proliferation, and cell cycle in gastric cancer cell lines. Furthermore, miR-137 might play a tumor suppressor role in gastric cancer by targeting KLF12 and MYO1C.
References
Ang TL, Fock KM. Clinical epidemiology of gastric cancer. Singap Med J. 2014;55(12):621–8.
Wu K, Li L, Li S. Circulating microRNA-21 as a biomarker for the detection of various carcinomas: an updated meta-analysis based on 36 studies. Tumour Biol. 2015;36(3):1973–81.
Kurashige J, Kamohara H, Watanabe M, Tanaka Y, Kinoshita K, Saito S, Hiyoshi Y, Iwatsuki M, Baba Y, Baba H. Serum microRNA-21 is a novel biomarker in patients with esophageal squamous cell carcinoma. J Surg Oncol. 2012;106(2):188–92.
Gao W, Lu X, Liu L, Xu J, Feng D, Shu Y. MiRNA-21: a biomarker predictive for platinum-based adjuvant chemotherapy response in patients with non-small cell lung cancer. Cancer Biol Ther. 2012;13(5):330–40.
Zhang HL, Yang LF, Zhu Y, Yao XD, Zhang SL, Dai B, Zhu YP, Shen YJ, Shi GH, Ye DW. Serum miRNA-21: elevated levels in patients with metastatic hormone-refractory prostate cancer and potential predictive factor for the efficacy of docetaxel-based chemotherapy. Prostate. 2011;71(3):326–31.
Kandalam MM, Beta M, Maheswari UK, Swaminathan S, Krishnakumar S. Oncogenic microRNA 17-92 cluster is regulated by epithelial cell adhesion molecule and could be a potential therapeutic target in retinoblastoma. Mol Vis. 2012;18:2279–87.
Valladares-Ayerbes M, Blanco M, Haz M, Medina V, Iglesias-Díaz P, Lorenzo-Patiño MJ, Reboredo M, Santamarina I, Figueroa A, Antón-Aparicio LM, Calvo L. Prognostic impact of disseminated tumor cells and microRNA-17-92 cluster deregulation in gastrointestinal cancer. Int J Oncol. 2011;39(5):1253–64.
Li Y, Xu Z, Li B, Zhang Z, Luo H, Wang Y, Lu Z, Wu X. Epigenetic silencing of miRNA-9 is correlated with promoter-proximal CpG island hypermethylation in gastric cancer in vitro and in vivo. Int J Oncol. 2014;45(6):2576–86.
Fiaschetti G, Abela L, Nonoguchi N, Dubuc AM, Remke M, Boro A, Grunder E, Siler U, Ohgaki H, Taylor MD, Baumgartner M, Shalaby T, Grotzer MA. Epigenetic silencing of miRNA-9 is associated with HES1 oncogenic activity and poor prognosis of medulloblastoma. Br J Cancer. 2014;110(3):636–47.
Du Y, Liu Z, Gu L, Zhou J, Zhu BD, Ji J, Deng D. Characterization of human gastric carcinoma-related methylation of 9 miRCpG islands and repression of their expressions in vitro and in vivo. BMC Cancer. 2012;12:249.
Wong TS, Man OY, Tsang CM, Tsao SW, Tsang RK, Chan JY, Ho WK, Wei WI, To VS. MicroRNA let-7 suppresses nasopharyngeal carcinoma cells proliferation through downregulating c-myc expression. J Cancer Res Clin Oncol. 2011;137(3):415–22.
Lee ST, Chu K, Oh HJ, Im WS, Lim JY, Kim SK, Park CK, Jung KH, Lee SK, Kim M, Roh JK. Let-7 microRNA inhibits the proliferation of human glioblastoma cells. J Neuro-Oncol. 2011;102(1):19–24.
Liang S, He L, Zhao X, Miao Y, Gu Y, Guo C, Xue Z, Dou W, Hu F, Wu K, Nie Y, Fan D. MicroRNA let-7f inhibits tumor invasion and metastasis by targeting MYH9 in human gastric cancer. PLoS One. 2011;6(4):e18409.
Xiu Y, Liu Z, Xia S, Jin C, Yin H, Zhao W, Wu Q. MicroRNA-137 upregulation increases bladder cancer cell proliferation and invasion by targeting PAQR3. PLoS One. 2014;9(10):e109734.
Zhang B, Liu T, Wu T, Wang Z, Rao Z, Gao J. microRNA-137 functions as a tumor suppressor in human non-small cell lung cancer by targeting SLC22A18. Int J Biol Macromol. 2015;74:111–8.
Li P, Ma L, Zhang Y, Ji F, Jin F. MicroRNA-137 down-regulates KIT and inhibits small cell lung cancer cell proliferation. Biomed Pharmacother. 2014;68(1):7–12.
Liu LL, SX L, Li M, Li LZ, Fu J, Hu W, Yang YZ, Luo RZ, Zhang CZ, Yun JP. FoxD3-regulated microRNA-137 suppresses tumour growth and metastasis in human hepatocellular carcinoma by targeting AKT2. Oncotarget. 2014;5(13):5113–24.
Zheng X, Dong J, Gong T, Zhang Z, Wang Y, Li Y, Shang Y, Li K, Ren G, Feng B, Li J, Tian Q, Tang S, Sun L, Li M, Zhang H, Fan D. MicroRNA library-based functional screening identified miR-137 as a suppresser of gastric cancer cell proliferation. J Cancer Res Clin Oncol. 2015;141(5):785–95.
Liu S, Cui J, Liao G, Zhang Y, Ye K, Lu T, Qi J, Wan G. MiR-137 regulates epithelial-mesenchymal transition in gastrointestinal stromal tumor. Tumour Biol. 2014;35(9):9131–8.
Cheng Y, Li Y, Liu D, Zhang R, Zhang J. miR-137 effects on gastric carcinogenesis are mediated by targeting cox-2-activated PI3K/AKT signaling pathway. FEBS Lett. 2014;588(17):3274–81.
Minocherhomji S, Ying S, Bjerregaard VA, Bursomanno S, Aleliunaite A, Wu W, Mankouri HW, Shen H, Liu Y, Hickson ID. Replication stress activates DNA repair synthesis in mitosis. Nature. 2015;528(7581):286–90.
Giefing M, Wierzbicka M, Rydzanicz M, Cegla R, Kujawski M, Szyfter K. Chromosomal gains and losses indicate oncogene and tumor suppressor gene candidates in salivary gland tumors. Neoplasma. 55(1):55–60.
Xu M, Jin H, CX X, Sun B, Mao Z, Bi WZ, Wang Y. miR-382 inhibits tumor growth and enhance chemosensitivity in osteosarcoma. Oncotarget. 2014;5(19):9472–83.
Nakamura Y, Migita T, Hosoda F, Okada N, Gotoh M, Arai Y, Fukushima M, Ohki M, Miyata S, Takeuchi K, Imoto I, Katai H, Yamaguchi T, Inazawa J, Hirohashi S, Ishikawa Y, Shibata T. Krüppel-like factor 12 plays a significant role in poorly differentiated gastric cancer progression. Int J Cancer. 2009;125(8):1859–67.
Greenberg MJ, Ostap EM. Regulation and control of myosin-I by the motor and light chain-binding domains. Trends Cell Biol. 2013;23(2):81–9.
Gillespie PG. Myosin I and adaptation of mechanical transduction by the inner ear. Philos Trans R Soc Lond Ser B Biol Sci. 2004;359(1452):1945–51.
Hedberg Oldfors C, Dios DG, Linder A, Visuttijai K, Samuelson E, Karlsson S, Nilsson S, Behboudi A. Analysis of an independent tumor suppressor locus telomeric to Tp53 suggested Inpp5k and Myo1c as novel tumor suppressor gene candidates in this region. BMC Genet. 2015;16:80.
Ihnatovych I, Sielski NL, Hofmann WA. Selective expression of myosin IC isoform a in mouse and human cell lines and mouse prostate cancer tissues. PLoS One. 2014;9(9):e108609.
Acknowledgment
This work was supported by Ningbo Municipal Natural Science Foundation (Grant Number: 2014A610219).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
None.
Rights and permissions
About this article
Cite this article
Du, Y., Chen, Y., Wang, F. et al. miR-137 plays tumor suppressor roles in gastric cancer cell lines by targeting KLF12 and MYO1C. Tumor Biol. 37, 13557–13569 (2016). https://doi.org/10.1007/s13277-016-5199-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-016-5199-3