Abstract
Genes important to stem cell progression have been involved in the genetics and clinical outcome of cancers. We investigated germ line variants in cancer stem cell (CSC) genes to predict susceptibility and efficacy of chemoradiotherapy treatment in gallbladder cancer (GBC) patients. In this study, we assessed the effect of SNPs in CSC genes (surface markers CD44, ALCAM, EpCAM, CD133) and (molecular markers NANOG, SOX-2, LIN-28A, ALDH1A1, OCT-4) with GBC susceptibility and prognosis. Total 610 GBC patients and 250 controls were genotyped by using PCR-RFLP, ARMS-PCR, and TaqMan allelic discrimination assays. Chemotoxicity graded 2–4 in 200 patients and tumor response was recorded in 140 patients undergoing neoadjuvant chemotherapy (NACT). Differences in genotype and haplotype frequency distributions were calculated by binary logistic regression. Gene-gene interaction model was analyzed by generalized multifactor dimensionality reduction (GMDR). Overall survival was assessed by Kaplan-Meier survival curve and multivariate Cox-proportional methods. ALCAM Ars1157Crs10511244 (P = 0.0035) haplotype was significantly associated with GBC susceptibility. In GMDR analysis, ALCAM rs1157G>A, EpCAM rs1126497T>C emerged as best significant interaction model with GBC susceptibility and ALDH1A1 rs13959T>G with increased risk of grade 3–4 hematological toxicity. SOX-2 rs11915160A>C, OCT-4 rs3130932T>G, and NANOG rs11055786T>C were found best gene-gene interaction model for predicting response to NACT. In both Cox-proportional and recursive partitioning ALCAM rs1157GA+AA genotype showed higher mortality and hazard ratio. ALCAM gene polymorphisms associated with GBC susceptibility and survival while OCT-4, SOX-2, and NANOG variants showed an interactive role with treatment response.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cancer is known as malignant neoplasia related to uncontrolled proliferation. Until now, there are over 200 different kinds of cancers that can affect the human body [1]. Among them, gallbladder cancer (GBC) is the most common biliary tract cancer in North Indian population, characterized by its high lethality, aggressive nature, and dismal prognosis [2]. For all advances in chemoradiation, the outcome of treatment with advanced GBC is still poor, because of chemoresistance and recurrence due to heterogeneity of tumor cells. Cancer stem cell (CSC) presents in a heterogeneous tumor with properties of tumor progression, chemoresistance, and recurrence [3].
Cancer stem cells are identified by expression of surface markers CD44, ALCAM, EpCAM, CD133, and molecular markers NANOG, SOX-2, LIN-28A, ALDH1A1, and OCT-4. Surface markers are transmembrane glycoprotein in nature and are involved in cell proliferation, invasion, and metastasis of cancer stem cell [4, 5]. CD44+CD133+ cells showed chemoresistance and CSCs like characteristics in gallbladder cancer cell lines [6, 7]. Our earlier reports have shown the important role of CD44 gene variants in GBC susceptibility [8]. ALCAM-ALCAM interconnections have a role in the development and maintenance of tissue architecture and tumor progression [9]. ALCAM considered as a breast cancer prognostic marker [10]. EpCAM+ cells have been possessed a tumor-initiating role [11]. EpCAM is overexpressed in different tumors, including colon, lung, pancreas, breast, and ovary [12].
Molecular marker ALDH1A1 enzyme belongs to dehydrogenases family of proteins that participates in cellular detoxification and differentiation through the oxidation of intracellular aldehydes [13] while Oct-4, Sox-2, and Nanog transcription factors responsible for pluripotency of stem cell. Oct-4, Sox-2, and Nanog induce expression in cooperative manner for maintaining pluripotency of cancer stem cells [14]. LIN28 encodes a microRNA-binding protein that binds to let-7 microRNA and inhibits production of the mature let-7 microRNA in stem cells [15].
Many studies have reported significant associations of CSC gene polymorphisms with susceptibility and treatment response in cancers [16–18]. Till now; there are a limited number of studies involving CSC genetic variants with GBC [8]. Therefore, in this study, we investigated 15 CSCs (surface and molecular marker) germline polymorphisms that have been previously associated with different cancers, to evaluate their influence on the risk and treatment outcomes of GBC in the North Indian population. In future, some cancer stem cell genetic variants may act as potential predictive and prognostic markers for GBC.
Materials and methods
Recruitment of subjects
In this study, 610 histologically confirmed gallbladder cancer patients and 250 controls were recruited after taking their informed consent. All healthy controls were age-, gender-, and ethnicity-matched and without gastrointestinal disorders and belonged to same geographical region. The study protocol was approved by the Ethics Committee of SGPGIMS. Two hundred GBC patients receiving adjuvant or neoadjuvant chemoradiotherapy, in a sequential manner for three cycles, were followed for treatment response. Radiotherapy was given by IMRT and 3D-CRT techniques. Chemotherapeutic drugs gemcitabine, 5-flurouracil, and cisplatin were given in combination. Two hundred patients were followed-up for drug toxicity according to the National Cancer Institute (NCI) Common Toxicity Criteria for Adverse Events (CTCAE), version 3.0 (http://ctep.cancer.gov). Tumor response was assessed in 140 NACT-treated patients, according to the Response Evaluation Criteria in Solid Tumors (RECIST1.1 criteria). Details of patient enrollment and treatment plan are shown in Fig. 1.
Patients with static and progressive disease were considered as nonresponders while with complete and partial pathological response were considered as responders. In chemotoxicity, grade 0–1 was considered as mild toxicity and grade 2–4 as moderate to severe. Hematological toxicity was in terms of anemia, leucopenia, and thrombocytopenia. We evaluated overall survival on the basis of patient’s histopathological and clinical stages in 199 patients. Treatment modality and clinical stage affect the survival pattern, so we divided 199 patients in three groups’ postoperative, metastatic, and locally advanced for survival analysis. Median follow-up survival period was 18 months.
Basic characteristics of study population and clinical data of follow up patients are given in Tables 1 and 2, respectively.
SNP selection
Most Informative tagger SNPs (Haploview software 4.2) were selected with >5 % MAF in GIH and CEU population by using the HapMap project database. The sample size was calculated by using QUANTO1.1. This study achieved 80 % power.
Genotyping
Venous blood (4 ml) was collected from patients, and the genomic DNA was extracted from peripheral blood using the standard salting out method [19]. Genotyping of the SNPs was carried out using the TaqMan allelic discrimination assay, ARMS-PCR, and PCR-RFLP. Details of genotyping methods are summarized in Table S1 (supplementary data).
Statistical evaluation
Descriptive statistics of patients were presented as mean and standard deviations for continuous measures and frequencies and percentages for categorical measures. Correlations between various genotypes and treatment outcomes were examined using binary logistic regression. The association was expressed with odds ratios (OR) or risk estimates with 95 % confidence intervals (CI). Statistical analysis was done using SPSS statistical analysis software, version 20.0. Haplotype analysis was done by using the SNPstat Software (bioinfo.iconcologia.net/SNPstats). Global P value represents a difference between the means associated with pair of haplotypes. To reduce chances of obtaining false-positive results (type I errors) Bonferroni correction was applied in subgroup analysis. The Bonferroni correction is an adjustment made to P values when several dependent or independent statistical tests are being performed simultaneously on a single dataset. To perform a Bonferroni correction, we divided the critical P value (0.05) by the number of comparisons being made (0.05/2 = 0.025). The statistical power of the study is then calculated based on this modified P value (0.025).
The functional effects of polymorphisms were determined by online F-SNP (http://compbio.Cs.Queens.CA/F-SNP/) database. F-SNP database provides integrated information about the functional effects of SNPs obtained from 16 bioinformatics tools and databases and provides output in terms of FS score. The functional effects are predicted and indicated at the splicing, transcriptional, translational, and posttranslational level. As such, the F-SNP database helps identify and focus on SNPs with potential pathological effect to human health.
Generalized multifactor dimensionality reduction (GMDR) analysis was done for the adjustment of quantitative and discrete covariants with gene-gene interactions [20]. GMDR replication of permutation determines accuracy of the P value assessed by permutation. We used 1000 replication of permutation to get significant P values.
The best gene-gene interaction model was obtained by reducing multi-locus genotypes into low-risk and high-risk groups on the basis of highest testing accuracy and permutation results, and cross-validation consistency (CVC) was considered to be statistically significant at the 0.05 level.
Survival curve was constructed using Kaplan-Meier method, and differences between the groups were tested by the log-rank method. The hazard ratio was calculated by Cox-proportional method at 95 % confidence interval. More intuitive survival model was generated by recursive partitioning (rpart version 3.1). Protein direct and functional interactions were determined by STRING database.
Results
Association of genetic variants with GBC susceptibility
In this study, we evaluated the frequency distribution of genetic polymorphisms in cancer stem cell surface markers CD44, ALCAM, EpCAM, and CD133 and molecular markers ALDH1A1, OCT-4, SOX-2, LIN-28A, and NANOG polymorphisms between healthy controls and GBC patients. All studied polymorphisms were in Hardy-Weinberg equilibrium in controls (P > 0.05).
The frequencies of ALCAM rs1157G>A and rs10511244T>C polymorphisms were found to be significantly associated with GBC risk, at genotypic [rs1157(AA), OR (CI) = 2.4 (1.09–5.29), P value = 0.03] and allelic levels [rs1157(A), OR (CI) = 1.42 (1.07–4.89), P value = 0.016; rs10511244 (C),OR (CI) = 1.42 (1.02–4.96), P value = 0.036], respectively (Tables 3 and S2).
To determine the effect of multiple single nucleotide polymorphisms within a gene, haplotype analysis was done. Those haplotypes, which did not cross rare haplotype frequency (<0.05 %), were excluded from analysis. Bonferroni correction was applied in subgroup analysis. TheArs1157Crs10511244 haplotype of ALCAM was significantly associated with increased risk in GBC patients [Table S3 OR (CI) = 2.40 (1.34–4.32), P value = 0. 0035], but the significance was lost after Bonferroni correction. ALCAM Ars1157Crs10511244 haplotype in GBC patients harboring gallstones [Table S4 OR (CI) = 3.71 (1.90–7.26), P value = 1e−04*] and GBC females [Table S5 OR (CI) = 3.62 (1.53–8.57), P value = 0. 0035*] remained significant after Bonferroni correction (P < 0.025).
Individual CD44 gene polymorphisms had no significant differences in frequency distribution between GBC cases and controls, but haplotype of CD44 Crs13347Ars353639Ars187116Crs187115 [OR (CI) = 0.51 (0.27–0.95)], P value = 0.033] was significantly associated with lower risk of GBC. Here also, the significance was lost after Bonferroni correction (Table S3).
There were no significant associations of EpCAM rs1126497T>C, EpCAM rs1421T>C, CD133 rs3130C>T, CD133 rs2240688T>G, ALDH1A1 rs13959A>G, OCT-4 rs3130932T>G, SOX-2 rs11915160A>C, LIN-28A rs4274112T>C, and NANOG rs11055786T>C polymorphisms with GBC risk in terms of overall case control frequency distribution, gender stratification, and gallstone status (Tables 3, S2, S3, S4, and S5).
Gene-gene interaction for GBC susceptibility
GMDR analysis was used to evaluate gene-gene interaction with GBC susceptibility by adjusting age, gender, tobacco, and gallstone status covariants.
We performed separate gene-gene interaction analysis for surface and molecular markers. In surface markers, ALCAM rs1157 and EpCAM rs1126497 [Table 4, OR (CI) = 1.8 (1.17–6.78), P value = 0. 005] was best significant interaction model for GBC susceptibility. In molecular markers OCT-4 rs3130932, LIN-28A rs4274112 [Table 4, OR (CI) = 1. 84 (1.18–5.88), P value = 0. 007] was best significant interaction model for GBC susceptibility.
Association of CSC genetic variants with treatment response and toxicity
Univariable analysis did not show significant association of variants in surface and molecular marker between responders and nonresponders of chemoradiotherapy treatment outcomes at allele, genotype, and haplotype levels (haplotypes, Table S6). Results were consistent after multivariable analysis. Hematological and gastrointestinal toxicity of treatment in GBC patients also did not associate with any particular CSC genetic variants (Tables S7 and S8).
The GMDR analysis was used to evaluate gene-gene interaction with clinical outcome by adjustment of the covariants tumor stage, age, gender, and treatment modality. The best combination model for clinical outcome is given in Table 4.
For surface markers, we did not get significant interaction model for treatment response. However, in molecular markers, the best significant model was NANOG rs11055786, OCT-4 rs3130932, and SOX-2 rs11915160 (Table 4, OR (CI) = 5.6 (1.71–18.3), P value = 0. 003] for treatment response. ALDH1A1 rs13959 (Table 4, OR (CI) = 3.0 (1.25–7.47), P value = 0.016) emerged as the best interaction model for hematological toxicity.
Associations between the CSCs genetic variants and GBC survival
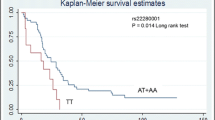
Kaplan-Meier survival curves were used to assess the associations between the CSCs polymorphisms and survival time. Survival was calculated separately in the three patient groups’ postoperative, metastatic, and locally advanced. The median survival period for metastatic was 9.5 months. Metastatic GBC cases with ALCAM rs1157GG genotype showed a higher survival rate than the individuals with GA+AA genotype (log-rank p = 0.026) (Fig. 2). In Cox-proportional hazard model ALCAM rs1157GA+AA genotype in GBC cases, associated with higher hazard ratio (HR = 1. 7, CI = 1.03–2.84, Table 5) as compared with GG genotype. Kaplan-Meier survival curve did not show any significant differences in other CSC polymorphisms’ survival rate between wild and variant genotypes. Model-based classical recursive partitioning is a statistical method that creates a decision tree with the split criteria of log-rank test that strives to correctly classify according survival times and the Kaplan-Meier graphs. Tree-structured survival analysis of our data represented that ALCAM (GA+AA) end node had highest hazard ratio and lowest survival period as compared to CD133rs3130C>T and OCT-4 rs3130932 (Figs. 3 and 4).
Discussion
Cancer stem cells are responsible for tumor growth, migration, invasion, aggressiveness, resistance, and pluripotency of a tumor. This study was conducted to determine the role of cancer stem cell gene variants in gallbladder cancer susceptibility and treatment outcomes.
CSC genetic variants and GBC susceptibility
Our results obtained by analyzing 610 GBC patients and 250 controls demonstrated that ALCAM rs1157G>A and rs10511244T>C polymorphisms were significantly associated with increased risk in GBC patients at genotype and haplotype analysis. On the other hand, CD44 Crs13347Ars353639Ars187116Crs187115 haplotype was significantly associated with a protective effect. The ALCAM and CD44 are cell surface adhesion molecules which mediate adhesion interactions between cell-cell and cell-substrates [9, 21]. Previous studies also showed ALCAM rs1157 homozygous variant to be associated with breast cancer susceptibility [22]. The ALCAM rs1157G>A and ALCAM rs10511244T>C are present in 3′UTR and intron1 region of the gene, respectively. By F-SNP, we hypothesized that the variant alleles of ALCAMrs1157 G>A and ALCAMrs10511244T>C may be involved in transcription regulation (Table 6). Variant allele may result in increased ALCAM expression which may disrupt adhesion interactions that disturb tissue architecture between cells [23].
In our study, CD44Crs13347Ars353639Ars187116Crs187115 haplotype conferred lower risk of GBC which reconfirms our earlier observation in smaller number of samples [8]. However, CD44 rs187116–rs187115 T-A haplotype has been reported to confer higher risk of gastric adenocarcinoma [24]. CD44rs187115 variant genotype was also correlated with risk of oral cancer development [25]. CD44 has been reported to be highly expressed in subserous gallbladder carcinoma [26] and CD44 protein variant overexpression was related to histologic dedifferentiation of gallbladder carcinoma [27]. Several mechanistic studies have shown dualistic nature of CD44, after its ligand hyaluronic acid interaction. In silico analysis using F-SNP predicted change in transcriptional regulation for rs13347, rs353639, and rs187115 SNPs (Table 6), supporting the influence of selected CD44 genetic variants on gene transcription and splicing mechanisms.
On applying logistic regression and haplotype analysis, we did not find significant association of EpCAM rs1126497T>C, EpCAM rs1421T>C, CD133 rs3130C>T, CD133 rs2240688T>G, ALDH1A1 rs13959A>G, OCT-4 rs3130932T>G, SOX-2 rs11915160A>C, LIN-28A rs4274112 T>C, and NANOG rs11055786T>C polymorphisms with overall frequency distribution, gender, and gallstone status. Some other studies showed that EpCAM rs1126497 CT+TT and OCT-4 rs3130932 T>G were associated with susceptibility to breast cancer risk, whereas no association was observed with EpCAM rs1421 [28, 29].
In genetic association studies involving low penetrance genes, effect of individual genes in disease risk is rather limited and gene-gene interaction have been proposed as important contributor in risk assessments. For gene-gene interaction analysis by GMDR with adjusting covariants, we found that SNPs ALCAM rs1157 and EpCAM rs1126497 surface marker as best significant interaction model with increased risk of GBC susceptibility. F-SNP predicted EpCAM rs1126497 functional role in splicing regulation. STRING 10 also suggested ALCAM and EpCAM protein- protein interaction (Fig. 5). So, there must be an interacting role of ALCAM and EpCAM in GBC susceptibility.
Likewise, GMDR analysis predicted interactive role of OCT-4 rs3130932 and LIN-28A rs4274112 molecular marker with GBC susceptibility. Lin-28a is a mRNA binding protein and facilitates posttranscriptional regulation of OCT-4 protein [30]. Our previous study also reported LIN-28A rs4274112 (AG+GG) association with lymph node metastases and OCT-4rs3130932 with hormone receptor positive tumors in breast cancer [31]. These findings uncover a new level of gene-gene interaction in gallbladder cancer susceptibility.
CSCs genetic variants and GBC treatment response
At both univariable and multivariable logistic regression and haplotype analyses, none of the CSC polymorphisms were significantly associated with GBC treatment response. Even after GMDR analysis, CSCs genetic variants for surface markers still did not significantly associate with treatment response and toxicity. In a previous study also, ALCAM rs1157 had no statistical significance with 5-FU-treated colon cancer recurrence [32] while in other study, it was shown to be associated with colon cancer recurrence [17]. CD44rs187116 and rs187115 T-A haplotype had lower risk to develop gastric adenocarcinoma recurrence [24]. The expression of CD44 with tumor prognosis is controversial. Certain studies have indicated that the overexpression of CD44 is correlated with elevated chemoradiotherapy resistance and increased risk of recurrence [33] whereas others have related poor prognosis to CD44 downregulation in tumor cells [34]. For EpCAM rs1126497T>C, rs1421T>C, our results with treatment response and toxicity are in tune with previous studies which also did not show any significant effect of these variants on prognosis in nonsmall cell lung cancer [35].
Variants for molecular marker genes showed an interactive association with treatment outcomes. In GMDR analysis, SNPs in SOX2 rs11915160, OCT4 rs3130932, and NANOG rs11055786 turned out to be the best interaction models for predicting poor response to NACT in GBC. The cooperative interaction sox-oct cis regulatory element is essential for nanog pluripotency. STRING 10 analysis showed interaction among SOX-2, OCT-4, and NANOG proteins (Fig. 6). Our earlier report also suggested SOX-2 rs11915160, OCT-4 rs3130932, and NANOG rs11055786 interactive role with treatment outcomes in breast cancer [31].
ALDH1A1 rs13959 elicited as the best model for higher grade 3–4 hematological toxicity. ALDH1A1 belongs to dehydrogenase family and plays a role in the detoxification of active cyclophosphamide metabolites. Our findings suggest that ALDH1A1 rs13959A>G genetic variations may affect the risk of severe hematological toxicity caused by myelosuppressive chemotherapy drugs. ALDH1A1 rs13959A>G is a synonymous polymorphism present in exon 3. F-SNP predicted its role in change in splicing regulation (Table 6). This variant allele could thus affect ALDH enzyme activity, thus resulting in an increased risk of chemotherapeutic drug toxicity. ALDH1A1 polymorphisms (rs3764435 C>A-rs63319C>A) A-A were also reported to be associated with grade 3–4 hematological toxicity in breast cancer [36].
CSC genetic variants and GBC survival
Survival analysis results showed that ALCAM rs1157 GA+AA genotype is associated with unfavorable survival in GBC metastasis cases. The Cox-proportional hazard regression analysis demonstrated that GA+AA genotype had higher hazard ratio. Swedish population study also showed similar results; ALCAM rs1157 homozygous minor allele carriers had worse survival in breast cancer [37]. CD44, EpCAM, CD133, NANOG, SOX-2, LIN-28A, OCT-4, and ALDH1A1 polymorphisms did not significantly associate with overall survival. But previously reported, CD44 rs187116G>A–rs187115T>C T-A haplotype and CD44rs13347 variant genotype associated with lowered survival rate in breast cancer patients [24, 38]. Survival decision tree terminal node determined ALCAM GA+AA as worse survival subgroup with higher hazard ratio.
These data collectively suggest important biological role of the CSC genetic variants in the susceptibility and prognosis of gallbladder cancer. However, functional studies are needed to elucidate the effects of the CSC genetic variants in various cancers. In future, cancer stem cells may prove to be important in defeating gallbladder cancer.
In conclusion, our findings suggest important role of CSC genetic variants in GBC susceptibility, prognosis, and survival outcomes.
References
Rebecca Siegel MJM, Zou Z, Jemal A. Cancer statistics. CA Cancer J Clin. 2014;64:29.
Misra S, Chaturvedi A, Misra NC, Sharma ID. Carcinoma of the gallbladder. Lancet Oncol. 2003;4:167–76.
Wicha MS, Liu S, Dontu G. Cancer stem cells: an old idea—a paradigm shift. Cancer Res. 2006;66:1883–90. discussion 1895-1886.
Bourguignon LY, Singleton PA, Zhu H, Diedrich F. Hyaluronan-mediated CD44 interaction with RhoGEF and Rho kinase promotes Grb2-associated binder-1 phosphorylation and phosphatidylinositol 3-kinase signaling leading to cytokine (macrophage-colony stimulating factor) production and breast tumor progression. J Biol Chem. 2003;278:29420–34.
Bourguignon LY. CD44-mediated oncogenic signaling and cytoskeleton activation during mammary tumor progression. J Mammary Gland Biol Neoplasia. 2001;6:287–97.
Baumann M, Krause M. CD44: a cancer stem cell-related biomarker with predictive potential for radiotherapy. Clin Cancer Res. 2010;16:5091–3.
Shi C, Tian R, Wang M, Wang X, Jiang J, Zhang Z, et al. CD44+ CD133+ population exhibits cancer stem cell-like characteristics in human gallbladder carcinoma. Cancer Biol Ther. 2010;10:1182–90.
Sharma KL, Yadav A, Gupta A, Tulsayan S, Kumar V, Misra S, et al. Association of genetic variants of cancer stem cell gene CD44 haplotypes with gallbladder cancer susceptibility in North Indian population. Tumour Biol. 2014;35:2583–9.
Bowen MA, Patel DD, Li X, Modrell B, Malacko AR, Wang WC, et al. Cloning, mapping, and characterization of activated leukocyte-cell adhesion molecule (ALCAM), a CD6 ligand. J Exp Med. 1995;181:2213–20.
Burandt E, Bari Noubar T, Lebeau A, Minner S, Burdelski C, Janicke F, et al. Loss of ALCAM expression is linked to adverse phenotype and poor prognosis in breast cancer: a TMA-based immunohistochemical study on 2,197 breast cancer patients. Oncol Rep. 2014;32:2628–34.
Yamashita T, Ji J, Budhu A, Forgues M, Yang W, Wang HY, et al. EpCAM-positive hepatocellular carcinoma cells are tumor-initiating cells with stem/progenitor cell features. Gastroenterology. 2009;136:1012–24.
Imrich S, Hachmeister M, Gires O. EpCAM and its potential role in tumor-initiating cells. Cell Adhes Migr. 2012;6:30–8.
Pereira F, Rosenmann E, Nylen E, Kaufman M, Pinsky L, Wrogemann K. The 56 kDa androgen binding protein is an aldehyde dehydrogenase. Biochem Biophys Res Commun. 1991;175:831–8.
Rodda DJ, Chew JL, Lim LH, Loh YH, Wang B, Ng HH, et al. Transcriptional regulation of nanog by OCT4 and SOX2. J Biol Chem. 2005;280:24731–7.
Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100.
Chou YE, Hsieh MJ, Chiou HL, Lee HL, Yang SF, Chen TY. CD44 gene polymorphisms on hepatocellular carcinoma susceptibility and clinicopathologic features. Biomed Res Int. 2014;2014:231474.
Gerger A, Zhang W, Yang D, Bohanes P, Ning Y, Winder T, et al. Common cancer stem cell gene variants predict colon cancer recurrence. Clin Cancer Res. 2011;17:6934–43.
Wang Q, Liu H, Xiong H, Liu Z, Wang LE, Qian J, et al. Polymorphisms at the microRNA binding-site of the stem cell marker gene CD133 modify susceptibility to and survival of gastric cancer. Mol Carcinog. 2015;54:449–58.
Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215.
Lou XY, Chen GB, Yan L, Ma JZ, Zhu J, Elston RC, et al. A generalized combinatorial approach for detecting gene-by-gene and gene-by-environment interactions with application to nicotine dependence. Am J Hum Genet. 2007;80:1125–37.
Goodison S, Urquidi V, Tarin D. CD44 cell adhesion molecules. Mol Pathol. 1999;52:189–96.
Jiang Y, Qin Z, Hu Z, Guan X, Wang Y, He Y, et al. Genetic variation in a hsa-let-7 binding site in RAD52 is associated with breast cancer susceptibility. Carcinogenesis. 2013;34:689–93.
Benchimol S, Fuks A, Jothy S, Beauchemin N, Shirota K, Stanners CP. Carcinoembryonic antigen, a human tumor marker, functions as an intercellular adhesion molecule. Cell. 1989;57:327–34.
Winder T, Ning Y, Yang D, Zhang W, Power DG, Bohanes P, et al. Germline polymorphisms in genes involved in the CD44 signaling pathway are associated with clinical outcome in localized gastric adenocarcinoma. Int J Cancer. 2011;129:1096–104.
Chou YE, Hsieh MJ, Hsin CH, Chiang WL, Lai YC, Lee YH, et al. CD44 gene polymorphisms and environmental factors on oral cancer susceptibility in taiwan. PLoS One. 2014;9:e93692.
Roa I, Villaseca M, Araya J, Roa J, de Aretxabala X, Ibacache G, et al. CD44 (HCAM) expression in subserous gallbladder carcinoma. Rev Med Chil. 2001;129:727–34.
Yanagisawa N, Mikami T, Mitomi H, Saegusa M, Koike M, Okayasu I. CD44 variant overexpression in gallbladder carcinoma associated with tumor dedifferentiation. Cancer. 2001;91:408–16.
Jiang L, Zhang C, Li Y, Yu X, Zheng J, Zou P, et al. A non-synonymous polymorphism Thr115Met in the EpCAM gene is associated with an increased risk of breast cancer in Chinese population. Breast Cancer Res Treat. 2011;126:487–95.
Katafigiotis S, Papamichos SI, Katopodi R, Papazisis K, Mylonaki E, Repana D, et al. A case-control study on the rs3130932 single nucleotide polymorphism in the OCT4B translation initiation codon in association with cancer state. Eur J Cancer Prev. 2011;20:248–51.
Qiu C, Ma Y, Wang J, Peng S, Huang Y. Lin28-mediated post-transcriptional regulation of Oct4 expression in human embryonic stem cells. Nucleic Acids Res. 2010;38:1240–8.
Tulsyan S, Agarwal G, Lal P, Mittal B. Significant association of combination of OCT4, NANOG, and SOX2 gene polymorphisms in susceptibility and response to treatment in North Indian breast cancer patients. Cancer Chemother Pharmacol. 2014;74:1065–78.
Szkandera J, Herzog S, Pichler M, Stiegelbauer V, Stotz M, Schaberl-Moser R, et al. Lgr5 rs17109924 is a predictive genetic biomarker for time to recurrence in patients with colon cancer treated with 5-fluorouracil-based adjuvant chemotherapy. Pharmacogenomics J. 2015.
Situ D, Long H, Lin P, Zhu Z, Wang J, Zhang X, et al. Expression and prognostic relevance of CD44v6 in stage I non-small cell lung carcinoma. J Cancer Res Clin Oncol. 2010;136:1213–9.
Stoll C, Baretton G, Soost F, Terpe HJ, Domide P, Lohrs U. Prognostic importance of the expression of CD44 splice variants in oral squamous cell carcinomas. Oral Oncol. 1999;35:484–9.
Yang Y, Fei F, Song Y, Li X, Zhang Z, Fei Z, et al. Polymorphisms of EpCAM gene and prognosis for non-small-cell lung cancer in Han Chinese. Cancer Sci. 2014;105:89–96.
Yao S, Sucheston LE, Zhao H, Barlow WE, Zirpoli G, Liu S, et al. Germline genetic variants in ABCB1, ABCC1 and ALDH1A1, and risk of hematological and gastrointestinal toxicities in a SWOG phase III trial S0221 for breast cancer. Pharmacogenomics J. 2014;14:241–7.
Varadi V, Bevier M, Grzybowska E, Johansson R, Enquist-Olsson K, Henriksson R, et al. Genetic variation in ALCAM and other chromosomal instability genes in breast cancer survival. Breast Cancer Res Treat. 2012;131:311–9.
Jiang L, Deng J, Zhu X, Zheng J, You Y, Li N, et al. Cd44 rs13347 c>t polymorphism predicts breast cancer risk and prognosis in Chinese populations. Breast Cancer Res. 2012;14:R105.
Acknowledgments
The authors would like to thank the Department of Biotechnology, Indian Council of Medical Research, and the Department of Science and Technology, Government of India, for financial support.
Conflicts of interest
None
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 56 kb)
Rights and permissions
About this article
Cite this article
Yadav, A., Gupta, A., Rastogi, N. et al. Association of cancer stem cell markers genetic variants with gallbladder cancer susceptibility, prognosis, and survival. Tumor Biol. 37, 1835–1844 (2016). https://doi.org/10.1007/s13277-015-3929-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-015-3929-6