Abstract
MicroRNAs have emerged as crucial regulators of tumorigenesis. However, it remains unknown whether miR-181a is involved in the pathogenesis of gastric cancer. In this study, we found that miR-181a is overexpressed in human gastric cancer tissues. Ectopic expression of miR-181a mimic promoted the proliferation, colony formation, migration, and invasion and inhibited the apoptosis of SGC-7901 gastric cancer cells, whereas ectopic expression of miR-181a inhibitor inhibited the malignant phenotypes of SGC-7901 cells. Site-directed mutagenesis and luciferase reporter assay demonstrated that miR-181a repressed KLF6 expression by targeting its 3′-UTR. Western blot analysis further showed that KLF6 protein was significantly decreased or increased when miR-181a mimic or inhibitor was transfected into SGC-7901 cells, respectively. In summary, these data suggest that KLF6 gene is a direct target of miR-181a and miR-181a functions as an oncomir in gastric cancer by repressing the expression of tumor suppressor KLF6.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gastric cancer is the fourth most common cancer worldwide, and it is estimated that there are 603,003 new cases per year among men and 330,290 new cases per year among women [1]. Despite recent advances in surgical technique, diagnostic method and new chemotherapy regimens, no effective targeting therapy is available for gastric cancer, mainly due to a lack of complete understanding of the molecular mechanisms underlying gastric cancer development. Multiple studies indicate that gastric cancer is a polygenic disease that arises via the dysregulation of many related genes [2]. Many well-characterized tumor suppressor genes and oncogenes have been analyzed to better define their respective roles in gastric cancer development and progression, but only a few consistent and frequent genetic alterations have been identified [3]. Germline mutations in the E-cadherin tumor suppressor gene, loss and somatic mutations of the p53 tumor suppressor gene, oncogenic activation of β-catenin, and K-ras mutations have been found in a subset of gastric cancers [3]. However, the gene regulatory mechanism involved in the development of gastric cancer is still unclear.
MicroRNAs (miRNAs) are small, approximately 22-nucleotide, noncoding RNAs that regulate the expression of several hundreds of genes by targeting their mRNAs post-transcriptionally [4]. MiRNAs bind to the partially complementary target sites in 3′-untranslated regions (3′-UTRs) of mRNAs, inducing direct mRNA degradation or translational inhibition [4]. MicroRNAs have emerged as important gene regulators and are recognized as key players in carcinogenesis by acting as oncogene or tumor suppressor gene [5–7]. Accumulating evidence suggests that miRNAs are aberrantly expressed and regulate the initiation, progression, and metastasis of a variety of cancers including gastric cancer. Human microRNA-181a (miR-181a), a member of the miR-181 family, has been reported to be associated with various human malignancies [8–10].
Kruppel-like factors (KLFs) are core zinc-finger transcription factors that regulate the differentiation and development, growth-related signal transduction, cell proliferation, apoptosis, and angiogenesis [11, 12]. Kruppel-like factor 6 (KLF6) is a member of KLF family and KLF6 gene mutations have been identified in hepatocellular carcinoma, prostate, colon cancers, and gastric cancer [13–16]. Furthermore, a functional single-nucleotide polymorphism (SNP) in the KLF6 gene has been identified recently to be associated with increased incidences of familial prostate and lung cancers [17, 18]. These findings suggest the potential role of KLF6 as a tumor suppressor gene.
However, it remains unknown whether miR-181a is involved in the pathogenesis of gastric cancer. In this study, we found that miR-181a is overexpressed in human gastric cancer tissues. In addition, ectopic expression of miR-181a promoted the proliferation, colony formation, migration and invasion and inhibited the apoptosis of gastric cancer cells. A luciferase reporter assay demonstrated that miR-181a repressed KLF6 expression by targeting its 3′-UTR, and Western blot analysis showed that KLF6 protein was significantly decreased when miR-181a mimic was transfected into SGC-7901 cells. These data suggest that KLF6 gene is a direct target of miR-181a and miR-181a promotes gastric cancer development by downregulating the expression of tumor suppress gene KLF6.
Materials and methods
Cell culture and tissue preparation
Human gastric adenocarcinoma cell lines SGC-7901, MGC-803, BGC-823 and HEK 293 T cells were purchased from the Cell Bank of Chinese Academy of Sciences. Cells were routinely cultured with RPMI-1640 medium supplemented with 10 % fetal bovine serum at 37°C in a humidified atmosphere with 5 % CO2. Nine specimens of adenocarcinoma and the adjacent non-cancerous tissues (10 cm away from the tumor and harboring no cancer cells detected by histology) were collected from either surgical resection or endoscopy biospy. All tissue samples were obtained with informed consent and the procedure was approved by the Medical Ethics Committee of Guangzhou Medical University.
Real-time reverse transcriptase quantitative PCR
Total RNA was extracted from cells and tissues samples with Trizol reagent (Invitrogen). The quality and quantity of the RNA samples were assessed by standard electrophoresis and spectrophotometric methods. The first-strand cDNA was synthesized using reverse transcription kit (EXIQON) according to the manufacturer's instruction. Real-time reverse transcriptase quantitative PCR (qRT-PCR) analysis were performed with locked nucleic acids (LNAs) linear primers (EXIQON) and SYBR Green I, and U6 small nuclear RNA (RNU6B; EXIQON) was used as normalizer. PCR primers used were as follows: Hsa-miR-181a forward: 5′-ACACTCCAGCTGGGAACATTCAACGCTGTCGG-3′; and reverse: 5′-CTCAACTGGTGTCGTGGA-3′; U6 forward: 5′-CTCGCTTCGGCAGCACA-3′;and reverse: 5′-AACGCTTCACGAATTTGCGT-3′. PCR was performed at 95°C for 10 min, followed by 40 cycles of 95°C for 10s and 60°C for 1 min. The relative expression ratio of miR-181a in gastric cancer tissues and cells was quantified by the 2−△△CT method [19].
Transfection with miR-181a inhibitor and mimic constructs
The anti-miR-181a (77 bp) was amplified with the following primers: Forward: 5′-GATCCACTCACCGACAGCGTTGAATGTTTTCGACTCACCGACAGCGTTGAATGTTTTCGACTCACCGACAGCGTTGAATGTTg-3′ and Reverse: 5′-AATTCAACATTCAACGCTGTCGGTGAGTCGAAAACATTCAACGCTGTCGGTGAGTCGAAAACATTCAACGCTGTCGGTGAGTg-3′, and cloned into pSIREN-RetroQZsGreen (Promega, Madison, WI, USA). The plasmid containing anti-miR-181a sequence was named miR-181a Inhibitor, and the control plasmid containing a scrambled sequence (Promega, Madison, WI, USA) was named Negative Control (NC). All constructs were verified by sequencer. To elevate miR-181a expression, a miR-181a mimic (RIBOBIO, Guangzhou, China) was designed, and a scrambled sequence was used as the Negative Control Inhibitor (NCI). For transfection, cells were seeded in 6-well plates at a density of 1 × 105 cells/well and transfected with 20 μM miR-181a mimic, miR-181a Inhibitor, NC or NCI using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instruction.
Cell growth assay
SGC-7901 cells were seeded in 96-well plates 1 day before transfection. Twenty-four hours after transfection, cells were trypsinized and seeded into 96-well culture plates at a density of 10,000 cells/well in growth medium supplemented with 10 % serum. The cells were harvested at different time points (24, 48, 72, and 96 h) for growth assay using the MTS kit (cellTiter96AQ, Promega, USA) following the manufacturer's protocol and the absorption was read at 490 nm.
Colony formation assay
Twenty-four hours after transfection, cells were trypsinized and seeded into 96-well plates with a density of 50, 100, or 200 per well (in triplicate). The cells were observed at every 24 h. When the cells grew to visible colonies, the colonies were washed once with PBS and fixed with 4 % paraformaldehyde for 20 min. Next, fixed colonies were washed again with PBS, and stained with crystal violet for 15 min. Finally, the crystal violet dye was washed with PBS. Automated enzyme-linked spot image analyzer (Elisporeader 400 PRO-X, BIOSYS) was used to read the plates.
Cell growth curve assay
Twenty-four hours after transfection, cells were trypsinized and seeded into 96-well plates for further culture (in triplicate). The cell viability was determined by trypan blue dye exclusion assay and the viable cells were counted on a hemacytometer. The cell growth curve was drawn based on the counting.
Cell migration and invasion assay
Cell migration assay was performed using 8 μm pore size 96-well MIC Transwell plates (Millipore). After transfection, the cells were resuspended in serum-free medium and 1 × 105 cells in 100 μl medium were added to the upper chamber while the lower chamber was filled with complete media as a chemoattractant. The cells were incubated for 24 h at 37°C, the cells on the upper surface of the membrane were removed by cotton swabs and the cells attached to the lower surface were fixed in ice-cold methanol for 10 min, and stained with 0.5 % crystal violet solution for 10 min. The number of migrated cells on the lower surface of the membrane was counted under a microscope in five fields (100×). For the invasion assay, the MIC plates were initially coated with matrigel (BD Biosciences, Bedford, MA, USA) diluted in serum free medium and the following procedures were the same as migration assay.
Flow cytometry analysis of apoptosis
Twenty-four hours after transfection, the cells were collected and washed with PBS twice and about 1–5 × 105 cells were resuspended in 400 μl Annexin V binding buffer (BD-Biosciences, San Jose, CA, USA). The cells were stained with 5 μl Annexin V FITC (BD-Biosciences) for 15 min at 4°C in the dark. The reaction was stopped by the addition of 10 μl propidium iodide (PI) and incubation for 5 min at 4°C in the dark. The samples were subjected to flow cytometry analysis within 1 h.
Western blot analysis
Total cell lysates were prepared using RIPA buffer (150 mM NaCl, 1 mM EDTA, 50 mM Tris–HCl pH 7.4, 1 % Triton X-100, 0.5 % deoxycholic acid, 0.1 % SDS) and the protein concentration was measured by BCA method. Equal amounts of proteins were separated by 10 % SDS-PAGE and blotted to PVDF membranes (Millipore, Billerica, MA, USA). The membranes were blocked with 5 % non-fat milk powder at room temperature for 2 h, then incubated with specific antibody for KLF6 or GAPHD (Abnova, USA) at 4°C o/n followed by incubation with secondary antibody (Southern Biotech, USA) for 1 h at room temperature. The membranes were developed using ECL kit (Pierce, Rockford, IL, USA) and exposed to X-ray film to visualize the images.
Luciferase reporter assay
The 3′-UTR of human KLF6 gene (containing two potential miR-181a-targeting sites at the positions 403 and 2,955 bp) was amplified with whole blood DNA as template using the following primers: 5′-CCGCTCGAG GGGAGCAGAGAGGTGGATCCTG-3′ (forward) and 5′-ATAAGAxATGCGGCCGC TGGCAGTGATGTCATCTTTTATTTTCTG-3′ (reverse). PCR product was cloned into the luciferase reporter vector PsiCHECKUM (Promega, Madison, WI, USA) and the recombinant plasmid was designated PsiCHECKUM-wt. Site-directed mutagenesis of the miR-181a target-site in the 3′-UTR of KLF6 was performed using site-directed mutagenesis kit (TaKaRa, Dalian, China). PsiCHECKUM-mut-1 plasmid with mutated binding site at the position 403 was constructed using the following primers: 5′-AGAGATGGAAAAAAAGTAGTTATCGATCTTTGTGTGTAAGGAGTATACC-3′ (forward) and 5′-GGTATACTCCTTACACACAAAGATCGATAACTACTTTTTTTCCATCTCT-3′ (reverse). PsiCHECKUM-mut-2 plasmid with mutated binding site at the position 2955 was constructed using the following primers: 5′-CAAGATTGTTATATTTCTGTAATCGATCATTTTTTATTGGAATAACATA-3′ (forward) and 5′-TATGTTATTCCAATAAAAAATGATCGATTACAGAAATATAACAATCTTG-3′ (reverse). PsiCHECKUM-mut-3 plasmid with mutated binding site at both positions was constructed similarly. All constructs were verified by sequencing.
For luciferase reporter assay, HEK 293 T cells at logarithmic phase were seeded in 24-well plates at 2 × 104 cells/well. After 24 h, the cells were transiently cotransfected with 0.5 μg wt. or mutant reporter plasmid and 1 μl miRNA mimic or inhibitor (50 μM) using lipofectamine 2000 (Invitrogen). Firefly and Renilla luciferase activities were measured using Dual-Luciferase Reporter Assay System (Promega) following the manufacturer's instructions and normalized for transfection efficiency by the control vector pRL-TK.
Statistical analysis
Statistical analysis was performed using SPSS 15.0 software (SPSS Inc, IL, USA). Data were expressed as the mean ± standard deviation from at least three independent experiments. Differences between groups were analyzed using Student's t test and Chi-square test. A two-tailed P value less than 0.05 was considered statistically significant.
Results
Expression of miR-181a and KLF6 in gastric cancer
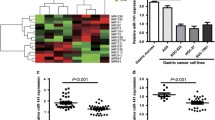
First, we detected the expression of miR-181a and KLF6 in gastric cancer tissues. qRT-PCR analysis showed that among the nine matched pairs of samples, the expression of miR-181a in eight samples of gastric cancer tissues was significantly higher than in the corresponding adjacent non-tumor tissues acquired 10 cm away from the tumor's margin (Fig. 1, upper panel). Moreover, Western blot analysis showed the lower expression level of KLF6 protein in gastric cancer tissues than in the corresponding adjacent non-tumor tissues (Fig. 1, lower panel). These results suggested the negative correlation between miR-181a and KLF6 expression in gastric cancer tissues. Next, we detected the expression of miR-181a in several gastric cancer cell lines such as SGC-7901, MGC-803, and BGC-823 by qRT-PCR (data not shown). Since the expression level of miR-181a is very high in SGC-7901 cells, we chose it as the cell model to further characterize the role of miR-181a in gastric cancer.
miR-181a is overexpressed while KLF6 is downregulated in gastric cancer tissues. Nine matched pairs of gastric cancer tissues and their corresponding adjacent non-tumor tissues (10 cm away from the tumor's margin) were subjected to qRT-PCR analysis of miR-181a level and Western blot analysis of KLF6 protein level. C cancer tissue; N the corresponding adjacent non-tumor tissue
miR-181a promotes the proliferation and clonogenicity of SGC-7901 cells
To assess the role of miR-181a in the growth of gastric cancer cells, we transfected miR-181a mimic or miR-181a inhibitor into SGC-7901 cells. MTS assay showed that miR-181a mimic promoted cell proliferation compared with miR-181a inhibitor, NC, NCI (Fig. 2a, *p < 0.05). Cell growth curve assay also showed that miR-181a mimic promoted cell growth while miR-181a inhibitor inhibited cell growth (Fig. 2b, *p < 0.05). Furthermore, clone formation assay showed that miR-181a mimic increased the clonogenicity of SGC-7901 cells while miR-181a inhibitor decreased the clonogenicity (Fig. 2c, *p < 0.05). Taken together, these data suggest that miR-181a promotes the proliferation and clonogenicity in SGC-7901 cells.
miR-181a promotes the proliferation and clonogenicity of SGC-7901 cells. a MTS assay showing the proliferation of SGC-7901 cells treated as indicated. b Cell growth curve assay showing the growth of SGC-7901 cells treated as indicated. c Clone formation assay showing the clonogenicity of SGC-7901 cells treated as indicated. The colonies were counted on day 7. d–h Representative flow histograms showing the apoptosis of SGC-7901 cells treated as indicated. i Quantification of apoptosis rate of SGC-7901 cells treated as indicated (n = 3), *p < 0.05
miR-181a inhibits the apoptosis of SGC-7901 cells
Next, we performed flow cytometry analysis to examine apoptosis of SGC-7901 cells transfected by miR-181a mimic or inhibitor. The results showed that compared with miR-181a inhibitor or NC or NCI groups, the apoptosis rate was significantly decreased in SGC-7901 cells transfected with miR-181a mimic (Fig. 2d–i, *p < 0.05). Furthermore, miR-181a inhibitor increased the apoptosis rate of SGC-7901 cells. These results indicate that miR-181a inhibits the apoptosis of SGC-7901 cells to promote cell proliferation and clonogenicity.
miR-181a promotes the migration and invasion of SGC-7901 cells
To investigate the role of miR-181a in gastric cancer metastasis, we transfected miR-181a mimic or miR-181a inhibitor into SGC-7901 cells and performed in vitro cell migration and invasion assay. The in vitro migration assay showed that SGC-7901 cells migrated faster after the transfection of miR-181a mimic but more slowly after the transfection of miR-181a inhibitor, compared to control cells (Fig. 3a,c, *p < 0.05). In addition, the in vitro invasion assay showed that the invasion ability of SGC-7901 cells was increased after the transfection of miR-181a mimic but decreased after the transfection of miR-181a inhibitor, compared to control cells (Fig. 3b,d, *p < 0.05). Collectively, these results suggest that miR-181a promotes the migration and invasion ability of SGC-7901 cells.
miR-181a promotes the migration and invasion of SGC-7901 cells. a In vitro migration assay was performed as described in “Materials and methods” section. Shown were representative images on the migration of SGC-7901 cells treated as indicated. b In vitro invasion assay was performed as described in “Materials and methods” section. Shown were representative images on the invasion of SGC-7901 cells treated as indicated. c Quantification of the migration of SGC-7901 cells treated as indicated (n = 3). d Quantification of the invasion of SGC-7901 cells treated as indicated (n = 3), *p < 0.05
miR-181a inhibits the expression of KLF6 by targeting its 3′-UTR
To elucidate the molecular mechanism by which miR-181a modulates the biological behaviors of SGC-7901 cells, we predicted the targets of miR-181a by using bioinformatics tools TargetScan, PicTar, and Miranda. All these programs predicted KLF6 as a target of miR-181a and two potential miR-181a targeting sites at the positions 403 and 2955 nt in the KLF6 3′-UTR were identified (Fig. 4a,b). To verify that KLF6 is a functional target of miR-181a, we cloned a reporter plasmid containing the wide-type 3′-UTR of KLF6 at the 3′position of the firefly luciferase reporter gene. In parallel, we constructed reporter plasmids in which the conserved target sequence at the positions 403 and 2955 nt were mutated individually or in combination, and transfected 293 cells with these constructs with miR-181a mimic or miR-181a inhibitor or NC or NCI. Luciferase activity was markedly diminished in cells transfected with miR-181a and wild type 3′-UTR reporter plasmid-transfected cells, compared to cells transfected with miR-181a mimics and mutant 3′-UTR reporter plasmids (Fig. 4c). Conversely, a significant increase in luciferase activity was observed after transfection with miR-181a inhibitor (Fig. 4c).
miR-181a inhibits the expression of KLF6 by targeting its 3′-UTR. a–b Schemes showing the two potential targeting sites of miR-181a in the 3′-UTR of KLF6. c Luciferase assay was performed as described in the “Materials and methods” section. The luciferase activities were quantified of the lysates from HEK 293 T cells treated as indicated (n = 3)
To confirm that miR-181a represses KLF6 expression in SGC-7901 cells, we performed Western blot analysis and found that the transfection of miR-181a mimic led to a significant decrease in KLF6 protein level, while the transfection of miR-181a inhibitor led to a significant increase in KLF6 protein level compared to the respective controls (Fig. 5a.b,*p < 0.05). In addition, RT-PCR analysis showed that the transfection of miR-181a mimic led to a significant decrease in KLF6 mRNA level (Fig. 5c, *p < 0.05). Taken together, these data provide evidence that miR-181a inhibits the expression of KLF6 by directly targeting the 3′UTR of KLF6.
miR-181a represses KLF6 expression in SGC-7901 cells. a Western blots showing the expression of KLF6 in SGC-7901 cells treated as indicated. Shown were representative blots from three independent experiments with similar results. GAPDH served as loading control. b Quantification of KLF6 protein level in SGC-7901 cells treated as indicated (n = 3). *p < 0.05. c RT-PCR analysis of KLF6 mRNA level in SGC-7901 cells treated as indicated (n = 3), *p < 0.05
Discussion
In recent years, many studies have shown that aberrant expression of miRNAs is implicated in human malignancies [5–7]. Identification of cancer-specific miRNAs as well as their targets is critical for understanding the roles of miRNAs in tumorigenesis and may be important for defining novel therapeutic targets [20].
In our preliminary miRNA microarray, we identified many dysregulated miRNAs in gastric cancer tissues, including the up-regulation of miR-181a. In this study, we focused on the potential oncomir miR-181a. MiR-181a was initially identified as a tumor-suppressing miRNA in human primary glioblastoma and as an oncomir in thymus cells [21, 22]. However, the role of miR-181a in gastric cancer remains elusive. In the present study, we provide several lines of evidence that miR-181a may function as an oncomir in gastric cancer. First, we found that miR-181a was upregulated in gastric cancer tissues compared with matched non-tumor tissues. Second, the in vitro assays on cell proliferation, growth, apoptosis, colony formation, migration, and invasion demonstrated that miR-181a mimic enhanced but miR-181a inhibitor inhibited the malignant phenotypes of SGC-7901 cells. Third, luciferase assay and Western blot analysis showed that miR-181a inhibited the expression of KLF6 by directly targeting its 3′ UTR. KLF6 has been shown to mediate the inhibition of proliferation by upregulating the cell-cycle inhibitor CDKN1A (p21WAF1/CIP1) through an interaction with cyclin D1 in a p53-independent manner, and KLF6 has been reported as a tumor suppressor in gastric cancer [13, 16]. It is reasonable to expect that miR-181a downregulates the expression of KLF6, leading to its loss of tumor suppressor function. This hypothesis satisfactorily explains the downreguation of KLF6 and overexpression of miR-181a in gastric cancer and is consistent with current opinion that KLF6 is a tumor suppressor and miR-181a is an oncomir.
KLFs are a large family characterized by a DNA-binding domain with conserved three tandem C2H2-type zinc finger motifs at the carboxy terminus which recognizes the GT/GC box or CACCC element sites on promoter/regulatory regions [23]. KLF family consists of 17 distinct members. Interestingly, a recent study reported that KLF8 was highly expressed in gastric cancer tissues and cell lines and lentivirus-mediated knockdown of KLF8 inhibited SGC7901 cell proliferation, promoted cell apoptosis, and inhibited the tumorigenicity of SGC7901 cells [24]. These results suggest the oncogenic role of KLF8 and further study is necessary to determine whether miR-181a could modulate the expression of other KLF members. Mounting evidence has highlighted a variety of KLF6 inactivation mechanisms relevant to tumorigenesis, including loss of heterozygosity, somatic mutation, transcriptional silencing through promoter hypermethylation, and dysregulated alternative splicing [15, 25–29]. In this aspect, our findings that KLF6 expression is inhibited by miR-181a provide new insight into the mechanism of loss of KLF6 function in cancer.
In conclusion, in this study, we showed that miR-181a was upregulated in gastric cancer. By using gain and loss of function approaches, we demonstrated that miR-181a promoted the proliferation and metastatic potential of gastric cancer cells. Furthermore, we identified tumor suppressor KLF6 as a direct target of miR-181a. These results suggest that miR-181a functions as an oncomir in gastric cancer and represents a potential molecular target for gastric cancer therapy.
References
Kamangar F, Dores GM, Anderson WF. Patterns of cancer incidence, mortality, and prevalence across five continents: defining priorities to reduce cancer disparities in different geographic regions of the world. J Clin Oncol. 2006;24:2137–50.
Tamura G. Alterations of tumor suppressor and tumor-related genes in the development and progression of gastric cancer. World J Gastroenterol. 2006;12:192–8.
Ushijima T, Sasako M. Focus on gastric cancer. Cancer Cell. 2004;5:121–5.
Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97.
Esquela-Kerscher A, Slack FJ. Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–69.
Petrocca F, Visone R, Onelli MR, Shah MH, Nicoloso MS, de Martino I, et al. E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell. 2008;13:272–86.
Kim YK, Yu J, Han TS, Park SY, Namkoong B, Kim DH, et al. Functional links between clustered microRNAs: suppression of cell-cycle inhibitors by microRNA clusters in gastric cancer. Nucleic Acids Res. 2009;37:1672–81.
Galluzzi L, Morselli E, Vitale I, Kepp O, Senovilla L, Criollo A, Servant N, Paccard C, Hupé P, Robert T, Ripoche H, Lazar V, Harel-Bellan A, Dessen P, Barillot E. Kroemer G.miR-181a and miR-630 regulate cisplatin-induced cancer cell death. Cancer Res. 2010;70:1793–803.
Shin KH, Bae SD, Hong HS, Kim RH, Kang MK. Park NH.miR-181a shows tumor suppressive effect against oral squamous cell carcinoma cells by downregulating K-ras. Biochem Biophys Res Commun. 2011;404:896–902.
Khella HW, White NM, Faragalla H, Gabril M, Boazak M, Dorian D, Khalil B, Antonios H, Bao TT, Pasic MD, Honey RJ, Stewart R, Pace KT, Bjarnason GA, Jewett MA, Yousef GM. Exploring the role of miRNAs in renal cell carcinoma progression and metastasis through bioinformatic and experimental analyses. Tumor Biol. 2012;33:131–40.
Bieker JJ. Kruppel-like factors: three fingers in many pies. J Biol Chem. 2001;276:34355–8.
Black AR, Black JD, Azizkhan-Clifford J. Sp1 and kruppel-like factor family of transcription factors in cell growth regulation and cancer. J Cell Physiol. 2001;188:143–60.
Chen C, Hyytinen ER, Sun X, Helin HJ, Koivisto PA, Frierson Jr HF, et al. Deletion, mutation, and loss of expression of KLF6 in human prostate cancer. Am J Pathol. 2003;162:1349–54.
Kremer-Tal S, Reeves HL, Narla G, Thung SN, Schwartz M, Difeo A, et al. Frequent inactivation of the tumor suppressor Kruppel-like factor 6 (KLF6) in hepatocellular carcinoma. Hepatology. 2004;40:1047–52.
Reeves HL, Narla G, Ogunbiyi O, Haq AI, Katz A, Benzeno S, et al. Kruppel-like factor 6 (KLF6) is a tumor-suppressor gene frequently inactivated in colorectal cancer. Gastroenterology. 2004;126:1090–103.
Cho YG, Kim CJ, Park CH, Yang YM, Kim SY, Nam SW, et al. Genetic alterations of the KLF6 gene in gastric cancer. Oncogene. 2005;24:4588–90.
Spinola M, Leoni VP, Galvan A, Korsching E, Conti B, Pastorino U, et al. Genome-wide single nucleotide polymorphism analysis of lung cancer risk detects the KLF6 gene. Cancer Lett. 2007;251:311–6.
Agell L, Hernandez S, de Muga S, Lorente JA, Juanpere N, Esgueva R, et al. KLF6 and TP53 mutations are a rare event in prostate cancer: distinguishing between Taq polymerase artifacts and true mutations. Mod Pathol. 2008;21:1470–8.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–8.
Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–8.
Ciafre SA, Galardi S, Mangiola A, Ferracin M, Liu CG, Sabatino G, et al. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun. 2005;334:1351–8.
Neilson JR, Zheng GX, Burge CB, Sharp PA. Dynamic regulation of miRNA expression in ordered stages of cellular development. Genes Dev. 2007;21:578–89.
Pearson R, Fleetwood J, Eaton S, Crossley M, Bao S. Kruppel-like transcription factors: a functional family. Int J Biochem Cell Biol. 2008;40:1996–2001.
Liu L, Liu N, Xu M, Liu Y, Min J, Pang H, Zhang N, Zhang H, Zhang H. Lentivirus-delivered Krüppel-like factor 8 small interfering RNA inhibits gastric cancer cell growth in vitro and in vivo. Tumor Biol. 2012;33:53–61.
Camacho-Vanegas O, Narla G, Teixeira MS, DiFeo A, Misra A, Singh G, et al. Functional inactivation of the KLF6 tumor suppressor gene by loss of heterozygosity and increased alternative splicing in glioblastoma. Int J Cancer. 2007;121:1390–5.
Song J, Kim CJ, Cho YG, Kim SY, Nam SW, Lee SH, et al. Genetic and epigenetic alterations of the KLF6 gene in hepatocellular carcinoma. J Gastroenterol Hepatol. 2006;21:1286–9.
Sangodkar J, Shi J, DiFeo A, Schwartz R, Bromberg R, Choudhri A, et al. Functional role of the KLF6 tumour suppressor gene in gastric cancer. Eur J Cancer. 2009;45:666–76.
Narla G, Difeo A, Reeves HL, Schaid DJ, Hirshfeld J, Hod E, et al. A germline DNA polymorphism enhances alternative splicing of the KLF6 tumor suppressor gene and is associated with increased prostate cancer risk. Cancer Res. 2005;65:1213–22.
DiFeo A, Narla G, Hirshfeld J, Camacho-Vanegas O, Narla J, Rose SL, et al. Roles of KLF6 and KLF6-SV1 in ovarian cancer progression and intraperitoneal dissemination. Clin Cancer Res. 2006;12:3730–9.
Acknowledgments
This work was partly supported by the grants from Health bureau of Guangzhou City (No.201102A212011) and from the Natural Science Foundation of Guangdong Province (No.10151006001000016).
Competing interests statement
The authors declare that they have no competing financial interests.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhang, X., Nie, Y., Du, Y. et al. MicroRNA-181a promotes gastric cancer by negatively regulating tumor suppressor KLF6. Tumor Biol. 33, 1589–1597 (2012). https://doi.org/10.1007/s13277-012-0414-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-012-0414-3