Abstract
MicroRNA 181a (miR-181a) was found dysregulated in a variety of human cancers and significantly associated with clinical outcome of cancer patients. However, the direct role of miR-181a has not yet been characterized in osteosarcoma progression. This study was aimed at investigating the effects of miR-181a on osteosarcoma cell biological behavior. First, the expression of miR-181a in osteosarcoma cell lines (MG63, HOS, SaOS-2, and U2OS) and a human osteoblastic cell line (hFOB1.19) was detected by qRT-PCR. Results showed that miR-181a was overexpressed in osteosarcoma cell lines compared to human osteoblastic cell line (hFOB1.19). To investigate the effects of miR-181a on proliferation, apoptosis, and invasion of osteosarcoma cells, we generated human osteosarcoma MG63 cells in which miR-181a was either overexpressed or depleted. The MG63 cell viability, cycle, apoptosis, and invasive ability were analyzed by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide staining, propidium iodide (PI) staining, Annexin V-FITC/PI double staining, and Transwell invasion experiment, respectively. The results showed that MG63 cell viability, proliferation, and invasive abilities were suppressed, and the apoptosis was enhanced in the group with underexpression of miR-181a. The viability, proliferation, and invasive abilities were improved, and the apoptosis was inhibited in the group with overexpression of miR-181a. The results from Western blotting indicated that miR-181a might be associated with the up-regulation of bcl-2 and matrix metalloproteinase 9 and the down-regulation of tissue inhibitor of metalloproteinases-3 and p21 in MG63 cells. Taken together, our results suggested that miR-181a might facilitate proliferation and invasion and suppress apoptosis of osteosarcoma cells, which might be a potential target for the treatment of osteosarcoma.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Osteosarcoma (OS) is the most common primary bone malignancy in the pediatric age group [1, 2]. Osteosarcoma is a tumor with a high propensity for local invasion, early metastasis, and relapse; about 20 % of patients present with lung metastases at initial diagnosis [3]. Treating metastatic osteosarcoma remains a challenge in oncology. In recent years, despite the use of multimodal therapies, including surgery and radio- and chemotherapy, the prognosis for patients with osteosarcoma is generally poor [2, 4]. The prognosis is unfavorable due to lack of effective treatment methods [5]. Progression to metastatic disease is the main cause of treatment failures. While, the molecular mechanisms behind osteosarcoma development and metastasis remain elusive. Therefore, broadening our understanding on the pathogenesis and biology of metastatic osteosarcoma tumors is a key element in improving treatment results, i.e., identifying potential therapeutic targets [2].
MicroRNAs (miRNAs) are a novel class of small non-coding RNAs which have eluded researchers for decades stealthily regulating many of the major biological processes in eukaryotic cells by regulating their target genes posttranscriptionally. Their target genes include numerous regulators of cell proliferation, differentiation, apoptosis, development, metabolism, and immunity [6, 7]. MiRNAs are precisely regulated in a tissue- and developmental-specific manner, but dysregulated in many human diseases, in particular cancers [8]. Emerging evidence has shown that miRNAs participate in human carcinogenesis as tumor suppressors or oncogenes and have prognostic value for patients with cancers [7]. Aberrant miRNA expression contributes to tumorigenesis and cancer progression [9]. Therefore, miRNAs are implicated in the initiation and progression of cancer, tissue invasion, and metastasis formation as well [6].
MicroRNA-181a (miR-181a) is a multifaceted miRNA that has been implicated in many cellular processes such as cell fate determination and cellular invasion [10]. In recent years, the miR-181a family was found dysregulated in a variety of human cancers and significantly associated with clinical outcome of cancerous patients [7]. Ciafre and his colleagues firstly reported that expression of miR-181a/b displayed a significantly down-regulation in primary glioblastomas and human glioblastoma cell lines compared to normal brain tissue [11]. Thereafter, as in glioblastoma, significant down-regulation of miR-181a was also observed in squamous lung cell carcinoma (SQCC) and non-small cell lung cancer (NSCLC) [12]. Shi and his colleagues showed that miR-181a functioned as tumor suppressors which triggered growth inhibition, induced apoptosis, and inhibited invasion in glioma cells. Down-regulation of miR-181a might be a critical factor that contributes to malignant appearance in human gliomas [13].
However, miR-181a was significantly overexpressed in MCF-7 breast cancer cells, hepatocellular carcinoma (HCC) cells, and human gastric cancer tissues [14–17]. Meng and his colleagues demonstrated that miR-181 was up-regulated in hepatocellular cancer stem cells (HSCs), and silencing of miR-181 led to a reduction in HSC motility and invasion [17]. The up-regulation of miR-181 was observed when oral squamous cell carcinoma (OSCC) progressed from leukoplakia, dysplasia, to invasive carcinoma, and overexpression of miR-181 was correlated with lymph node metastasis, vascular invasion, and a poor survival [18]. MiR-181a is overexpressed in human gastric cancer tissues. Ectopic expression of miR-181a mimic promoted the proliferation, colony formation, migration, and invasion and inhibited the apoptosis of SGC-7901 gastric cancer cells [15].
Therefore, it is still unclear that miR-181a acts as a tumor suppressor or as an oncogene. Current data indicate that the dysregulated of miR-181a is associated with a variety of human cancers. Jones and Hu reported that miR-181a is overexpressed in osteosarcoma tissue and cells [19, 20]. However, the effects of miR-181a on the biological behavior of osteosarcoma cells are rarely reported. This study intended to examine miR-181a expression in osteosarcoma cell lines (MG63, HOS, SaOS-2, and U2OS) and a human osteoblastic cell line (hFOB1.19) by qRT-PCR analysis and to observe changes in the cell viability, cycle, apoptosis, and invasive ability of MG-63 cells following the increased and reduced expression of miR-181a in the human osteosarcoma MG-63 cell line. These data might provide scientific information for prognosis prediction and targeted therapy for osteosarcoma.
Materials and methods
Reagents
All cell culture components were purchased from Gibco-BRL (Gaithersburg, MD). MG63, HOS, SaOS-2, U2OS, and a human osteoblastic cell line hFOB1.19 were purchased from American Type Culture Collection (Rockville, MD). LipofectamineTM 2000 was purchased from Invitrogen Life Technologies (Carlsbad, CA). MirVana miRNA Isolation Kit was from Ambion (Austin, TX). Taq Man MicroRNA Reverse Transcription Kit, TaqMan MicroRNA Assay Kit, and TaqMan Universal PCR Master Mix were purchased from Applied Biosystems (Foster City, CA). MiR-181a mimic and inhibitor were purchased from RiboBio Co. (Guangzhou, China). Protein extraction buffer, Annexin V-FITC, PI, crystal violet, and RNAse A were from Sigma (St. Louis, MO). Polyvinylidenedifluoride (PVDF) membranes were from Millipore Inc. (Bedford, MA). The electrogenerated chemiluminescence (ECL) chemiluminescence kit was from Pierce (Rockford, IL). Matrigel was from Collaborative Research, Inc. (Bedford, MA). The Transwell invasion chamber was from Costar Corp (Cambridge, MA). The antibodies used in this study include: rabbit anti-tissue inhibitor of metalloproteinase 3 (TIMP3), bcl-2 and p21 polyclonal antibodies (Abcam, Cambridge, MA), rabbit anti-human matrix metalloproteinase 9 (MMP9) polyclonal antibodies (Abnova Corp., Taipei, Taiwan), rabbit anti β-actin polyclonal antibody (Abbiotec Corp., San Diego, CA), and horseradish peroxidase-conjugated goat anti-rabbit (Invitrogen, Carlsbad, CA).
Methods
Cell culture
Osteosarcoma cell lines (MG63, HOS, SaOS-2, and U2OS) were cultured in RPMI 1640 supplemented with 10 % fetal bovine serum (FBS) and 1 % penicillin/streptomycin at 37 °C. hFOB1.19 human osteoblasts were grown in DMEM/Ham’s F-12 containing 10 % FBS and Geneticin (400 μg/mL) at 34 °C in a humidified 5 % CO2 incubator.
miRNA transfection
To enforce miR-181a expression or inhibit miR-181a expression in MG63 cells, MG63 cells were transfected with miR-181a mimics and inhibitors, which served as the overexpression group and the underexpression group, respectively. MG63 cells without any treatment were used as blank group. One day before transfection, cells at about 40 to 60 % confluency were changed to the antibiotic-free media. On the next day, the cells were transfected with different 50 nM miR-181a mimics and inhibitors using Lipofectamine™ 2000 reagent according to the manufacturer’s protocol.
Real-time RT-PCR for miRNAs
Total RNA was isolated from MG63 cells using the mirVana miRNA Isolation kit according to the manufacturer’s protocol. MiRNA qRT-PCR was performed using the TaqMan MicroRNA Reverse Transcription Kit, TaqMan Universal PCR Master Mix, and TaqMan MicroRNA Assay primers for human miR-181a. All reactions were analyzed using StepOne Real-Time PCR System. The levels of miRNA were normalized to U6 controls. For miRNA expression quantification, each reverse transcription (RT) reaction consisted of 50 ng of purified total RNA, 1× RT buffer, dNTPs (each at 0.375 mM), 5 U μl−1 MultiScribe reverse transcriptase, 50 nM stem-loop RT primer, and 0.38 U μl−1 RNase inhibitor. RT reactions were incubated at 16 °C for 30 min, 42 °C for 30 min, and 85 °C for 5 min. Real-time PCR reactions were performed in quadruplicate in 20 μl volumes. The real-time reaction mix consisted of 1.33 μl RT product, 1 μl of 20× TaqMan microRNA assay mix, and 10 μl TaqMan 2× Universal PCR Master Mix. Quantitative miRNA expression data were acquired and analyzed using an Applied Biosystems 7500 real-time PCR system. The cycle threshold (Ct) values, corresponding to the PCR cycle number at which fluorescence emission reaches a threshold above baseline emission, were determined and the relative miRNA expression was calculated using the 2−ΔΔCt method [21].
Western blotting
Total protein was extracted from MG63 cells and quantified using a BCA assay kit. Protein samples (40 μg/sample) were separated by SDS-PAGE and transferred to a PVDF membrane. The membrane was blocked for 1 h in TBS solution with 5 % skimmed milk at room temperature and probed with primary antibody at 4 °C overnight. Then, the membrane was washed in TBST for 3 × 5 min and probed with corresponding secondary antibody for 2 h at room temperature. Then, autoradiography was conducted with ECL chemiluminescence reagents. The relative expression of the target protein was valuated with the gray value ratio of target protein content to β-actin (target protein/β-actin) content.
Cell viability assay
To investigate the effect of miR-181a on proliferation of OS cells, MG63 cells, in which miR-181a was either overexpressed or inhibited, were evaluated by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. MG63 cells were seeded into 96-well plates and allowed to adhere overnight. Ten microliters of MTT (5 mg mL−1) was added and incubated with MG63 cells for another 4 h. Then, media were removed and 100 μL DMSO was added to all wells and mixed thoroughly to dissolve the dark blue crystals. The absorbance at 570 nm was measured on a plate reader and a 690-nm measurement was used as a reference. The relative cell proliferation (percent) is calculated by the equation as described in previous study [22] and the experiment was repeated three times.
Cell cycle assay
MG63 cells were cultured in serum-free medium for 1 day to complete synchronization and changed to complete medium for another day. Then, the cells were detached by exposure to trypsin, washed, and fixed in 70 % cold ethanol overnight at −20 °C. The next day, fixed cells were washed and then treated with 1 mg/mL RNAse (DNAse-free) for 30 min at 37 °C. Propidium iodine was added to the solution at a final concentration of 100 μg/mL and incubated with MG63 cells at room temperature for 30 min. The cell cycle was evaluated by flow cytometry and the experiment was repeated three times.
Apoptosis assay
Briefly, MG63 cells were harvested, washed, and resuspended in Annexin V binding buffer. Then, the MG63 cells were stained with Annexin V-FITC in the dark at room temperature for 10 min, centrifuged, and gently resuspended in Annexin V binding buffer. Finally, 10 μL propidium iodide staining solution was added and gently mixed, and MG63 cells were kept on ice in the dark and immediately subjected to flow cytometry analysis. The experiment was in repeated thrice.
Invasion assay
The Transwell invasion chamber was washed with serum-free medium, and then, 20 μL Matrigel (1 mg/mL) was added to evenly cover the surface of the polycarbonate membrane (8-μm pore size) to create the Matrigel membrane. The chamber was divided into upper and lower chambers. For invasion assays, MG63 cells (4 × 105) were serum starved overnight and seeded in starvation medium on the top chamber. The bottom chamber contained 10 % FBS in RPMI 1640 medium which acted as chemoattractant. After 48 h incubation, cells from the top chamber were removed by cotton swab and invading cells were fixed with 4 % formaldehyde for 15 min and then stained with a crystal violet solution for 10 min. Images of the invading cells were photographed using an inverted microscope and total cell numbers were counted and quantified by ImageJ software. The results are presented as the mean ± SD, and the experiment was repeated three times for each group.
Statistical analysis
The SPSS 17.0 software was used to analyze the related data with one-way ANOVA. The results were considered to be statistically significant if p < 0.05.
Results
MiR-181a is overexpressed in osteosarcoma cell lines
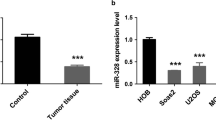
To address the expression of miR-181a in osteosarcoma cells, four osteosarcoma cell lines (MG63, HOS, SaOS-2, and U2OS) and a human osteoblastic cell line hFOB1.19 were used to detect the expression of miR-181a by qRT-PCR. The expression of miR-181a was significantly up-regulated in osteosarcoma cell lines compared to that in normal human osteoblastic cell line hFOB1.19 (p < 0.05), as shown in Fig. 1.
Among these osteosarcoma cell lines, MG63 cells were used to study further. The result from qRT-PCR analysis indicated that miR-181a displayed significant up-regulation in overexpression group and significant down-regulation in underexpression group compared to blank group (p < 0.01). These data demonstrated that we effectively enforce or inhibit miR-181a expression in MG63 cells (Fig. 1).
MiR-181a improves MG63 cell viability
The results from MTT assay suggested that the viability of MG63 cells from overexpression group was significantly higher than that in blank group, and that the viability of MG63 cells from underexpression group was significantly lower than that in the blank group (p < 0.05) (Fig. 2). These results indicated that miR-181a might be related to the improvement of MG63 cell viability.
MiR-181a accelerates MG63 cell cycle progression
The results from cell cycle assay indicated that overexpression group had less MG63 cells in G0/G1 phase (p < 0.05), and underexpression group had more MG63 cells in G0/G1 phase than the blank group (p < 0.05). Furthermore, overexpression group had more MG63 cells in S and G2M phases (p < 0.05), and underexpression group had less MG63 cells in S and G2M phases than blank group (p < 0.05). Proliferation index (PI) = (S + G2M) / (G0G1 + S + G2M). The proliferation index was higher in the overexpression group (p < 0.05) and was lower in the underexpression group than that in the blank group (p < 0.05) (shown in Fig. 3). Based on the above results, we could conclude that miR-181a might promote MG63 cell proliferation.
MiR-181a negatively regulates MG63 cell apoptosis
The results from flow cytometry analysis demonstrated that the number of apoptotic MG63 cells was significantly lower in overexpression group and significantly higher in underexpression group than that in blank group (p < 0.05) (Fig. 4). These data suggested that miR-181a might negatively regulate MG63 cell apoptosis
MiR-181a improves MG63 cell invasion
The results from Transwell invasion chamber experiments showed that the number of invading MG63 cells was significantly higher in overexpression group and was significantly lower in knockdown group than that in blank group (p < 0.05) (Fig. 5). These data indicated that miR-181a might improve MG63 cell invasiveness.
The effects of miR-181a on the expression of TIMP3, p21, bcl-2, and MMP9 in MG63 cells
The viability, proliferation, and invasive abilities were improved, and the apoptosis was inhibited in MG63 cells with overexpression of miR-181a. Therefore, these proteins, p21 protein (a cyclin-dependent kinase inhibitor), bcl-2 (proto-oncogene), MMP9 (associated with tumor invasion and metastasis), and Western blotting analysis indicated that the expression of bcl-2 and MMP9 displayed an up-regulation in overexpression group and a down-regulation in knockdown group than those in the blank group (p < 0.05). However, TIMP3 and p21 were at a lower level in overexpression group and at a higher level in knockdown group than those in blank group (p < 0.05) (Fig. 6). These results indicated that miR-181a might be associated with the up-regulation of bcl-2 and MMP9 and the down-regulation of TIMP3 and p21 in MG63 cells.
The effects of miR-181a on the expression of bcl-2, MMP9, p21, and TIMP3 in MG63 cells. I bcl-2, MMP9, p21, and TIMP3 protein expression in MG63 cells. II Relative expression of bcl-2, MMP9, p21, and TIMP3 in MG63 cells. *p < 0.05 compared to blank group. These data were analyzed by one-way ANOVA. All experiments were repeated three times with three replicates each
Discussion
miRNAs have attracted attention because of their key regulatory functions in many biological events, including differentiation and tumorigenesis. In recent years, the dysregulated of miR-181a was associated with a variety of human cancers [7]. MiR-181a displayed a significant down-regulation in primary glioblastomas, SQCC, and NSCLC [11, 12]. However, miR-181a was significantly overexpressed in MCF-7 breast cancer cells, human gastric cancer tissues, and HCC cells [14–17]. Therefore, miR-181a displayed perplexing function in tumorigenesis and development.
In this study, we found that miR-181a displayed higher expression levels in osteosarcoma cell lines (MG63, HOS, SaOS-2, and U2OS) compared to noncancerous osteoblastic cell line hFOB1.19. The expression model of miR-181a in osteosarcoma cell lines in this study was consistent with that in osteosarcoma tissue, which showed that miR-181a significantly up-regulated in osteosarcoma tissue [19]. Zhang and his colleagues demonstrated that ectopic expression of miR-181a mimic could promote the proliferation and colony formation of gastric cancer cells [15]. Depletion of miR-181a inhibited tumor growth of HCC cells in nude mice [23]. Therefore, we speculated that miR-181a might positively regulate tumor cell proliferation, migration, and invasion. In order to explore the role of miR-181a in osteosarcoma cells, the expression of miR-181a was enforced or inhibited in MG63 cells. The results of MTT assay indicated that MG63 cell viability was significantly lower in overexpression group and significantly higher in underexpression group than that in blank group, which suggested that miR-181a could improve MG63 cell viability. The results of cell cycle assay indicated that miR-181a could improve MG63 cell proliferation, which was possibly due to an increase in growth-promoting factors or a reduction in growth inhibitory factors in the downstream of the miR-181a target genes. The p21 protein, a cyclin-dependent kinase inhibitor, is a key regulator of the cell growth. p21 promotes cell cycle arrest primarily at the G1/S transition of the cell cycle [24]. Therefore, p21 was examined in our study. The results showed that the expression of p21 was suppressed in overexpression group and enhanced in underexpression group, which suggested that the overexpression of miR-181a might be a major event in cancer pathogenesis, in part due to loss of its ability to up-regulate p21, leading to a failure to induce cell cycle arrest. However, further study is required to elucidate the exact mechanism. Our results also showed that miR-181a could inhibit MG63 cell apoptosis, which was consistent with that in gastric cancer cell lines, which showed that ectopic expression of miR-181a could inhibit the apoptosis of SGC-7901 gastric cancer cells [15]. Bcl-2 is unique among proto-oncogenes, being localized to mitochondria and extending cell survival by blocking programmed cell death [25]. Therefore, we examined the expression of bcl-2 and found that bcl-2 was up-regulated in the overexpression group, which suggested that the overexpression of miR-181a might promote the up-regulation of bcl-2 in MG63 cells. However, the exact mechanisms still require further elucidation. The above results implied that miR-181a might promote the growth and proliferation of MG63 cells and suppress the apoptosis of MG63 cells.
Recent studies have shown that miR-181a plays an important role in the process of tumor metastasis and invasion [15, 18]. MiR-181a may enhance lymph node metastasis through regulating migration, which could potentially be exploited as a putative biomarker for patients with OSCC [18]. Therefore, we investigated the effect of miR-181a on the invasion ability of MG63 cells in vitro. In this study, we found that the number of invading MG63 cells was significantly higher in overexpression group and significantly lower in knockdown group than that in blank group, which suggested that miR-181a might positively regulate MG63 cell invasion. Ectopic expression and depletion of miR-181a demonstrated that miR-181a could enhance MMP9 activity and promote growth, clonogenic survival, migration, and invasion of HCC cells that could be reversed by modulating TIMP3 level, which was a tumor suppressor and a validated miR-181a target [23]. Therefore, we examined TIMP3 and MMP9 in this study. The results showed that TIMP3 displayed lower level expression in overexpression group and higher level expression in underexpression group, while MMP9 displayed higher level expression in overexpression group and lower level expression in underexpression group. Based on these data, we supposed that miR-181a contributed to tumor cell invasion possibly by abrogating TIMP3-induced inhibition of MMP9 in MG63 cells.
In view of the above, we inferred that miR-181a acted as an oncogene and might be involved in the improvement of growth, proliferation, and invasion of osteosarcoma cells, as well as suppression of apoptosis of osteosarcoma cells, which might be a potential target for the treatment of osteosarcoma. However, further research is still needed to provide a good understanding of the function and mechanism of miR-181a in osteosarcoma.
References
Endo-Munoz L, Evdokiou A, Saunders NA. The role of osteoclasts and tumour-associated macrophages in osteosarcoma metastasis. Biochim Biophys Acta. 2012;1826(2):434–42.
Poletajew S, Fus L, Wasiutynski A. Current concepts on pathogenesis and biology of metastatic osteosarcoma tumors. Ortop Traumatol Rehabil. 2011;13(6):537–45.
Harting MT, Blakely ML. Management of osteosarcoma pulmonary metastases. Semin Pediatr Surg. 2006;15(1):25–9.
Yao C, Wei JJ, Wang ZY, Ding HM, Li D, Yan SC, et al. Perifosine induces cell apoptosis in human osteosarcoma cells: new implication for osteosarcoma therapy? Cell Biochem Biophys. 2013;65(2):217–27.
Cui J, Wang W, Li Z, Zhang Z, Wu B, Zeng L. Epigenetic changes in osteosarcoma. Bull Cancer. 2011;98(7):E62–68.
Gaal Z, Olah E. MicroRNA-s and their role in malignant hematologic diseases. Orv Hetil. 2012;153(52):2051–9.
Lin S, Pan L, Guo S, Wu J, Jin L, Wang JC, et al. Prognostic role of microRNA-181a/b in hematological malignancies: a meta-analysis. PLoS One. 2013;8(3):e59532.
Wang Z, Yao H, Lin S, Zhu X, Shen Z, Lu G, et al. Transcriptional and epigenetic regulation of human microRNAs. Cancer Lett. 2013;331(1):1–10.
Baer C, Claus R, Plass C. Genome-wide epigenetic regulation of miRNAs in cancer. Cancer Res. 2013;73(2):473–7.
Neel JC, Lebrun JJ. Activin and TGFbeta regulate expression of the microRNA-181 family to promote cell migration and invasion in breast cancer cells. Cell Signal. 2013;25(7):1556–66.
Ciafre SA, Galardi S, Mangiola A, Ferracin M, Liu CG, Sabatino G, et al. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun. 2005;334(4):1351–8.
Gao W, Yu Y, Cao H, Shen H, Li X, Pan S, et al. Deregulated expression of miR-21, miR-143 and miR-181a in non small cell lung cancer is related to clinicopathologic characteristics or patient prognosis. Biomed Pharmacother. 2010;64(6):399–408.
Shi L, Cheng Z, Zhang J, Li R, Zhao P, Fu Z, et al. hsa-mir-181a and hsa-mir-181b function as tumor suppressors in human glioma cells. Brain Res. 2008;1236:185–93.
Miller TE, Ghoshal K, Ramaswamy B, Roy S, Datta J, Shapiro CL, et al. MicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27Kip1. J Biol Chem. 2008;283(44):29897–903.
Zhang X, Nie Y, Du Y, Cao J, Shen B, Li Y. MicroRNA-181a promotes gastric cancer by negatively regulating tumor suppressor KLF6. Tumour Biol. 2012;33(5):1589–97.
Ji J, Yamashita T, Budhu A, Forgues M, Jia HL, Li C, et al. Identification of microRNA-181 by genome-wide screening as a critical player in EpCAM-positive hepatic cancer stem cells. Hepatology. 2009;50(2):472–80.
Meng F, Glaser SS, Francis H, DeMorrow S, Han Y, Passarini JD, et al. Functional analysis of microRNAs in human hepatocellular cancer stem cells. J Cell Mol Med. 2012;16(1):160–73.
Yang CC, Hung PS, Wang PW, Liu CJ, Chu TH, Cheng HW, et al. miR-181 as a putative biomarker for lymph-node metastasis of oral squamous cell carcinoma. J Oral Pathol Med. 2011;40(5):397–404.
Jones KB, Salah Z, Del Mare S, Galasso M, Gaudio E, Nuovo GJ, et al. miRNA signatures associate with pathogenesis and progression of osteosarcoma. Cancer Res. 2012;72(7):1865–77.
Hu H, Zhang Y, Cai XH, Huang JF, Cai L. Changes in microRNA expression in the MG-63 osteosarcoma cell line compared with osteoblasts. Oncol Lett. 2012;4(5):1037–42.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−delta delta C(T)) method. Methods. 2001;25(4):402–8.
Wang HJ, Ruan HJ, He XJ, Ma YY, Jiang XT, Xia YJ, et al. MicroRNA-101 is down-regulated in gastric cancer and involved in cell migration and invasion. Eur J Cancer. 2010;46(12):2295–303.
Wang B, Hsu SH, Majumder S, Kutay H, Huang W, Jacob ST, et al. TGFbeta-mediated upregulation of hepatic miR-181b promotes hepatocarcinogenesis by targeting TIMP3. Oncogene. 2010;29(12):1787–97.
Narla G, Friedman SL, Martignetti JA. Kruppel cripples prostate cancer: KLF6 progress and prospects. Am J Pathol. 2003;162(4):1047–52.
Hockenbery D, Nunez G, Milliman C, Schreiber RD, Korsmeyer SJ. Bcl-2 is an inner mitochondrial membrane protein that blocks programmed cell death. Nature. 1990;348(6299):334–6.
Acknowledgments
This study was supported by The “Six Talent Peaks Program” of Jiangsu Province of China.
Conflicts of interest
None
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Jianwei, Z., Fan, L., Xiancheng, L. et al. MicroRNA 181a improves proliferation and invasion, suppresses apoptosis of osteosarcoma cell. Tumor Biol. 34, 3331–3337 (2013). https://doi.org/10.1007/s13277-013-0902-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-013-0902-0