Abstract
Kinesin superfamily protein 2A (KIF2A), an M‑type nonmotile microtubule depolymerase, has received attention for its role in carcinogenesis and prognostic value in several types of cancer. In this study, we evaluated the expression of KIF2A and its potential and robustness to predict clinical outcomes in colorectal cancer (CRC) patients. The messenger RNA (mRNA) expression of KIF2A was determined in 20 pairs of cancerous and adjacent nontumor tissues by real-time polymerase chain reaction. KIF2A immunohistochemistry was performed on tissue microarray (TMA), composed of 182 CRC and 179 matched adjacent nontumor tissues from surgery, 23 chronic colitis, 43 low-grade, and 18 high-grade intraepithelial neoplasias acquired through intestinal endoscopic biopsy. Univariate and multivariate Cox regression models were used to perform survival analyses. Both KIF2A mRNA and protein product exhibited CRC tissue-preferred expression, when compared with benign tissues. The high KIF2A expression was significantly correlated to TNM stage (P = 0.046) and tumor status (T) (P = 0.007). In univariate and multivariate analyses, high KIF2A expression showed a major prognostic value regarding 5-year survival. The influences of KIF2A expression on the survival were further proven by Kaplan–Meier survival analysis. This study demonstrated CRC tissue-preferred expression pattern of the KIF2A and suggested that high KIF2A expression might serve as an independent maker for poor prognosis in CRC patients.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
As the second common causes of cancer deaths, colorectal cancer (CRC) is one of top contributors to cancer-related mortality and morbidity [1, 2]. Several countries and regions (i.e., Australia and New Zealand, Europe, and North America) have been known to have highest incidence rates of CRC. Nevertheless, a trend toward a rapidly increasing incidence of CRC has been found in some low-risk areas including several countries within Eastern Asia [3, 4]. Notwithstanding rapid advances in treatments including surgery, chemotherapy, and targeted therapy, the prognosis for CRC patients is far from satisfying. A major difficulty in enhancing clinical outcomes is the fact that clinicians currently have no adequate knowledge and tools to predict individual patients’ responses to treatments. Therefore, it is imperative to discover novel biomarkers that can predict prognosis of CRC in order for the clinician to choose appropriate and optimal therapies for the CRC patients. Thus far, countless efforts have been made to pinpoint reliable prognostic biomarkers for CRC based on tumor biology [5]. Consequently, various factors, including genetic signatures (e.g., microsatellite instability) [6–8], gene mutations (e.g., BRAF V600E) [9, 10], and miRNA (e.g., miR-451, miR-625, miR-29c) [11–13], have exhibited association with favorable or poor CRC prognosis, to some extent. Nevertheless, many issues such as reproducibility and specificity are warranted to be addressed. There remains much work to be done to identify high-quality CRC prognostic markers.
Kinesin superfamily protein 2A (KIF2A), together with KIF2B, mitotic centromere-associated kinesin (MCAK, also known as KIF2C), constitute the kinesin-13 family [14]. These family members are M‑type nonmotile microtubule depolymerases and play central role in regulating microtubule dynamics during mitotic progression. The spindle, a microtubule-based structure, is a requirement for accurate chromosome segregation in both the mitotic and meiotic cell cycles [15, 16]. KIF2A is a microtubule minus end-depolymerizing motor, and it is essential in assembling normal bipolar spindles [17]. With the ablation of KIF2A, through small interfering RNA (siRNA) or antibody, cells end up forming monopolar spindles, which may result in chromosome gain or loss in daughter cells [17, 18]. As a result, cell cycle progression is also halted [17, 18]. Interestingly, several lines of evidence has indicated that KIF2A may be implicated in carcinogenesis [19–21] and the development of drug resistance [22]. For instance, it was demonstrated that upregulation of KIF2A was related to the progression and metastasis of squamous cell carcinoma of the tongue [19]. Thus far, the expression of KIF2A in CRC and its prognostic role have not been explored in CRC. With this in mind, we examined expression of KIF2A in CRC and evaluated its association with progression, invasion, and metastasis of CRC.
Materials and methods
Clinical tissue samples
One hundred and eighty-two CRC patients were recruited in this study, consisting 116 men and 66 women. All enrolled patients received surgery for CRC in the Affiliated Hospital of Nantong University between January 2005 and December 2010. All CRC patients were histopathologically confirmed by at least two pathologists. Patients would be precluded if they were treated with chemotherapy or radiotherapy before surgery. The age range of patients was between 17 and 90 years, with a mean age of 65.21 years. The following clinical data of each patient were also acquired: gender, tumor site, histological type, tumor differentiation, TNM stage (T, N, and M stand for tumor status, lymph node metastasis, and distant metastasis), and preoperative carcinoembryonic antigen (CEA) level. Informed consent was provided by patients prior to surgical operation. Primary CRC tissues and adjacent normal colorectal tissues were collected with the approval of the Ethics Committee of the Nantong University. After quick removal of the necrotic and ulcerated parts of samples, a fraction of sample sets underwent formalin-fixation and paraffin embed, while remaining tissues were snap-frozen in liquid nitrogen until further utilization. Additionally, for the comparison of KIF2A expression in cancerous and noncancerous tissues, 23 chronic colitis, 43 low-grade, and 18 high-grade intraepithelial neoplasias obtained by intestinal endoscopic biopsy were also included to increase sample size.
qRT-PCR analysis
To investigate the difference in the KIF2A gene expression levels between cancerous and normal tissues, total RNA was extracted from fresh CRC cancer tissues and matched, tumor-adjacent normal tissue samples (n = 20). The LightCycler FastStart DNA Master SYBR Green I Kit (Roche Diagnostics, Tokyo, Japan) was used to carry out One-Step quantitative real-time polymerase chain reaction (qRT-PCR) analysis, following previously published procedure [23]. The primers for KIF2A gene amplification were as follows: forward primer 5′-GCCGAATACATCAAGCAAT-3′ and reverse primer 5′-CTCTCCAGGTCAATCTCTT-3′, which generated a 109-bp amplicon. Moreover, a PCR assay with primers specific for the housekeep gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene was run in parallel to normalize KIF2A gene expression (forward primer 5′-TGCACCACCAACTGCTTAGC-3′ and reverse primer 3′-GGCATGGACTGTGGTCATGAG-5′). Reverse transcription and Taq activation were conducted by sequential incubation for 30 min at 42 °C and 2 min at 94 °C, respectively. The amplification parameters were 95 °C for 20 s, 56 °C for 20 s, and 72 °C for 30 s for 35 cycles. All samples were run in triplicate.
TMA construction and IHC analysis
Tissue Microarray System (Quick-Ray, UT06, UNITMA, Korea) was used to construct tissue microarrays (TMAs) with 182 CRC tissue specimens and corresponding tumor-adjacent normal tissues in the Department of Clinical Pathology, Nantong University Hospital, Jiangsu, China. Briefly, we captured core tissue biopsies (2 mm in diameter) from each donor paraffin-embedded section and put them in recipient paraffin blocks in order. TMA specimens were then sliced into 4-μm sections and positioned on super frost-charged glass microscope slides. Immunohistochemistry (IHC) analysis was performed according to standard protocol [24] with a mouse antihuman KIF2A monoclonal primary antibody (5 μg/ml, ab55383, Abcam) and a biotinylated antimouse secondary antibody. Briefly, slides sequentially underwent all staining procedure, including deparaffinization, rehydration through graded alcohols, blockade of endogenous peroxidase activity, antigen retrieval, and incubation with antibodies. A slide in which primary antibody was omitted served as a negative control. After incubation with antibodies, slides then continued to undergo staining procedures with horseradish peroxidase, 3,3-diaminobenzidine chromogen solution, and counterstain (hematoxylin). The KIF2A staining intensity of each slide was assessed and scored by two independent pathologists and fell into four categories: 0 (negative), 1 (weakly positive), 2 (moderately positive), and 3 (strongly positive). Moreover, the percentage of positive cells were also scored, with 0 representing for 0–20 %, 1 for 21–50 %, 2 for 51–75 %, and 3 for 76–100 % of positive cells. The final staining score was generated by the product of the percentage and intensity score. Finally, we dichotomized the continuous KIF2A staining scores into low and high expression levels using specific cutoff value, which was determined using the X-tile software program (The Rimm Lab at Yale University; http://www.tissuearray.org/rimmlab) as a statistically significant KIF2A expression score regarding overall survival (OS) [25].
Statistical analysis
KIF2A messenger RNA (mRNA) expression in fresh CRC compared with matching tumor-adjacent tissues was analyzed using the Wilcoxon nonparametric signed-rank test. The strength of the associations of clinicopathologic variables with KIF2A expression was determined using χ 2 tests. The Kaplan–Meier Survival curves were plotted. Univariate and multivariate Cox regression models were adopted to evaluate prognostic significance of variables. The statistical significance level was set at P < 0.05. All statistics were carried out using STATA Version 12.0 (Stata Corporation, College Station, TX, USA).
Results
Evaluation of KIF2A mRNA expression by qPCR
In order to ascertain whether KIF2A mRNA expression differs between CRC and normal tissues, we isolated total RNA from 20 CRC tissues and matched tumor-adjacent tissues. One-step qRT-PCR analysis revealed that KIF2A mRNA expression was significantly higher in cancer tissues than in tumor-adjacent normal tissues (Fig. 1).
Differential expression of KIF2A mRNA in colorectal cancer (CRC) and tumor-adjacent normal tissues. KIF2A mRNA expression levels in CRC (Ca) and tumor-adjacent tissue (N) were examined using One-Step quantitative reverse transcription-polymerase chain reaction (qRT-PCR). Expression of KIF2A mRNA was normalized to the expression levels of GAPDH. KIF2A mRNA was preferentially expressed in CRC tissues over matched tumor-adjacent normal tissues (P = 0.027)
IHC for KIF2A expression in CRC
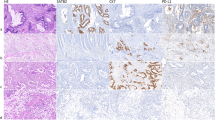
Given the significantly enhanced KIF2A mRNA expression in CRCs compared with tumor-adjacent tissues, we further explored KIF2A expression in the protein level, by performing IHC analysis on TMA comprising CRC and matched tumor-adjacent tissues from 182 patients with CRC. We also examined KIF2A expression in 23 chronic colitis, 43 low-grade intraepithelial neoplasias, and 18 high-grade intraepithelial neoplasias. Images in Fig. 2 exhibited demonstrative IHC staining for KIF2A in CRC tissues. IHC staining indicated that KIF2A protein was mainly confined to the cytoplasm of CRC cells. TMA data analysis using X-tile software program yielded a cutoff value of 6 for KIF2A regarding overall CRC survival. Tissues with KIF2A staining score (the product of the percentage and intensity score) ≤6 or >6 were considered low or high expression, respectively. Overall, only small proportion of chronic colitis (17.39 %, 4/23) and matched tumor-adjacent tissues (12.85 %, 23/179) displayed high KIF2A expression, whereas high expression of this protein was detected in 39.53 % (17/43) of low-grade intraepithelial neoplasia, 77.22 % (13/18) of high-grade intraepithelial neoplasia, or 43.41 % (79/182) of CRCs (P < 0.001) (Table 1). These finding substantiated that the distribution pattern of KIF2A protein in CRC and normal tissues was in line with that of its mRNA expression.
Representative KIF2A immunohistochemical (IHC) staining of tissue microarray cores in the following order: a1, a2 colorectal cancer (case 1) with moderately positive staining; b1, b2 colorectal cancer (case 2) with strongly positive staining; c1, c2 adjacent noncancerous tissues, weakly positive; d1, d2 high-grade intraepithelial neoplasia, strongly positive; e1, e2 low-grade intraepithelial neoplasia, weakly positive; and f1, f2 chronic colitis, negative. Original magnification × 40 (bar = 500 μm) in a1, b1, c1, d1, e1, f1 and × 400 (bar = 50 μm) in a2, b2, c2, d2, e2, f2. Red arrows indicated positive KIF2A staining
Association of KIF2A expression with demographic and clinic pathological parameters of CRC
The associations between KIF2A expression and a number of demographic and clinical pathological variables of CRC patients were evaluated, including gender, age, tumor location, histological type, tumor differentiation, TNM stage, tumor status (T), lymph node metastasis (N), and preoperative CEA level (Table 2). We found that high cytoplasmic KIF2A expression was significantly associated with TNM stage (P = 0.046) and tumor status (T) (P = 0.007). Larger percentage of stage II or III + IV CRCs exhibited high KIF2A expression than that of stage 0-I CRCs. Similarly, the proportion of tumor with high KIF2A expression increased with the degree of lymph node metastasis (25.58 % of CRCs with Tis + T1 + T2 vs. 48.92 % of CRCs with T3, 4b) (Table 2). These results suggested that CRCs with high cytoplasmic KIF2A expression were prone to progress to a more advanced stage than those with low KIF2A expression. Nevertheless, null association was observed between KIF2A expression and the remaining clinical variables.
Survival analysis
Univariate Cox regression analyses demonstrated that KIF2A expression (P < 0.001), tumor differentiation (P = 0.002), tumor TNM stage (P < 0.001), T (P = 0.001), N (P < 0.001), and preoperative CEA levels (P = 0.001) significantly affected 5-year survival of patients with CRC. Multivariate Cox regression analyses further showed that high cytoplasmic KIF2A expression (P = 0.001), poor tumor differentiation (P = 0.001), high preoperative CEA levels (P < 0.001), and advanced tumor TNM stage (P = 0.012) were independent prognostic factors for poor overall survival (Table 3).
In Kaplan–Meier survival analysis, patients with high KIF2A expression had worse survival than those with low KIF2A expression during all time points of the follow-up period (log rank, overall, 5 years, P < 0.001) (Fig. 3). Likewise, tumor differentiation, TNM stage, and preoperative CEA levels also individually influenced survival, with well-differentiated tumors, early TNM stage, and low preoperative CEA levels predicting a favorable prognosis.
Kaplan–Meier survival curves of CRC patients. a Overall survival rate in patients with high KIF2A expression in cancer tissues (1) was significantly decreased in comparison with those with no or low KIF2A expression (0). b Patients with poorly differentiated tumors (2) tended to have significantly lower overall survival rate than that patients with moderately to highly differentiated tumors (1). c Advanced TNM stage (II (2); III + IV (3)) significantly worsened overall survival when compared with early TNM stage (0-I (1)). d Patients with high preoperative CEA level (1) displayed significantly poorer prognosis than those with low preoperative CEA level (0)
Discussion
CRC is one of most frequently diagnosed cancer worldwide. CRC incidence rates continue to ascend in low-risk countries, especially in Czech Republic and Japan [3, 4], partially due to alterations in dietary patterns, a trend toward obesity, and an augmented prevalence of smoking [26, 27]. In the most of high-risk countries (e.g., Canada and Australia), rates are decreasing or remain steady, and the USA is the only exception with significantly decreasing incidence rate mainly because of early detection and management of precancerous lesions [3, 4, 28]. These epidemiology data suggests the high prevalence and the importance of improving early detection, treatment, and prognosis of CRC.
CRC patients are often subject to overtreatment due to failure to distinguish low-risk subgroup from all cancer patients. Clinical factors (e.g., TMN and tumor grade) that a clinician has relied on to predict prognosis for decades are likely necessary, but not sufficient. The mission of “precision medicine,” also known as “personalized medicine,” is to tailor available treatments to the individual patient based on traditional clinical staging systems and pathology in conjunction with molecular diagnostics (e.g., genetic alterations and biomarkers). CRC is a complex and heterogeneous disease; thus far, no single biomarker with high accuracy and robustness is available to identify this subset of patients. Therefore, we performed this study, attempting to enhance the capacity of predicting clinical outcome by identifying new biomarkers.
In the present study, qRT-PCR analysis revealed that expression level of KIF2A mRNA in CRC tissues was significantly higher than in normal tumor-adjacent tissues. Moreover, IHC staining performed on TMA also substantiated increased KIF2A protein product in CRC tissues in comparison with matched tumor-adjacent tissues. We also provided additional evidence of the overexpression of KIF2A protein in cancerous tissues. We further confirmed increased prevalence of high KIF2A expression in cancerous tissues (low- and high-grade intraepithelial neoplasia) as compared to noncancerous tissues (chronic colitis). Survival analysis elucidated that high KIF2A protein expression significantly decreased life span of patients with CRC. Moreover, poor tumor differentiation, advanced TNM stage, and high preoperative CEA levels also independently denoted adverse clinical outcomes of CRC, which was in line with previous studies.
Although few investigations have explored the role of KIF2A in cancers, there are some published findings in accordance with our results. Wang et al. studied the role of KIF2A in carcinogenesis and progression of breast cancer and found that KIF2A expression in breast cancers exceeded its expression in normal adjacent tissues, and KIF2A expression negatively correlated with the survival time of breast cancer patients [20]. Likewise, high expression of KIF2A was more frequently detected in the squamous cell carcinoma of the oral tongue (SCCOT) than in the corresponding adjacent tissues [19]. Moreover, the elevated KIF2A expression correlated with progressive phenotype of SCCOT [19]. Numerous studies indicated that a number of kinesin proteins were deregulated in a variety of human cancers, including KIF2A, MCAK, KIF4A, and KIF14 [14, 29]. However, the underlying mechanism by which overexpression of KIF2A contributes to cancer progression remains unclear.
KIF2A is a microtubule (MT)-based motor protein (a member of the kinesin-13 family) and MT depolymerase, which is known to be involved in mitosis and cytokinesis, particularly for spindle formation [30]. MTs, the essential dynamic structural elements of the cytoskeleton, are well-known for their crucial roles in mitosis, cell migration, cell signaling, and trafficking [31]. Therefore, it is biologically plausible that overexpression of KIF2A may be engaged in carcinogenesis by regulating the status and function of MTs. Ganem et al. found that KIF2A-knockdown cells were not able to progress through the cell cycle, and these cells misguidedly yielded dysfunctional monopolar spindles, instead of normal bipolar spindles. Consequently, chromosome mis-segregation occurred in cells [17]. The KIF2A deficiency-induced monopolar spindle formation and cell proliferation inhibition were also verified in different human cell lines and Xenopus eggs [18, 30]. In terms of cancer cells, KIF2A silencing significantly suppressed proliferation of breast cancer cell line MDA-MB-231 cells [20]. The same study also indicated that cancer cells treated with KIF2A-siRNA exhibited significantly reduced migration and invasion capacity as compared to control cells [20]. KIF2A silencing was also shown to impair migratory ability of tongue squamous cell carcinoma cell line Tca8113 cells [19]. These findings suggested that KIF2A might regulate some activities of cells. Nonetheless, thus far, there is no direct in vitro evidence of how overexpression of KIF2A influences cancer cell behaviors.
Interestingly, it was recently reported that siRNA-mediated KIF2A silencing inhibited phosphatidylinositol-3-kinase (PI3K)/AKT pathway in Tca8113 cells and led to cell apoptosis [21]. PI3K is a lipid kinase. Once stimulated, PI3K sequentially activates protein kinase B (Akt) as well as downstream signaling cascades to regulate multiple important cellular events, such as proliferation, survival, apoptosis, and migration [32, 33]. Excessive activity of PI3K/AKT signaling pathway has been implicated in carcinogenesis of a variety of human cancers [34]. Collectively, these findings suggested that KIF2A might promote tumor growth and invasion partially through stimulating PI3K/AKT signaling pathway. In spite of the interesting findings, limitations of this study were needed to be addressed. We lacked in vitro data to verify our results and did examine the function of KIF2A in the CRC cell lines. The potential mechanisms underlying the oncogenic effects of KIF2A in CRC are also needed to be clarified in the future.
In summary, the current study demonstrated that KIF2A was significantly more highly expressed in cancer tissue than cancer-adjacent normal tissues. High KIF2A expression was significantly associated with advanced CRCs and might serve as an unfavorable prognostic biomarker for CRC. In the future, larger well-designed studies are warranted to verify the prognostic and therapeutic value of KIF2A in CRC.
References
Haggar FA, Boushey RP. Colorectal cancer epidemiology: incidence, mortality, survival, and risk factors. Clin Colon Rectal Surg. 2009;22:191–7.
Bosetti C, Bertuccio P, Malvezzi M, Levi F, Chatenoud L, Negri E, et al. Cancer mortality in europe, 2005–2009, and an overview of trends since 1980. Ann Oncol : official journal of the European Society for Medical Oncology / ESMO. 2013;24:2657–71.
Center MM, Jemal A, Smith RA, Ward E. Worldwide variations in colorectal cancer. CA Cancer J Clin. 2009;59:366–78.
Center MM, Jemal A, Ward E. International trends in colorectal cancer incidence rates. Cancer Epidemiol Biomarkers Prev : Publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2009;18:1688–94.
Sideris M, Papagrigoriadis S. Molecular biomarkers and classification models in the evaluation of the prognosis of colorectal cancer. Anticancer Res. 2014;34:2061–8.
Lee JK, Chan AT. Molecular prognostic and predictive markers in colorectal cancer: current status. Curr Colorectal Cancer Rep. 2011;7:136–44.
Markovic S, Antic J, Dimitrijevic I, Zogovic B, Bojic D, Svorcan P, et al. Microsatellite instability affecting the t17 repeats in intron 8 of hsp110, as well as five mononucleotide repeats in patients with colorectal carcinoma. Biomark Med. 2013;7:613–21.
Domingo E, Ramamoorthy R, Oukrif D, Rosmarin D, Presz M, Wang H, et al. Use of multivariate analysis to suggest a new molecular classification of colorectal cancer. J Pathol. 2013;229:441–8.
Eklof V, Wikberg ML, Edin S, Dahlin AM, Jonsson BA, Oberg A, et al. The prognostic role of kras, braf, pik3ca and pten in colorectal cancer. Br J Cancer. 2013;108:2153–63.
Lochhead P, Kuchiba A, Imamura Y, Liao X, Yamauchi M, Nishihara R, et al. Microsatellite instability and braf mutation testing in colorectal cancer prognostication. J Natl Cancer Inst. 2013;105:1151–6.
Li HY, Zhang Y, Cai JH, Bian HL. Microrna-451 inhibits growth of human colorectal carcinoma cells via downregulation of pi3k/akt pathway. Asian Pac J Cancer Prev : APJCP. 2013;14:3631–4.
Lou X, Qi X, Zhang Y, Long H, Yang J. Decreased expression of microrna-625 is associated with tumor metastasis and poor prognosis in patients with colorectal cancer. J Surg Oncol. 2013;108:230–5.
Yang IP, Tsai HL, Huang CW, Huang MY, Hou MF, Juo SH, et al. The functional significance of microrna-29c in patients with colorectal cancer: a potential circulating biomarker for predicting early relapse. PLoS One. 2013;8:e66842.
Ishikawa K, Kamohara Y, Tanaka F, Haraguchi N, Mimori K, Inoue H, et al. Mitotic centromere-associated kinesin is a novel marker for prognosis and lymph node metastasis in colorectal cancer. Br J Cancer. 2008;98:1824–9.
Kline-Smith SL, Walczak CE. The microtubule-destabilizing kinesin xkcm1 regulates microtubule dynamic instability in cells. Mol Biol Cell. 2002;13:2718–31.
Manning AL, Ganem NJ, Bakhoum SF, Wagenbach M, Wordeman L, Compton DA. The kinesin-13 proteins kif2a, kif2b, and kif2c/mcak have distinct roles during mitosis in human cells. Mol Biol Cell. 2007;18:2970–9.
Ganem NJ, Compton DA. The kini kinesin kif2a is required for bipolar spindle assembly through a functional relationship with mcak. J Cell Biol. 2004;166:473–8.
Gaetz J, Kapoor TM. Dynein/dynactin regulate metaphase spindle length by targeting depolymerizing activities to spindle poles. J Cell Biol. 2004;166:465–71.
Wang CQ, Qu X, Zhang XY, Zhou CJ, Liu GX, Dong ZQ, et al. Overexpression of kif2a promotes the progression and metastasis of squamous cell carcinoma of the oral tongue. Oral Oncol. 2010;46:65–9.
Wang J, Ma S, Ma R, Qu X, Liu W, Lv C, et al. Kif2a silencing inhibits the proliferation and migration of breast cancer cells and correlates with unfavorable prognosis in breast cancer. BMC Cancer. 2014;14:461.
Wang K, Lin C, Wang C, Shao Q, Gao W, Song B, et al. Silencing kif2a induces apoptosis in squamous cell carcinoma of the oral tongue through inhibition of the pi3k/akt signaling pathway. Molec Med Rep. 2014;9:273–8.
Schimizzi GV, Currie JD, Rogers SL. Expression levels of a kinesin-13 microtubule depolymerase modulates the effectiveness of anti-microtubule agents. PLoS One. 2010;5:e11381.
Han L, Jiang B, Wu H, Wang X, Tang X, Huang J, et al. High expression of cxcr2 is associated with tumorigenesis, progression, and prognosis of laryngeal squamous cell carcinoma. Med Oncol. 2012;29:2466–72.
Sun R, Wang X, Zhu H, Mei H, Wang W, Zhang S, et al. Prognostic value of lamp3 and tp53 overexpression in benign and malignant gastrointestinal tissues. Oncotarget. 2014;5:12398–409.
Huang J, Zhang J, Li H, Lu Z, Shan W, Mercado-Uribe I, et al. Vcam1 expression correlated with tumorigenesis and poor prognosis in high grade serous ovarian cancer. Am J Transl Res. 2013;5:336–46.
de Kok IM, Wong CS, Chia KS, Sim X, Tan CS, Kiemeney LA, et al. Gender differences in the trend of colorectal cancer incidence in singapore, 1968–2002. Int J Color Dis. 2008;23:461–7.
Garcia-Alvarez A, Serra-Majem L, Ribas-Barba L, Castell C, Foz M, Uauy R, et al. Obesity and overweight trends in Catalonia, Spain (1992–2003): gender and socio-economic determinants. Public Health Nutr. 2007;10:1368–78.
Austin H, Henley SJ, King J, Richardson LC, Eheman C. Changes in colorectal cancer incidence rates in young and older adults in the united states: what does it tell us about screening. Cancer Causes Control : CCC. 2014;25:191–201.
Rath O, Kozielski F. Kinesins and cancer. Nat Rev Cancer. 2012;12:527–39.
Zhu C, Zhao J, Bibikova M, Leverson JD, Bossy-Wetzel E, Fan JB, et al. Functional analysis of human microtubule-based motor proteins, the kinesins and dyneins, in mitosis/cytokinesis using rna interference. Mol Biol Cell. 2005;16:3187–99.
Honore S, Pasquier E, Braguer D. Understanding microtubule dynamics for improved cancer therapy. Cell Molec Life Sci : CMLS. 2005;62:3039–56.
McCubrey JA, Steelman LS, Chappell WH, Abrams SL, Franklin RA, Montalto G, et al. Ras/raf/mek/erk and pi3k/pten/akt/mtor cascade inhibitors: how mutations can result in therapy resistance and how to overcome resistance. Oncotarget. 2012;3:1068–111.
Chappell WH, Steelman LS, Long JM, Kempf RC, Abrams SL, Franklin RA, et al. Ras/raf/mek/erk and pi3k/pten/akt/mtor inhibitors: rationale and importance to inhibiting these pathways in human health. Oncotarget. 2011;2:135–64.
Bader AG, Kang S, Zhao L, Vogt PK. Oncogenic pi3k deregulates transcription and translation. Nat Rev Cancer. 2005;5:921–9.
Acknowledgments
This study was supported by the Technological Innovation and Demonstration of Social Undertakings Projects (HS2014049) of Nantong, Jiangsu, China, and the Translational Medicine Research (TDFzh2014001) from the Affiliated Hospital of Nantong University, Jiangsu, China.
Conflicts of interest
None.
Author information
Authors and Affiliations
Corresponding author
Additional information
Xiangjun Fan and Xudong Wang contributed equally to this work.
Rights and permissions
About this article
Cite this article
Fan, X., Wang, X., Zhu, H. et al. KIF2A overexpression and its association with clinicopathologic characteristics and unfavorable prognosis in colorectal cancer. Tumor Biol. 36, 8895–8902 (2015). https://doi.org/10.1007/s13277-015-3603-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-015-3603-z