Abstract
Over the past decade, the importance of non-protein-coding functional elements in the human genome has emerged from the water and been identified as a key revelation in post-genomic biology. Since the completion of the ENCODE (Encyclopedia of DNA Elements) and FANTOM (Functional Annotation of Mammals) project, tens of thousands of pseudogenes as well as numerous long non-coding RNA (lncRNA) genes were identified. However, while pseudogenes were initially regarded as non-functional relics littering the human genome during evolution, recent studies have revealed that they play critical roles at multiple levels in diverse physiological and pathological processes, especially in cancer through parental-gene-dependent or parental-gene-independent regulation. Herein, we review the current knowledge of pseudogenes and synthesize the nascent evidence for functional properties and regulatory modalities exerted by pseudogene-transcribed RNAs in human cancers and prospect the potential as molecular signatures in cancer reclassification and tailored therapy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
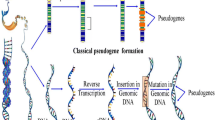
In the past decade, benefiting from the improvement of large-scale sequencing technique and bioinformatics methods, the completion of the Human Genome Project ENCODE (Encyclopedia of DNA Elements) [1, 2] and FANTOM (Functional Annotation of Mammals) consortia [3] has highlighted the prevalence of non-protein-coding functional elements in human genome. Following the sequencing of the whole human genome, GENCODE, using the next-generation sequencing and the study of genetic landmarks indicative of transcription, has revealed that there are 60,498 total genes (Version 23, March 2015 freeze, GRCh38). Among of these 60,498 genes, ENCODE only defines 19,797 genes as protein-coding genes, with almost all of the other genes being classified as long non-coding RNA, small non-coding RNA genes, and pseudogenes (http://www.gencodegenes.org/stats/current.html). Recently, accumulating evidence has revealed that lncRNAs play important roles in the process of carcinogenesis and tumor progression through chromatin remodeling, epigenetic modification, and sponging miRNAs. Since their discovery in 1977, pseudogenes have been considered as non-functional genomics fossils or biologically inconsequential [4]. Generally, pseudogenes are derived from unfaithful gene duplications or retrotransposition of processed mRNAs back into the genome, and they are divided into three main categories: processed pseudogenes, unprocessed pseudogenes, and unitary pseudogenes based on how they were generated from their ancestral gene [5]. Accordingly, the unprocessed pseudogenes are generated by segmental duplications and disabled by mutations, and they typically have promoter, intron, and exons; even these elements might lose their function. The second major class of pseudogenes is processed pseudogenes that is derived from the reverse transcription of a parental gene’s mRNA and therefore are limited to a single exon in structure [6]. The unitary pseudogenes only constitute a small fraction of all pseudogenes and are a subclass of unprocessed pseudogenes; they do not have an identifiable parental gene in the genome where they reside [7, 8]. However, one of them, the XIST gene, has been widely investigated for its key roles in X-chromosome inactivation [9–11].
Although pseudogenes were once regarded as “genomic fossil” or “junk genes” [12, 13], recent studies have revealed the multilayered biological function of some pseudogenes in multiple cellular processes [14], especially their involvement in human cancers [15]. In addition, increasing evidence showed that pseudogenes play important roles in post-transcriptional or transcriptional regulations of gene expression by function as antisense RNA, endogenous small-interference RNA (endo-siRNA or esiRNA), endogenous competitors for miRNA, RNA-binding protein (RBP), or translational machinery (Fig. 1) [7, 16]. Moreover, some pseudogenes transcribed non-coding RNAs more than 200 nt in length cloud be included in lncRNA classification. In spite of that, the investigation of pseudogene function has remained limited for a long time, and the recent improvement of genome-wide platforms has led to the discovery that many pseudogenes are transcriptionally active and their disorder-transcribed RNAs contribute to several human diseases including cancers. In this review, we discuss the functions and clinical relevance of pseudogene-expressed RNAs in cancer and hope that this will increase our understanding of the regulatory functions and mechanisms of pseudogenes in cancer development.
Mechanisms of pseudogenes mediated regulation in cancer cells. a The pseudogenes act as competing endogenous RNA for microRNA sponge. b The pseudogene acts as a decoy for mRNA. c The pseudogene generates into endogenous siRNAs. d The pseudogene acts as a decoy for protein. e The pseudogene recruit histone modification protein EZH2 and G9a to target gene promoter, thereby regulating their transcription
Disorder pseudogene-expressed RNAs in cancers
Previously, studies have shown that the pseudogenes participate in ceRNA interactions and contribute to tumorigenesis; however, it is difficult to distinguish pseudogenes from their parental genes due to their high homology to parental genes, which hindered attempts to study pseudogenes on a large scale. Recently, Shanker et al. developed a bioinformatics pipeline for the identification of transcribed pseudogenes based on next-generation sequencing data of cancer samples and identified 2082 pseudogene transcripts. Among them, 218 expressed only in cancer samples, and a breast cancer-specific pseudogene-ATP8A2-ψ was further validated and found to be restricted to breast tumor with luminal histology. Moreover, subsequent gain- and loss-of-function assays in vitro revealed the oncogenic role of ATP8A2-ψ in breast cancer cells [17]. Meanwhile, Han et al. developed a similar computational pipeline and detected 9925 pseudogene transcriptions in 7 cancer types from The Cancer Genome Atlas (TCGA) RNA-seq data, and they found that many pseudogene transcripts are tissue- and/or cancer-specific. Importantly, the study firstly systematically revealed the potential of pseudogenes as subtype and prognostic biomarkers in cancers [18]. In addition, Joshua and colleagues identify a set of 440 pseudogenes that are transcribed in breast cancer, and 309 of them exhibit significant differential expression among breast cancer subtypes [19]. The progression in sequencing accuracy and coverage depth and advance on the detection efficacy of these RNA-based bioinformatics pipelines is facilitating the researches in this regard. As of following, we will list and discuss some important pseudogene-expressed RNA (Table 1) roles and their underlying mechanisms or regulated pathways in cancer development and progression.
The PTEN pseudogene PTENP1
In 2010, Poliseno et al. revealed that RNA molecules that share MREs can regulate each other by competing for microRNA binding, and they hypothesize that protein-coding mRNAs and non-coding RNAs would communicate with each other in a microRNA-dependent manner through a MRE language [40, 41]. Specifically, they identified several pseudogene transcripts exerting regulatory control of their ancestral cancer gene’s expression by competing for microRNAs, and one of these pseudogene transcripts is PTENP1 [20]. PTENP1, a processed pseudogene residing at 9p13.3, is highly homologous to tumor suppressor gene PTEN with only 18 mismatches throughout the coding sequence and a missense mutation of the initiator methionine codon prevents its RNA translation [42]. There are evidence showing that PTEN is post-transcriptionally regulated by numerous miRNAs in cancer cells, and the conserved seed matches for the PTEN-targeting miR-17, miR-21, miR-214, miR-19, and miR-26 families are also found in the PTENP1 sequence within the high-homology region. Furthermore, PTENP1 was found to possess a regulatory function of PTEN through acting as a decoy for PTEN-related miRNAs and competing for these miRNAs, while loss of PTENP1 expression released these miRNAs, which instead targeted PTEN and reduced its protein levels [20]. In addition, PTENP1 over-expression repressed the tumorigenic properties of HCC cells by decoying miR-17, miR-19b, and miR-20a, which would target PTEN, PHLPP and such autophagy genes as ULK1, ATG7, and p62 [22]. Meanwhile, PTENP1 is found to be downregulated in clear-cell renal cell carcinoma tissues and cells due to methylation, and PTENP1 suppressed cancer progression by functions as a competing endogenous RNA (ceRNA) through decoying miR-21 [23]. However, deletion of the PTENP1 and PTEN locus was found in melanoma, which suggested that potential roles of PTENP1 go beyond acting as decoy for PTEN-related miRNAs [43]. Interestingly, Johnsson et al. found that PTENP1 also encodes two asRNA isoforms: PTENP1 asRNA alpha and beta, which indicated that the regulatory function of PTENP1 is more complex. The alpha isoform shares the greatest sequence with PTEN and recruits the DNMT3a and EZH2 to the PTEN promoter to suppress its transcription; however, the beta isoform asRNA forms RNA–RNA interactions with the PTENpg1 sense transcript and stabilizes PTENP1 sense, which consequently affecting its ability to sponge PTEN-related miRNAs [44]. Although the involvement of PTENP1 asRNA has not yet been investigated in human cancers, and it is likely that PTENP1 may also play both transcriptional and post-transcriptional regulatory roles in cancer cells.
The BRAF pseudogene
Although in vitro experiments over the past few years demonstrated that pseudogenes contribute to cell transformation and tumorigenesis through several mechanisms, however, in vivo evidence for potential roles of pseudogenes in cancer development is lacking. Recently, Florian et al. generated a transgenic allele containing murine B-Raf pseudogene Braf-rs1 and found that mice engineered to over-express murine B-Raf pseudogene develop an aggressive malignancy resembling human diffuse large B cell lymphoma. Moreover, they revealed that the in vivo proto-oncogenic function of murine B-Raf pseudogene is partly dependent on its regulation of B-Raf and through decoying miR-134, miR-543, and miR-653. Similarly, its human ortholog-BRAFP1 also elicits oncogenic activity at least in part as ceRNAs that elevate BRAF expression and MAPK activation. Furthermore, BRAFP1 over-expression increased BRAF and pERK levels as well as proliferation of human cells, while BRAFP1 silencing reduced proliferation of OCI-Ly18, H1299, and PC9 cells and elicited a significant effect on BRAF expression. Moreover, luciferase-report assays showed that four human miRNAs (miR-30a, miR-182, miR-876, and miR-590) were able to repress both BRAFP1-and BRAF-luciferase reporters, which indicated that BRAFP1 may be an oncogenic ceRNA in human cancer [24].
The OCT4 pseudogenes
Unlike the PTEN and BRAF pseudogene, it has been expanded to a total number of six OCT4 pseudogenes since the first OCT4-related pseudogene was reported [45]. OCT4 (POU5F1) is a transcription factor that plays critical roles in maintaining pluripotency and self-renewal of stem cells [46]. Recent transcriptional investigations have revealed that several of its pseudogenes are transcribed, and one of them OCT4pg1 is initially reported to be putative cancer susceptibility gene and is over-expressed in prostatic carcinoma by Kastler et al. [47]. In addition, OCT4pg1 (POU5F1B) is also reported to be amplified and expressed at a high level in gastric cancer, and its amplification is associated with a poor prognosis in gastric cancer patients as well as confers an aggressive phenotype on GC cells. Importantly, over-expression of OCT4pg1 promoted GC cells colony formation in vitro as well as both tumorigenicity and tumor growth in vivo, and knockdown of OCT4pg1 expression confirmed the role for OCT4pg1 in the promotion of cancer cell growth and tumor growth. Moreover, OCT4pg1 over-expression upregulated various growth factors in GC cells as well as exhibited angiogenic, mitogenic, and antiapoptotic effects in GC xenografts [34]. Besides, another OCT4 pseudogene-OCT4-pg4 is also involved in carcinogenesis. OCT4-pg4 is abnormally activated in hepatocellular carcinoma (HCC), and its expression level is positively correlated with that of OCT4, and survival analysis suggests that a high OCT4-pg4 level is significantly correlated with poor prognosis of HCC patients. Moreover, mechanism investigation revealed that OCT4-pg4 functions as a competing endogenous RNA to protect OCT4 transcript from being inhibited by miR-145, thus promoting HCC cell growth and tumorigenicity [35]. Meanwhile, mouse Oct4P4 lncRNA could form a complex with the SUV39H1 HMTase to direct the imposition of H3K9me3 and HP1a to the promoter of the ancestral Oct4 gene, leading to its silencing and reduced mESC self-renewal [48]. Unlike OCT4-pg1 and OCT4-pg4, OCT4pg5 could encode an antisense RNA that acts as a negative regulator of OCT4 and Oct4 pseudogenes 4 and 5 expressions. The Oct4-pg5 antisense RNA could recruit Ezh2 and G9a to the promoter of Oct4, which in turn leads to the trimethylation of histone 3 Lys27 (H3K27me3) modifications and silencing of Oct4 transcription. Moreover, knockdown of PURA and NCL had a negative effect on Oct4 mRNA levels, which may through interaction with Oct4-pg5 asRNA and sequester it away from targeted loci [36].
The HMGA1 pseudogenes
The HMGA1 is one of the high-mobility group A (HMGA) family, which are nuclear proteins that participate in the organization of nucleoprotein complexes and contribute to chromatin structure, replication, and gene transcription. The HMGA1 gene codes for two proteins-HMGA1a and HMGA1b that bind to DNA and organize chromatin architecture, interacting with several transcription factors and regulating the gene transcription. HMGA1 over-expression is a feature of human cancer, and their expression levels point out a poor prognosis of the cancer patients [49]. Recently, Esposito et al. used bioinformatics analysis to investigated HMGA1 pseudogenes in cancer settings, and they identified and characterized two processed pseudogenes HMGA1P6 and HMGA1P7 that are placed at 13q12.12 and 6q23.2, respectively. Moreover, HMGA1P6 and HMGA1P7 were over-expressed in human anaplastic thyroid carcinomas that are highly aggressive, but not in differentiated papillary carcinomas that are less aggressive. Consistently, over-expression of HMGA1P6 and HMGA1P7 promoted 8505c cells proliferation, cell cycle progression, migration, and invasion, while knockdown of their expression impaired cell growth and increased the number of cells in the sub-G1 phase, inhibited cell migration and invasion and induced cell apoptosis. Bioinformatic analysis revealed that HMGA1P6 and HMGA1P7 contain sequences that can be targeted by miRNAs (miR-15, miR-16, miR-26a, miR-214, miR-548c-3p, and miR- 761) that target the HMGA1 gene, and luciferase reporter assays revealed that miR-15, miR-16, miR-214, and miR-761 could directly bind to HMGA1P6 and HMGA1P7 sequences. Importantly, over-expression of HMGA1P6 or HMGA1P7 drastically reduced the effects exerted by miRNA on the levels of both the HMGA1 transcript and proteins, supporting that HMGA1P6 and HMGA1P7 act as decoys for HMGA1-targeting miRNAs to regulate HMGA1 levels. Finally, the expression of HMGA1P6 or HMGA1P7 was significantly correlated with HMGA1 protein levels thereby implicating their over-expression in cancer progression [28].
The TUSC2P pseudogene
The tumor suppressor candidate-2 (TUSC2), also known as Fus-1, is a novel tumor suppressor gene that functions as a “gatekeeper” in the molecular pathogenesis of cancer, which may function as a proapoptotic factor and is involved in the release of cytochrome c from the inner membrane of the mitochondria. When analyzing the sequence of the TUSC2 3′UTR, one pseudogene of TUSC2-TUSC2P was identified, which shared 89 % homology with the 3′UTR of TUSC2. Interestingly, many miRNAs were found to have common binding sites for both TUSC2 and TUSC2P, including miR-661, miR-299-3p, miR-93, miR-17, miR-608, and miR-502. TUSC2P can bind to and antagonize these endogenous miRNAs, thereby modulating TUSC2, TIMP2, and TIMP3 expression, while absence of TUSC2P, those miRNA can bind and inhibit the translation of TUSC2, TIMP2, and TIMP3 mRNAs through RNA-induced silencing complex. Moreover, ectopic over-expression of TUSC2P and the TUSC2 3′UTR inhibits cancer cell proliferation, migration, invasion, and induces cell death, suggesting that the TUSC2P may thus be used as combinatorial miRNA inhibitors and might have clinical applications [25].
The INTS6P1 and VEGFR-1 pseudogenes
As the ceRNA paradigm has refocused the attention on pseudogenes, Peng et al. identified the putative tumor suppressor INTS6 and its pseudogene INTS6P1 in HCC through the whole genome microarray expression. INTS6 and INTS6P1 were downregulated in HCC tissues compared with normal tissues, while miR-17-5p was found to be upregulated in same HCC tissues. Moreover, INTS6 and INTS6P1 over-expression impaired cell proliferation in vitro and tumor growth in vivo, inhibited cell migration and induced cell apoptosis in HCC. In addition, increased miR-17-5p expression induced downregulation of INTS6, as well as INTS6P1 while inhibition of miR- 17-5p induced de-repression and subsequent upregulation of INTS6 and INTS6P1. Lastly, the mechanistic experiments revealed that INTS6P1 and INTS6 are reciprocally regulated through competition for miR-17-5p in HCC cells [31]. Interestingly, the plasma INTS6P1 levels were also significantly decreased in HCC patients compared with non-HCC patients, which indicating that INTS6P1 may be used as a novel plasma-based biomarker and might improve the accuracy of HCC screening [50].
Similarly, Ye et al. report their finding of an actively transcribed VEGFR1/FLT1 pseudogene that is transcribed bidirectionally (FLT1P1) in human colorectal cancer cells. Knockdown of FLT1P1 expression by RNA interference (RNAi) markedly inhibited CRC cells proliferation and xenograft tumor growth in vivo. Furthermore, mechanistic investigation showed that expression of FLT1P1 antisense transcript not only inhibited the VEGFR1 expression but also inhibited non-cognate VEGF-A expression through interacting with miR-520a in CRC cells [39].
The PPM1K and AOC4P pseudogenes
As aforementioned, most of the pseudogenes regulated underlying target gene expression by function as ceRNA or interacting with microRNAs. However, Wang et al. identified a novel tumor suppressive pseudogene termed amine oxidase, copper containing 4, pseudogene (AOC4P), whose expression was significantly downregulated in HCC samples and negatively correlated with advanced clinical stage, capsule, and vessel invasion. Meanwhile, decreased AOC4P expression is correlated with poor prognostic outcomes and may serve as an independent prognostic factor for HCC patients. Moreover, functional assays showed that AOC4P over-expression significantly reduced cell proliferation, migration, and invasion through inhibiting the epithelial-mesenchymal transition (EMT). In vivo experiments confirmed the ability of AOC4P to inhibit tumor growth and metastasis. RNA immunoprecipitation assays demonstrated that AOC4P could bind to vimentin protein and promote its degradation [32].
More interestingly, Chan et al. used bioinformatics methods for identifying pseudogene-derived esiRNA through a genome-wide survey, and they identified a partial retrotranscript pseudogene PPM1KP that contained inverted repeats capable of folding into hairpin structures that can be processed into two esiRNAs. Moreover, these esiRNAs were significantly downregulated in HCC tumor tissues, and over-expression of PPM1KP decreased cell growth and clonogenic activity. Bioinformatics analysis predicted that PPM1K-specific esiRNAs are expected to regulate cognate gene PPM1K and NEK8 through association with multiple target sites. Additionally, PPM1K and NEK8 were downregulated in PPM1K-overexpressing cells, and expression of NEK8 can counteract the growth inhibitory effects of PPM1K. These findings suggested that PPM1KP can exert tumor suppressor activity independent of its parental gene by generation of esiRNAs that regulate human cell growth [38].
Others
In addition to the above pseudogenes, there are also some other pseudogenes that contribute to cancer development. Zheng et al. reported that ectopic expression of pseudogene CYP4Z2P 3′UTR in breast cancer cells increased the expression of VEGF-A without affecting cell proliferation in vitro but could enhance proliferation, tube formation of HUVEC, and promote angiogenesis in vivo models [27]. Further study indicated that increased CYP4Z2P-3′UTR expression promotes tumor angiogenesis in breast cancer may be partly via miRNA-dependent activation of PI3K/Akt and ERK1/2 [26]. Moreover, downregulation of pseudogene TPTE2P1 inhibits migration and invasion of gallbladder cancer cells [37], while upregulation of SUMO1 pseudogene 3 (SUMO1P3) in gastric cancer is associated with patients poor prognosis [33].
Conclusion
Although pseudogenes have been considered as non-functional relics littering the genome for a long time, it is clear that many pseudogenes are transcribed now. Recently, a handful of investigations have highlighted their involvement during pathogenesis of diseases such as cancer. A growing body of evidence indicates that thousands of pseudogenes are transcribed as sense transcripts, but only a few of them were found to regulate gene expression through acting as sponges/decoys for miRNAs and proteins or pseudogene asRNAs-mediated regulation. To date, only a few intriguing reports reveal the involvement of pseudogene in human cancers and their underlying mechanisms, while the biological function of the great majority of the tens of thousands of annotated pseudogenes currently remains unknown and investigations of them have proved challenging.
In this review, we highlighted the critical roles of some important pseudogenes in human cancer, and revealed that some of them may be potential therapeutic targets. Although only a small number of pseudogenes have been well characterized in human cancers, the research of pseudogenes is expanding quickly. Therefore, more functional investigations are needed in order to better understand their exact roles in tumorigenesis process, and it is meaningful to elucidate underlying molecular mechanisms and pathways of these pseudogenes. Despite accumulating evidence supporting the potential therapeutic value of pseudogenes for cancers, the regulators involved in pseudogene dysregulation and underlying mechanisms are still not well-known. Additionally, how pseudogenes cross-talk with epigenetic machineries in the pathogenesis of cancer needs to be further investigated. Therefore, integration of pseudogenes into cancer biology will deepen our understanding of the mechanisms of this deadly disease, and some specific pseudogenes may be translated into clinical applications for diagnosis, prognosis, or treatment of cancer patients.
References
Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22(9):1775–89. doi:10.1101/gr.132159.111.
Pei B, Sisu C, Frankish A, Howald C, Habegger L, Mu XJ, et al. The GENCODE pseudogene resource. Genome Biol. 2012;13(9):R51. doi:10.1186/gb-2012-13-9-r51.
Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309(5740):1559–63. doi:10.1126/science.1112014.
Jacq C, Miller JR, Brownlee GG. A pseudogene structure in 5S DNA of Xenopus laevis. Cell. 1977;12(1):109–20.
Li W, Yang W, Wang XJ. Pseudogenes: pseudo or real functional elements? J Genet Genomics. 2013;40(4):171–7. doi:10.1016/j.jgg.2013.03.003.
Kazazian Jr HH. Processed pseudogene insertions in somatic cells. Mob DNA. 2014;5:20. doi:10.1186/1759-8753-5-20.
Grander D, Johnsson P. Pseudogene-expressed RNAs: emerging roles in gene regulation and disease. Curr Top Microbiol Immunol. 2015. doi:10.1007/82_2015_442.
Goodhead I, Darby AC. Taking the pseudo out of pseudogenes. Curr Opin Microbiol. 2015;23:102–9. doi:10.1016/j.mib.2014.11.012.
Brown CJ, Hendrich BD, Rupert JL, Lafreniere RG, Xing Y, Lawrence J, et al. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71(3):527–42.
Hall LL, Byron M, Sakai K, Carrel L, Willard HF, Lawrence JB. An ectopic human XIST gene can induce chromosome inactivation in post differentiation human HT-1080 cells. Proc Natl Acad Sci U S A. 2002;99(13):8677–82. doi:10.1073/pnas.132468999.
Kota SK, Roy Chowdhury D, Rao LK, Padmalatha V, Singh L, Bhadra U. Uncoupling of X-linked gene silencing from XIST binding by DICER1 and chromatin modulation on human inactive X chromosome. Chromosoma. 2015;124(2):249–62. doi:10.1007/s00412-014-0495-4.
Zhang Z, Harrison PM, Liu Y, Gerstein M. Millions of years of evolution preserved: a comprehensive catalog of the processed pseudogenes in the human genome. Genome Res. 2003;13(12):2541–58. doi:10.1101/gr.1429003.
Proudfoot N. Pseudogenes. Nature. 1980;286(5776):840–1.
Groen JN, Capraro D, Morris KV. The emerging role of pseudogene expressed non-coding RNAs in cellular functions. Int J Biochem Cell Biol. 2014;54:350–5. doi:10.1016/j.biocel.2014.05.008.
Xiao-Jie L, Ai-Mei G, Li-Juan J, Jiang X. Pseudogene in cancer: real functions and promising signature. J Med Genet. 2015;52(1):17–24. doi:10.1136/jmedgenet-2014-102785.
Chan WL, Chang JG. Pseudogene-derived endogenous siRNAs and their function. Methods Mol Biol. 2014;1167:227–39. doi:10.1007/978-1-4939-0835-6_15.
Kalyana-Sundaram S, Kumar-Sinha C, Shankar S, Robinson DR, Wu YM, Cao X, et al. Expressed pseudogenes in the transcriptional landscape of human cancers. Cell. 2012;149(7):1622–34. doi:10.1016/j.cell.2012.04.041.
Han L, Yuan Y, Zheng S, Yang Y, Li J, Edgerton ME, et al. The Pan-Cancer analysis of pseudogene expression reveals biologically and clinically relevant tumour subtypes. Nat Commun. 2014;5:3963. doi:10.1038/ncomms4963.
Welch JD, Baran-Gale J, Perou CM, Sethupathy P, Prins JF. Pseudogenes transcribed in breast invasive carcinoma show subtype-specific expression and ceRNA potential. BMC Genomics. 2015;16:113. doi:10.1186/s12864-015-1227-8.
Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465(7301):1033–8. doi:10.1038/nature09144.
Tay Y, Kats L, Salmena L, Weiss D, Tan SM, Ala U, et al. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell. 2011;147(2):344–57. doi:10.1016/j.cell.2011.09.029.
Chen CL, Tseng YW, Wu JC, Chen GY, Lin KC, Hwang SM, et al. Suppression of hepatocellular carcinoma by baculovirus-mediated expression of long non-coding RNA PTENP1 and microRNA regulation. Biomaterials. 2015;44:71–81. doi:10.1016/j.biomaterials.2014.12.023.
Yu G, Yao W, Gumireddy K, Li A, Wang J, Xiao W, et al. Pseudogene PTENP1 functions as a competing endogenous RNA to suppress clear-cell renal cell carcinoma progression. Mol Cancer Ther. 2014;13(12):3086–97. doi:10.1158/1535-7163.MCT-14-0245.
Karreth FA, Reschke M, Ruocco A, Ng C, Chapuy B, Leopold V, et al. The BRAF pseudogene functions as a competitive endogenous RNA and induces lymphoma in vivo. Cell. 2015;161(2):319–32. doi:10.1016/j.cell.2015.02.043.
Rutnam ZJ, Du WW, Yang W, Yang X, Yang BB. The pseudogene TUSC2P promotes TUSC2 function by binding multiple microRNAs. Nat Commun. 2014;5:2914. doi:10.1038/ncomms3914.
Zheng L, Li X, Gu Y, Lv X, Xi T. The 3′UTR of the pseudogene CYP4Z2P promotes tumor angiogenesis in breast cancer by acting as a ceRNA for CYP4Z1. Breast Cancer Res Treat. 2015;150(1):105–18. doi:10.1007/s10549-015-3298-2.
Zheng L, Li X, Gu Y, Ma Y, Xi T. Pseudogene CYP4Z2P 3′UTR promotes angiogenesis in breast cancer. Biochem Biophys Res Commun. 2014;453(3):545–51. doi:10.1016/j.bbrc.2014.09.112.
Esposito F, De Martino M, Petti MG, Forzati F, Tornincasa M, Federico A, et al. HMGA1 pseudogenes as candidate proto-oncogenic competitive endogenous RNAs. Oncotarget. 2014;5(18):8341–54.
Esposito F, De Martino M, Forzati F, Fusco A. HMGA1-pseudogene overexpression contributes to cancer progression. Cell Cycle. 2014;13(23):3636–9. doi:10.4161/15384101.2014.974440.
Han YJ, Ma SF, Yourek G, Park YD, Garcia JG. A transcribed pseudogene of MYLK promotes cell proliferation. FASEB J. 2011;25(7):2305–12. doi:10.1096/fj.10-177808.
Peng H, Ishida M, Li L, Saito A, Kamiya A, Hamilton JP, et al. Pseudogene INTS6P1 regulates its cognate gene INTS6 through competitive binding of miR-17-5p in hepatocellular carcinoma. Oncotarget. 2015;6(8):5666–77.
Wang TH, Lin YS, Chen Y, Yeh CT, Huang YL, Hsieh TH, et al. Long non-coding RNA AOC4P suppresses hepatocellular carcinoma metastasis by enhancing vimentin degradation and inhibiting epithelial-mesenchymal transition. Oncotarget. 2015;6(27):23342–57.
Mei D, Song H, Wang K, Lou Y, Sun W, Liu Z, et al. Up-regulation of SUMO1 pseudogene 3 (SUMO1P3) in gastric cancer and its clinical association. Med Oncol. 2013;30(4):709. doi:10.1007/s12032-013-0709-2.
Hayashi H, Arao T, Togashi Y, Kato H, Fujita Y, De Velasco MA, et al. The OCT4 pseudogene POU5F1B is amplified and promotes an aggressive phenotype in gastric cancer. Oncogene. 2015;34(2):199–208. doi:10.1038/onc.2013.547.
Wang L, Guo ZY, Zhang R, Xin B, Chen R, Zhao J, et al. Pseudogene OCT4-pg4 functions as a natural micro RNA sponge to regulate OCT4 expression by competing for miR-145 in hepatocellular carcinoma. Carcinogenesis. 2013;34(8):1773–81. doi:10.1093/carcin/bgt139.
Hawkins PG, Morris KV. Transcriptional regulation of Oct4 by a long non-coding RNA antisense to Oct4-pseudogene 5. Transcription. 2010;1(3):165–75. doi:10.4161/trns.1.3.13332.
Lv W, Wang L, Lu J, Mu J, Liu Y, Dong P. Downregulation of TPTE2P1 inhibits migration and invasion of gallbladder cancer cells. Chem Biol Drug Des. 2015;86(4):656–62. doi:10.1111/cbdd.12533.
Chan WL, Yuo CY, Yang WK, Hung SY, Chang YS, Chiu CC, et al. Transcribed pseudogene psiPPM1K generates endogenous siRNA to suppress oncogenic cell growth in hepatocellular carcinoma. Nucleic Acids Res. 2013;41(6):3734–47. doi:10.1093/nar/gkt047.
Ye X, Fan F, Bhattacharya R, Bellister S, Boulbes DR, Wang R, et al. VEGFR-1 pseudogene expression and regulatory function in human colorectal cancer cells. Mol Cancer Res. 2015;13(9):1274–82. doi:10.1158/1541-7786.MCR-15-0061.
Cazalla D, Yario T, Steitz JA. Down-regulation of a host microRNA by a Herpesvirus saimiri noncoding RNA. Science. 2010;328(5985):1563–6. doi:10.1126/science.1187197.
Jeyapalan Z, Deng Z, Shatseva T, Fang L, He C, Yang BB. Expression of CD44 3′-untranslated region regulates endogenous microRNA functions in tumorigenesis and angiogenesis. Nucleic Acids Res. 2011;39(8):3026–41. doi:10.1093/nar/gkq1003.
Fujii GH, Morimoto AM, Berson AE, Bolen JB. Transcriptional analysis of the PTEN/MMAC1 pseudogene, psiPTEN. Oncogene. 1999;18(9):1765–9. doi:10.1038/sj.onc.1202492.
Poliseno L, Haimovic A, Christos PJ, Vega YSMEC, Shapiro R, Pavlick A, et al. Deletion of PTENP1 pseudogene in human melanoma. J Investig Dermatol. 2011;131(12):2497–500. doi:10.1038/jid.2011.232.
Johnsson P, Ackley A, Vidarsdottir L, Lui WO, Corcoran M, Grander D, et al. A pseudogene long-noncoding-RNA network regulates PTEN transcription and translation in human cells. Nat Struct Mol Biol. 2013;20(4):440–6. doi:10.1038/nsmb.2516.
Pain D, Chirn GW, Strassel C, Kemp DM. Multiple retropseudogenes from pluripotent cell-specific gene expression indicates a potential signature for novel gene identification. J Biol Chem. 2005;280(8):6265–8. doi:10.1074/jbc.C400587200.
Pesce M, Scholer HR. Oct-4: gatekeeper in the beginnings of mammalian development. Stem Cells. 2001;19(4):271–8. doi:10.1634/stemcells.19-4-271.
Kastler S, Honold L, Luedeke M, Kuefer R, Moller P, Hoegel J, et al. POU5F1P1, a putative cancer susceptibility gene, is overexpressed in prostatic carcinoma. Prostate. 2010;70(6):666–74. doi:10.1002/pros.21100.
Scarola M, Comisso E, Pascolo R, Chiaradia R, Maria Marion R, Schneider C, et al. Epigenetic silencing of Oct4 by a complex containing SUV39H1 and Oct4 pseudogene lncRNA. Nat Commun. 2015;6:7631. doi:10.1038/ncomms8631.
Fusco A, Fedele M. Roles of HMGA proteins in cancer. Nat Rev Cancer. 2007;7(12):899–910. doi:10.1038/nrc2271.
Lui KY, Peng HR, Lin JR, Qiu CH, Chen HA, Fu RD, et al. Pseudogene integrator complex subunit 6 pseudogene 1 (INTS6P1) as a novel plasma-based biomarker for hepatocellular carcinoma screening. Tumour Biol. 2015. doi:10.1007/s13277-015-3899-8.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (No.81472198), the Key Clinical Medicine Technology Foundation of Jiangsu Province (No.BL2014096), and the Medical Key Talented Person Foundation of the Jiangsu Provincial Developing Health Project (No.RC2011080).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflicts of interest
None
Additional information
Xuefei Shi and Fengqi Nie contributed equally to this work.
Rights and permissions
About this article
Cite this article
Shi, X., Nie, F., Wang, Z. et al. Pseudogene-expressed RNAs: a new frontier in cancers. Tumor Biol. 37, 1471–1478 (2016). https://doi.org/10.1007/s13277-015-4482-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-015-4482-z