Abstract
The purpose of this study is to provide a new method for detecting fetal QRS complexes from non-invasive fetal electrocardiogram (fECG) signal. Despite most of the current fECG processing methods which are based on separation of fECG from maternal ECG (mECG), in this study, fetal heart rate (FHR) can be extracted with high accuracy without separation of fECG from mECG. Furthermore, in this new approach thoracic channels are not necessary. These two aspects have reduced the required computational operations. Consequently, the proposed approach can be efficiently applied to different real-time healthcare and medical devices. In this work, a new method is presented for selecting the best channel which carries strongest fECG. Each channel is scored based on two criteria of noise distribution and good fetal heartbeat visibility. Another important aspect of this study is the simultaneous and combinatorial use of available fECG channels via the priority given by their scores. A combination of geometric features and wavelet-based techniques was adopted to extract FHR. Based on fetal geometric features, fECG signals were divided into three categories, and different strategies were employed to analyze each category. The method was validated using three datasets including Noninvasive fetal ECG database, DaISy and PhysioNet/Computing in Cardiology Challenge 2013. Finally, the obtained results were compared with other studies. The adopted strategies such as multi-resolution analysis, not separating fECG and mECG, intelligent channels scoring and using them simultaneously are the factors that caused the promising performance of the method.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Electronic analysis of fetal heart rate (FHR), which has been introduced for clinical use since 40 years ago, gives wide knowledge about the intrauterine oxygenation and it is now one of the methods for monitoring embryos [1] and pregnant women from 24 to 26 weeks of gestation in modern obstetric units [1–4]. Although there are significant advances in electrocardiogram (ECG) signal processing techniques, analysis of fetal electrocardiogram (fECG) still needs a lot of improvement to become a credible alternative to the usual Doppler ultrasound monitoring technique. Heart defect is one of the most common birth diseases and defects. It is the major cause of birth defects associated death [5, 6]. Congenital heart defects are established in the early stages of fetus formation [7]. Therefore, monitoring of fetal heart activity can provide important information to detect cardiac abnormalities in fetal heart formation in the early stages of pregnancy. However, in spite of the enormous potential applications, non-invasive fetal electrocardiography has not yet made enough satisfaction for fetal disease detection. This is mainly due to the powerful maternal ECG (mECG) interference, muscle contractions and motion artifacts that are added to the fECG signal [8, 9].
FECG processing methods in the literature can be categorized by their methodologies: adaptive filtering, linear decomposition, and nonlinear decomposition.
Adaptive filtering is a common approach for mECG cancelation and fECG extraction. This filtering approach consists of training an adaptive filter for eliminating maternal ECG by using one or more maternal reference channels [10], or training the filter to extract the fECG directly [11, 12]. Several adaptive filters namely, Kalman filter [13], least mean square (LMS) and recursive least square (RLS) [14] can be used for extracting fECG and cancelling mECG.
The second category is linear decomposition in which signals are decomposed into different components using proper basis functions that are somehow in coherence with the time, frequency, or scale characteristics of the fetal components. Wavelet decomposition [15] and matching pursuits [16] are among these methods. The third category is nonlinear decomposition. In the linear decomposition approach, there is a limitation that fECG and other interferences or noises are not always ‘linearly separable’. In the third approach, nonlinear transforms are used in order to separate the signal and noise. The nonlinear transforms require some initial information regarding the desired and undesired parts of the signal such as thoracic ECG channel in which only mECG exists, and it can be used for separating the desired (fECG) and undesired (mECG) parts of the signal [7, 17]. Some of the linear and nonlinear decomposition techniques for fECG extraction are: blind or semi-blind source separation (BSS) [18, 19], singular value decomposition (SVD) [7, 20], principal component analysis (PCA) [21] and wavelet transform (WT). Since in this study WT is employed for FHR extraction and fetal QRS detection, this method is briefly recalled here.
In the WT method, singularities that are obtained from an abdominal signal are detected with the modulus maxima in the wavelet domain. However, extracted amplitudes from fECG signal in this method are not accurate [22]. Nevertheless, the wavelet method is the best tool to detect featured points on ECG signal. Li et al. [23] used this method for the first time. Later, Zhen et al. [24] used biorthogonal wavelet coefficients to detect QRS complex.
Wavelet decomposition was also combined with blind source separation methods for extracting and denoising fECG. Jafari et al. [25] addressed the problem of fetal ECG extraction using BSS in the wavelet domain. They showed that the major advantage of this method is that it can be particularly beneficial when mixing environment is noisy and time varying. The other major advantage of wavelet is that it is a multi-resolution transform and produces a time–frequency decomposition of the signal which makes it a useful tool for detecting the components of various time series by detecting related time–frequency components, and fECG is one of the obvious candidates for this type of analysis [26]. Furthermore, even if FIR filtering may seem appropriate for some tasks, as for preprocessing and denoising, wavelet transform outperforms simple FIR denoising since it introduces less event distortion [27].
Reviewing the literature of the fetal ECG shows that in most of the current works one of the major parts is fECG extraction from the mixture of maternal and fetal ECG signal. This study will show that even without separating fECG and mECG good results can be obtained if some other specific characteristics are employed, such as intelligent selection of the best lead based on stochastic techniques, combination of all available channels and using discrete wavelet transform. A new approach is presented for improving the performance of the method which is based on two rules. These rules can help remove the noises which are confused with fetal R-peaks (fR-peaks). Based on geometric features, fECG signal is divided into three categories. Because of the specific shape of each category, separated strategies are considered to find the fetal heartbeat. Despite various geometric structure of fECG, the proposed approach has promising accuracy in all types of fECG.
Materials
Discrete wavelet transform (DWT)
The basic structure of wavelet bases may be valued by considering generation of an orthonormal wavelet basis for function \(g \in L^{2}\) (the space of square integrable real functions). The method of Daubechies is the most often adopted in studies and applications of wavelets in statistics, bilateral orthonormal functions or parent wavelets: the scaling function, φ(father wavelet), and the mother wavelet, \(\psi\) which are written in Eq. 1.
For some constants \({\text{j}}_{ 0\,} \, \in \,{\text{Z}}\), where Z is the set of integers. The 2j/2 term maintains uniqueness norm of the basis function at different scales, j is scaling parameter and k is the translation parameter. A unit growth in j in Eq. 1 has no effect on scaling function (\(\varphi_{\text{j0k}}\) has constant size), but packs oscillations of \(\psi_{\text{jk}}\) into half the size (doubles its scale or resolution). A unit growth in k in Eq. 1 changes the location of both \(\varphi_{\text{j0k}}\) and \(\psi_{\text{jk}}\), the former by a fixed amount (2−j0) and the latter by an amount proportional to its size (2−j). Given the wavelet root, a function \(g \in L^{2}\) is then signified in a matching wavelet series as in Eq. 2.
with \({\text{c}}_{\text{j0k}} \, = \left\langle {{\text{g,}}\phi_{\text{j0k}} } \right\rangle\) and \({\text{w}}_{\text{jk}} \, = \left\langle {{\text{g,}}\psi_{\text{jk}} } \right\rangle\) where \(\left\langle {.,.} \right\rangle\) is the standard l2 inner product of two functions:
The wavelet extension in Eq. 2 signifies the function g as a series of consecutive estimates. Given an array of function value \({\text{g}} = \left[{\text{g}}\left({\text{t}}_{1}\right), {\text{g}}\left({\text{t}}_{2}\right), \ldots,{\text{g}}\left({\text{t}}_{\text{n}} \right) \right]^{\text{T}}\) of equally spaced points ti, the DWT of g is given by Eq. 3.
where d is an n × 1 vector comprising both discrete scaling constants uj0,k and discrete wavelet constants dj,k and also W is an orthogonal n × n matrix related to orthonormal chosen wavelet basis. Both uj0,k and dj,k are related to their continuous doublets cj0,k and Wj,k via the equation \({{{\text{c}}_{{{\text{j0,k}}\,}} \approx \,{\text{u}}_{\text{j0,k}} } \mathord{\left/ {\vphantom {{{\text{c}}_{{{\text{j0,k}}\,}} \approx \,{\text{u}}_{\text{j0,k}} } {\sqrt {\text{n}} }}} \right. \kern-0pt} {\sqrt {\text{n}} }}\) and \({{{\text{w}}_{{{\text{j,k}}\,}} \approx \,{\text{d}}_{\text{j,k}} } \mathord{\left/ {\vphantom {{{\text{w}}_{{{\text{j,k}}\,}} \approx \,{\text{d}}_{\text{j,k}} } {\sqrt {\text{n}} }}} \right. \kern-0pt} {\sqrt {\text{n}} }}\). The factor \(\sqrt {\text{n}}\) rises because of the difference between continuous and discrete orthonormality conditions. It should be notted that coefficients cj0,k are called approximations and coefficients of Wj,k are called details. Furthermore, DWT can be realized az low pass and high pass filters. The approximation coefficients are obtained from the output of low pass filter (h(t)) and detailed coefficients are obtained from the output of high pass filter (g(t)) [26, 28–30]. Figure 1 demonstrates structure of DWT in high and low frequency, and the red rectangle associated to the approximation part (Ai, i is the level of decomposition) which is divided to approximation part of the next level (Ai+1) and detail part of the next level (Di+1) in each step.
Databases
Three different datasets used for testing the proposed method are DaISy [31], PhysioNet Noninvasive fECG (PNIFECGDB) [32] and PhysioNet/Computing in Cardiology Challenge 2013 [33]. A general description of the used databases is shown in Table 1. The recordings which do not have reference annotations have been annotated by an expert cardiologist. Since PNIFECGDB was annotated manually, the 26 recordings in which the fetal QRS complexes are visible are selected.Footnote 1 In some of the selected recordings fetal beats can be observed only after denoising. The second minute of 26 selected recordings was extracted and used for evaluation. In PhysioNet/Computing in Cardiology Challenge 2013 dataset six recordings (a33, a38, a52, a54, a71, a74) have some missed reference annotations, so they were eliminated and only 69 records of set A were used.
Evaluation protocol
The evaluation of QRS detectors’ performances is usually based on the beat-to-beat comparison between the reference beats and the detected beats. The classical matching window for reliable candidate points is 150 ms in adults [34]. However, for evaluation of performance of fetal QRS detectors, a matching window of 50 ms is commonly used, e.g. Guerrero-Martinez et al. [35] and Zaunseder et al. [36]. To assess the performance of the proposed method sensitivity (Se), positive diagnostic value, (PDV) and accuracy (Acc) (Eqs. 4, 5 and 6) can be calculated as:
where TP is true positive (fR-peak was correctly detected), FP is false positive (artifact was detected as fR-peak), and FN is false negative (fR-peak was not detected by the proposed method).
Data classification
Depending on geometric features of fECG signals, the signals can be categorized into three groups. Because of different observed geometrical structures, utilization of the frequency characteristics of the signal will help to recognize fetal QRS complex more easily. In the first category, fetal QRS complexes may just be observed in one or two channels and even weak noises will increase the possibility of detection error. Also, some of the beats are so small so that they are difficult to be observed (Fig. 2a). The second category is the best kind for detecting fetal beats easily and more accurately because of medium amplitude of the fetal complexes which will not be confused with noises or maternal complexes (Fig. 2b). As it can be seen, in the third category of signals (Fig. 2c) fetal QRS complex has high amplitude and so geometric detection of maternal beat is difficult. Here the combination of geometry and frequency domain analysis of signal is inevitable. In addition, the discipline of maternal heartbeat along with frequency difference between mother and fetal heartbeats can help to find maternal QRS complexes exactly. If the initial detection of fetal R-peaks, finds few fetal beats in a 1-min abdominal signal with the initial thresholds, the signal belongs to the first category. It means the fetal heartbeats have small amplitude that cannot be detected through the initial thresholds, so they should be changed. After initial detection of maternal R-peaks, if there are many maternal beats which have less RR-interval than the minimum maternal RR-interval, the signal belongs to the third category; otherwise it belongs to the second category of abdominal signals. Since all the AECG recordings are taken in clinical setting and resting state of mother, the minimum maternal RR-interval is considered as 0.5 s. This classification will make the algorithm robust to geometric structure differences of fetal QRS complexes.
Categories of fECG. The green rectangles are maternal QRS complexes, and the red ellipses are fetal QRS complexes. Fetal R-peaks have the lowest amplitude in the first category (a), medium amplitude in the second one (b), and high amplitude in the third category, (a), (b) and (c) belong to record b54, a26 and b49 of Challenge 2013 database
Method
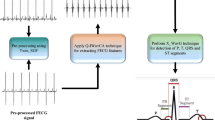
The method presented in this paper has four main parts. The first part is preprocessing that plays a significant role in improving the performance of the algorithm. This part which is shown as the step 1 in Fig. 3 increases the visibility of fetal heartbeats among various noises. The second part is finding maternal R-peaks which is shown as step 2. The performance of this step directly affects other steps. The third part is to perform necessary processing on available channels for finding the best channel and scoring them (step 3–6). First, in step 3 the maternal QRS range is found. Afterwards, in step 4 the high energy zones out of maternal QRS domain are detected. Then, in step 5, the high energy zones which are not related to the fetal heartbeats are removed, and in step 6 the processed channels are scored. The last part is to perform the final processing and producing the final annotations (steps 7 and 8). In step 7, the channels are combined for detecting the fetal R-peaks according to their scores, and in step 8 the missed beats are predicted. The flowchart of the algorithm demonstrates the details in Fig. 3.
Preprocessing and denoising
At the first step of preprocessing, the non-value data is eliminated and then estimated. The non-value data is NaN which stands for not a number and is a numeric data type value representing an undefined value. For instance, division by zero is not defined as a number value and consequently is represented by NaN. In AECG data the most NaNs are generated during recording the data, and some exemplary reasons are loose electrode contact and defective sensors [37]. For elimination and estimation of non-value data, they are imputed using nearest-neighbor method. Since the used datasets include a multichannel and synchronous fECG signal, this technique is practical. This method removes the non-value data and replaces with the corresponding value of the nearest-neighbor column. The nearest-neighbor column is a column whose Euclidean distance is closest to the column of NaN. If the value of the nearest-neighbor column is NaN itself, the next nearest-neighbor column is employed [38]. If Xab is a non-value data and Xij presents the samples of data (i and j are channel and sample number, respectively), the Euclidean distance function dE(x, y) is defined as in Eq. 7.
where a and b are channel and sample numbers of the non-value data, respectively. M is the number of available channels, and N is the length of signal. The Xij corresponds to the minimum value of the Euclidean distance function dE (x, y) which itself is not non-value data and will be considered as a substitute for Xab. Then, for removing the power line noise a band stop FIR filter was applied on four different harmonics. Using discrete wavelet transform (daubechies6, level 10), and removing the reconstructed signals in approximations of level 10 and details of levels 7, 8, 9 and 10, the base line and other low frequency noises were eliminated. The elimination of detail parts of levels 7 and 8 improves the algorithm performance in comparison with traditional use in which they are considered. The last step of preprocessing is denoising using wavelet technique based on the estimation of noises of wavelet coefficients [39].
Maternal R-peaks detection
There are two main reasons that wavelet transformation cannot be used alone to identify the maternal QRS complex. The first reason is that in the method presented in this paper it is not necessary to separate the fetal and maternal complexes. Therefore, fetal and maternal heartbeats are investigated together, and this is one of the characteristics of the proposed method. The second reason is that, there is no significant structural differences between wavelet transform of maternal and fetal QRS complexes as shown in Fig. 4. Therefore, maternal heartbeats cannot be detected through wavelet transform. Nevertheless, there are significant geometric structure differences between maternal and fetal heartbeats (as shown in Fig. 2). Thereby, here the geometric structure of maternal signal such as beats order and QRS complex amplitude which are different from fetal characteristics can help maternal QRS complexes to be easily identified based on threshold method. The optimal threshold was searched for and set at 2.2 times bigger than the mean of the absolute value of the entire signal (Eq. 9); and the zones with bigger amplitude of the threshold are considered as maternal R-peaks. Therefore, a decision function, \(\hat{D}\), for i = 1, 2,…, M (length of the signal) is calculated using Eq. 8.
where Si is the preprocessed signal and TH is the defined threshold. The mentioned technique would be appropriate for the first and second categories of fetal ECGs. However, in the third category, fetal QRS complex has high amplitude and concurrent use of time and frequency domain techniques is necessary. As fetal QRS complex has greater frequency than maternal complex, this frequency difference could be helpful in discrimination of fetal and maternal QRS complexes. In this group, for decreasing the error of maternal complex detection, the fetal complexes which are recognized incorrectly as maternal complexes should be refined. Here, a discrete wavelet transform on level 10 with the function ‘daubecheis6’ is applied on fECG signal. Then, the signal which is related to the first level of detail coefficients of wavelet is reconstructed. The reconstructed signal is named as signal-D1. After identifying maternal QRS complexes, one of the two closer beats than mother’s minimum RR-interval in rest condition (“Data classification” section) which has more energy in the same time in signal-D1 is recognized as fetal beat. At this stage, fetal beats should not be considered and only maternal R-peak locations are saved at an array named MR-loc.
Finding maternal QRS complex domain
As mentioned in “Maternal R-peaks detection” section, the method does not separate maternal and fetal ECGs. Identifying maternal R wave in previous step can help us know the maternal QRS complex range which is 50 ms before and after maternal R-peak locations. The output of this stage is named MQRS-range. For further analyzing, this range is not considered and just the periods of the signal which are out of MQRS-range will be processed.
Finding high-energy zones out of MQRS-range from signal-D1
At this stage, the areas with high energy are detected in signal-D1which are candidates for fetal R-peaks. The zones of signal-D1which have bigger amplitude than defined thresholds (TH) are considered as high-energy zones. The TH is 2.2 times of the mean of the absolute value of the entire signal-D1. The high-energy zones which are outside the maternal QRS complex domain (MQRS-range) are acquired. To meet this end, a decision function, \(\tilde{D}\), for i = 1, 2,…, M (length of the signal) is defined as in Eq. 10.
where Si is the preprocessed signal and \(d_{i}^{1}\) is the sample i of signal-D1.
The \(d_{i}^{1}\) signal is comprised of four major parts. The first one is related to the maternal heartbeats. The second one is related to the fetal heartbeats. The third part is related to the noises with the same or higher amplitude than fetal QRS complexes. The last part is related to the noises with smaller amplitude than fetal QRS complexes. Therefore, the areas of signal-D1 with considerable amplitude and out of MQRS-range are possibly related to fetal beats or they are artifacts with same or higher amplitude of fatal heartbeats. The vectors resulted from this process for the channels are named as high-energy-points-j (j = channel number).
Removing non-fetal complexes and artifacts
As it was mentioned, not all the high-energy zones derived from the previous stage are fetal beats and some of them are noises. For instance, some of them can be artifacts or the noises with the same or higher energy than fetal beats, which have higher amplitude than the given threshold. To increase confidence, they should be checked again. The detected high-energy zones which are candidate fetal R-peaks are checked based on two assumptions, and if any of the high-energy zones meets at least one of these assumptions they are considered as fetal R-peaks. Firstly, since the fetal heartbeats are on a regular basis and fetal QRS complexes come together in a certain discipline, this arrangement can be useful to identify fetal beats correctly. Therefore, the high-energy regions with a certain distance are kept and the rest are deleted (as it can be seen in Fig. 4). This certain distance is fetal RR interval (FRR-interval). By calculating the FRR-interval of various fECGs, it was found that this certain distance is in the range of 350–480 ms. The range of fetal RR-interval plays a key role in the current method, as this range determines which detected high-energy zones are related to the fetal beats and which are related to the artifacts. The second assumption is that at least one and maximum two fetal beats exist between two consecutive maternal R-peaks. If between two consecutive maternal R-peaks there are two high-energy zones whose distance is between 350 and 480 ms, they are considered as fetal R-peaks. If between two consecutive maternal R-peaks there is one high-energy zone, it is considered as a fetal beat. If the number of detected fetal R-peaks in 1 min of fECG signal is less than 60 beats (for other durations it is scaled), it means the current signal places in the first category of the fECG signal so the selected threshold is not small enough to detect more beats. Therefore, the coefficient of the threshold is reduced to 0.8 and the steps of 4.4 and 4.5 are repeated. This process is applied on available channels of fECG signals. The resulting vectors of this process are named as FR-loc1-i (i = channel number).
Channel scoring
One of the novel techniques that have been used in this paper is channel scoring. Channels with higher scores have higher priority, and the channel which has the highest score is considered as the reference channel. Two criteria are considered for scoring the channels.
The first criterion is noise distribution. The channel with less noise scattering has better signal quality. Distribution of noise is expressed by standard deviation of detail coefficients of the discrete wavelet transform applied to the preprocessed signal.
For the second criterion, any channel which has greater length of the vector FR-loc1-i (obtained from the “Removing non-fetal complexes and artifacts” section) is in priority because the points existing in FR-loc1-i vector represent fetal heartbeats. If the length of FR-loc1-i vector is larger, it means that in the initial scan a larger number of fetal heart beats have been detected which itself can be an indication of the channel quality in fetal beats visibility, and it shows that the channel has stronger fECG signal.
Score from the first criterion is called Scr1. As the current method uses 4 channels, the score 3 is assigned to the channel with the minimum distribution of noise. Consequently, for the channel with the highest noise distribution the score is zero.
The score from the second criterion is called Scr2. The score 3 is assigned to the channel whose FR-loc1-i array has maximum length and for the channel whose FR-loc1-i array has minimum length the score 0 is considered. If the number of available channels is three the maximum value for the Scr1 and Scr2 will be 2 instead of 3. The final score (Eq. 11) is the sum of theScr1 and the Scr2.
The channel which has greater Final-Score is in priority for any further processing. In other words, channels are ranked based on their Final-Score. The first criterion measures the noise existence and the second one measures better visibility of fetal heartbeats. In order to validate this method, it was applied on the 100 recordings of set B of PhysioNet/CinC Challenge 2013 dataset, and the accuracy of 89 % was obtained. It means that in 89 % of data, the best channel suggested by this method was the same as the best channel selected manually.
Combination of initial detected fetal R-peaks of each channels based on priority given by their score
The initial scan of fetal beats in “Removing non-fetal complexes and artifacts” section does not contain all of the fetal QRS complexes because of two reasons. Firstly, some of the fetus beats are overlapped with maternal QRS complex domains which were not considered at any stages. Secondly, some fetal beats have not been considered due to existence of noise or lack of proper FRR interval. For example, the beat existing between two beats which are not detected because of noise existence or overlapping with the maternal QRS complexes is a missed beat because of improper FRR interval. At this step, some of the missed beats can be detected with the help of all available leads.
The combination of the leads can find the missed beats which are not detected because of noise, signal quality or lack of FRR interval. However, the beats overlapped onto maternal QRS complexes cannot be found through combination of the leads as they are not detected in any of the channels. The combination is based on the channels ranking. All the detected fetal beats existing in the best lead are considered. Then, the missed beats are searched in the second ranked channel one by one. If any of missed beats were not found, they are searched in the third ranked channel and if any of them were not detected again, they are searched in the last ranked lead. If any of the missed beats was not detected in any channels it will be predicted in the next section. This process is shown in Fig. 5. The output array is named as FR-loc-1-combine.
Prediction of the location of missed fetal R-peaks
All the fetal QRS complexes which overlap with maternal QRS complexes are detected by prediction. Besides, the fetal beats which are not detected in any available leads are predicted in this step. The possible locations of these eliminated beats are predicted based on the current detected beats order.
Using the average FRR interval of the existing recorded fetal beats, the location of remaining heartbeats can be predicted. Between the two far beats that the later beat comes 0.6 s or more after the former one, there is at least one or more missed beats. The number of missed beats is calculated by dividing the intervals of the two far beats by the FRR interval average of the existing recorded fetal beats. The predicted fetal heart beat locations are named as FR-loc2. For finalizing the fetal heartbeat locations, the fetal R-peaks locations, resulted from “Combination of initial detected fetal R-peaks of each channels based on priority given by their score” and “Prediction of the location of missed fetal R-peaks”, are combined and sorted and finally the total fetal R-peak locations array (named Fetal-R) is produced.
Applying to the datasets
In this section, the proposed approach is applied to the mentioned datasets in “Databases” section. In this study the thoracic signals are not used, and only the abdominal signals are utilized. For consistency, only four abdominal channels of the records are employed. In recordings containing three abdominal channels, the results are obtained using three available channels. Furthermore, records’ sampling frequency is changed to 1000 Hz by decimation or interpolation if the sampling rate is not 1 kHz. The procedure of measuring the performance of the methods was mentioned in “Evaluation protocol” section
Results
In this section the proposed method was applied to the described datasets. The results for fetal R-peak detection were evaluated by an expert cardiologist, who calculated three quantitative results: FP, FN, and TP. Indices of test performance can be derived from these parameters such as sensitivity, positive diagnostic value, and accuracy which were described in previous sections. Table 2 illustrates the exhausted average results of evaluation of the algorithm, which was applied to the mentioned datasets, and Table 3 compares the current study with previous ones.
The processed recordings of the PhysioNet Noninvasive FECG contain 3986 fR-peaks from which 38 were not detected (0.95 %). The number of detected fetal R-peaks was 3982 beats from which 34 were false detections (0.85 %) and 3948 (99.15 %) were detected correctly. The DaISy database contains five abdominal channels in which there are 22 fetal beats in 10 s. The presented technique detected 21 fetal beats correctly and one fetal beat was detected 0.02 s later than the allowable matching window. Therefore, the accuracy reduced to 91.3. In these short signals, only one beat that is not detected correctly, will significantly reduce the accuracy. The 69 processed recordings of the PhysioNet/CinC Challenge 2013 contain 9791 fR-peaks from which 796 fR-peaks were not detected (8.13 %). The number of detected fetal R-peaks was 10,023 beats from which 976 were false detections (9.74 %) and 9047(90.26 %) were detected correctly.
Discussion
The training set of the PhysioNet/Computing in Cardiology Challenge 2013 [33] database (set A) contains various fECG signals with different qualities. In some signals, the fetal beats cannot be observed, and in some other cases, the fetal heartbeats can only be observed after denoising. In some signals, the big parts of all the available channels are noisy in which fetal R-peaks are detected by prediction (“Prediction of the location of missed fetal R-peaks” section) based on other detected beats’ FRR interval. Nevertheless, some other signals were excellent in which Se and PDV are 100 %. Thereby, this database is a good one to be used by researchers who work on fetal ECG analysis to evaluate different algorithms which do not need thoracic signals and check the algorithms robustness. As this dataset contains different signals with a broad range of quality, the accuracy reduces to 83.62 % with Se 91.91 % and PDV 90.26 %. If the signals do not have noises which are present on all the channels and the fetal beats can be observed after denoising, the current method will have high performance; such as the accuracy for the PhysioNet Noninvasive fECG Database which is 98.21 %. According to the Table 3, the wavelet based techniques have good performance. As there is no benchmark database for noninvasive fetal ECG analysis [48], a direct comparison between proposed methods in Table 3 is not feasible.
The final fetal R-peaks are comprised of three parts. The first part is related to the fetal R-peaks which are obtained by the first scan in the reference channel by detecting the high energy zones of signal-D1 (“Finding high-energy zones out of MQRS-range from signal-D1” section) and refining them (“Removing non-fetal complexes and artifacts” section). The second part is related to the fetal R-peaks which are obtained by combining the channels via the priority given by their scores (“Combination of initial detected fetal R-peaks of each channels based on priority given by their score” section), and the third part is related to the fetal R-peaks that their locations are predicted (“Prediction of the location of missed fetal R-peaks” section). Table 4 shows the number of fetal R-peaks in each part and the detection accuracy of each part when 69 1-min recordings of PhysioNet/CinC Challenge 2013, set A were used. According to Table 4, 65.4 % of fetal R-peaks are detected by the first scan in reference channel with accuracy of 92.95, 6.4 % of fetal R-peaks are detected by combinational use of available channels with accuracy of 83.18 and 28.20 % of fetal R-peaks are detected by predicting the missed fetal heartbeats with accuracy of 85.64 %.
In this study three or four abdominal channels were used regardless of the number of abdominal leads. It should be noted that some recordings of PhysioNet Noninvasive fECG Database have three abdominal channels. If there are more or fewer than four channels, then all the available leads are ranked and used. This is an important feature over the blind source separation techniques which require large number of channels (usually between 8 and 16) [14, 49]. Furthermore, it is a valuable advantage due to the fact that placement of high number of electrodes on the mother’s body is possible only under clinical settings; while under nonclinical and mobile settings it is difficult or even impractical. Scoring and ranking the channels are major advantages of this study which can be used in other studies in which the best channel should be selected for fECG extraction and analysis. Scoring and ranking of long duration recordings (more than 1 min duration) requires more processing time, while it is suggested that a random 1 min of the signal be extracted and used for scoring and finding the best channel which carries stronger fECG signal.
To remove non-fetal complexes and high-energy disturbances (“Removing non-fetal complexes and artifacts” section) two assumptions were adopted and implemented for the first time. As the fECG and mECG are examined together, utilization of these assumptions is inevitable. It should be mentioned that these assumptions were developed by analyzing 17,494 fetal heartbeats. The number of candidate fetal R-peaks which were obtained by the first scan in the reference channel by detecting the high energy zones of signal-D1 (“Finding high-energy zones out of MQRS-range from signal-D1” section) using 69 1-min recordings of PhysioNet Challenge 2013, Set A was 8681 from which 1859 were false detections (FP) with detection accuracy of 79.05 %. After refining the candidate fetal R-peaks for finalization, using the mentioned assumptions (“Removing non-fetal complexes and artifacts” section) the number of detections reduced to 6554 from which 519 were FP with detection accuracy of 92.95 %. This difference in false detections and accuracy illustrates how these two assumptions have improved the performance of the proposed method. It should be mentioned in the case of congenital cardiac defects or even in healthy ones, the fetal heart rate may contain irregularities such as extra systolic heartbeats, so some heartbeats may not observe one of these assumptions.
The noise handling procedure of the proposed method has a major positive effect on the total performance of the method. Some noises such as power line interference, baseline oscillations and electromyographic contamination are handled by the method and not by the recording devices. The P-wave and the T-wave of the mECG have been filtered out because of elimination of the levels 9, 8, and 7 of the details parts of the discrete wavelet transform in the preprocessing stage (Fig. 4). This higher cut-off improved the performance of the method.
Conclusion
This study presents a method to detect fetal QRS complexes from non-invasive fetal electrocardiogram (fECG) signals. Employing both time and frequency domain techniques caused more accurate detection of fetal heartbeats. The proposed method is a good candidate for real time implementation and production of low-cost, portable devices since in this method fECG is not separated from mixture signal and there is no need to thoracic leads, thus, the required computational operations have been significantly reduced. Furthermore, the method is robust to the number of available channels which means fetal QRS complexes can be detected in AECG recorded by fewer electrodes. This study has some other main features which are briefly listed. Intelligent scoring of the channels for finding the best channel which is related to the signal quality based on noise distribution and fetal R-peak visibility, simultaneous combination of channels with the priority of their scores, and being robust to geometric structural differences of fECG are the advantages which lead to better performance of the method. On the other hand, there are some negative aspects which should be considered. The fetal beats which overlap with the maternal QRS complex cannot be observed and checked, so their locations should be predicted. Furthermore, analyzing fECG waveform information is limited to only fetal QRS complexes which are not overlapped with maternal QRS complexes. In addition, detection of the fetal P-waves or T-waves is impossible as they are eliminated in preprocessing stage, and if they were not omitted by the preprocessing stage, in some fetal heartbeats the detection of fetal P-waves and T-waves would be hard because of existence of maternal P and T-waves; while FHR is calculated in a fast, but accurate procedure. Finally, not separating the mECG and fECG has reduced the required processing and time for FHR extraction.
Notes
The 26 records of PhysioNet Noninvasive fetal ECG which were used for evaluation are: ecgca154, 192, 244, 274, 290, 300, 323, 368, 392, 410, 444, 585, 597, 649, 659, 733, 746, 748, 771, 811, 826, 840, 848, 886, 906, and 986.
Abbreviations
- ECG:
-
Electrocardiogram
- fECG:
-
Fetal electrocardiogram
- mECG:
-
Maternal electrocardiogram
- FHR:
-
Fetal heart rate
- fR-peak:
-
Fetal R-peak
- BSS:
-
Blind or semi-blind source separation
- SVD:
-
Singular value decomposition
- PCA:
-
Principal component analysis
- WT:
-
Wavelet transform
- DWT:
-
Discrete wavelet transform
- LMS:
-
Least mean square
- RLS:
-
Recursive least square
- Se:
-
Sensitivity
- PDV:
-
Positive diagnostic value
- ACC:
-
Accuracy
- TP:
-
True positive
- FP:
-
False positive
- FN:
-
False negative
References
Gardosi J, Maeda K (1995) Intrapartum surveillance: recommendations on current practice and overview of new developments. FIGO study group on the assessment of new technology. International federation of gynecology and obstetrics. Int J Gynecol Obstet 49(2):213–221
American College of Obstetricians and Gynecologists (ACOG) (2005) ACOG practice bulletin. Clinical management guidelines for obstetrician-gynecologists, number 70, December 2005. Int J Gynecol Obstet 106(6):1453–1460
RCOG (2001) Royal College of Obstetricians and Gynaecologists. Evidence-based clinical guideline number 8. The use of electronic fetal monitoring. London: Royal College of Obstetricians and Gynaecologists
Rooth G, Huch A, Huch R (1987) Figo news. Guidelines for the use of fetal monitoring. Int J Gynecol Obstet 25:159
Congenital Heart Defects (2005) March of Dimes. 2005. http://www.marchofdimes.com/professionals/14332_1212.asp
Minino AM, Heron MP, Murphy SL, Kochanek KD (2007) Deaths: Final data for 2004. National Vital Statistics Reports 2007
Sameni R, Clifford GD (2010) A review of fetal ECG signal processing; issues and promising directions. J Open Pacing Electrophysiol Ther J 3:4–20
Outram NJ, Ifeachor EC, van Eetvelt PWJ, Curnow JSH (1995) Techniques for optimal enhancement and feature extraction of fetal electrocardiogram. J IEE Proc-Sci Meas Technol 142:482–489
Mollakazemi MJ, Atyabi SA, Ghaffari A, Jalali A (2014) A novel technique for analyzing noisy noninvasive fetal electrocardiogram signals. In: Proceeding of computing in cardiology 2014
Farvet AG (1968) Computer matched filter location of fetal R waves. J Med Biol Eng 6:467–475
Park Y, Lee K, Youn D, Kim N, Kim W, Park S (1992) On detecting the presence of fetal R-wave using the moving averaged magnitude difference algorithm. J IEEE Trans Biomed Eng 39:868–871
Khamene A, Negahdaripour S (2000) A new method for the extraction of fetal ecg from the composite abdominal signal. J IEEE Trans Biomed Eng 47:507–516
Niknazar M, Rivet B, Jutten C (2013) Fetal ECG extraction by extended state Kalman filtering based on single-channel recordings. J Biomed Eng IEEE Trans 60(5):1345–1352
Behar J, Johnson A, Clifford GD, Oster J (2014) A comparison of channel fetal ECG extraction methods. J Ann Biomed Eng 42(6):1340–1353
Akay M, Mulder E (1996) Examining fetal heart-rate variability using matching pursuits. J IEEE Eng Med Biol Mag 15:64–67
Schreiber T, Kaplan DT (1996) Nonlinear noise reduction for electrocardiograms. J Chaos 6:87–92
Richter M, Schreiber T, Kaplan DT (1998) Fetal ECG extraction with nonlinear state-space projections. J IEEE Trans Biomed Eng 45:133–137
Zhang ZYZ (2006) Extraction of temporally correlated sources with its application to noninvasive fetal electrocardiogram extraction. J Neurocomput 69(7–9):894–899
Li Y, Yi Z (2008) An algorithm for extracting fetal electrocardiogram. J Neurocomput 71(7-9):1538–1542
Ouali MA, Chafaa K (2013) Separation of composite maternal ECG using SVD decomposition. In: Computer applications technology (ICCAT), 2013 international conference pp 1–4
Moraru L, Sameni R, Schneider U, Haueisen J, Schleußner E, Hoyer D (2011) Validation of fetal auditory evoked cortical responses to enhance the assessment of early brain development using fetal MEG measurements. J Physiol Meas 32(11):847–868
Niknazar M (2013) Extraction and denoising of fetal ECG signals. Ph.D. Thesis, University of Grenoble, Grenoble, France, 2013. http://theses.eurasip.org/theses/534/extraction-and-denoising-of-fetal-ecg-signals/download
Li CW, Zheng CX, Tai CF (1995) Detection of ECG characteristic points using wavelet transforms. J IEEE Trans Biomed Eng 42(1):22–28
Zhen J, Zheng XY, Luo J, Li Z (2008) Detection of QRS complexes based on biorthogonal spline wavelet. J Shenzhen Univ Sci Eng 25(2):167–172
Jafari MG, Chambers JA (2005) Fetal electrocardiogram extraction by sequential source separation in the wavelet domain. J IEEE Trans Biomed Eng 52(3):390–400
Mallat SG (1989) A theory for multiresolution signal decomposition: the wavelet representation. J IEEE Trans Pattern Anal Mach Intell 11(7):674–693
De Ridder S, Neyt X, Pattyn N, Migeotte PF (2011) Comparison between EEMD, wavelet and FIR denoising: Influence on event detection in impedance cardiography. Annual International Conference of the IEEE, Boston, Engineering in Medicine and Biology Society, EMBC, 2011, pp 806–809
Vettereli M (2001) Wavelets, approximation and compression. IEEE Signal Process Mag 18(5):59–73
Rioul O, Vetterli M (1991) Wavelets and signal processing. IEEE Signal Process Mag 8(4):14–38
Rashmi P (2012) Removal of artifacts from electrocardiogram. Master’s thesis, national institute of technology Rourkela Odisha 2012. http://ethesis.nitrkl.ac.in/4122/1/Rashmi_Thesis.pdf
De Moor B (2010) Database for the identification of systems (DaISy); [online]. http://homes.esat.kuleuven.be/~tokka/daisydata.html/
Amaral LAN, Glass L, Hausdorff JM, Ivanov PCh, Mark RG, Mietus JE, et al. (2000) PhysioBank, PhysioToolkit, and PhysioNet: components of a new research resource for complex physiologic signals. J Circ 12(101):e215–e220. http://physionet.org/physiobank/database/nifecgdb/
Silva I, Behar J, Clifford GD, Moody GB (2013) Noninvasive fetal ECG: the PhysioNet/Computing in Cardiology Challenge 2013. Computing in Cardiology 2013, Zaragoza
ANSI/AAMI/ISO EC57 (1998/(R)2008) Testing and reporting performance results of cardiac rhythm and ST segment measurement algorithms
Guerrero-Martinez J, Martinez-Sober M, Bataller-Mompean M, Magdalena-Benedito J (2006) New algorithm for fetal QRS detection in surface abdominal records. Comput Cardiol 2006:441–444
Zaunseder S, Andreotti F, Cruz M, Stepan H, Schmieder C, Malberg H, Jank A (2012) Fetal QRS detection by means of Kalman filtering and using the Event Synchronous Canceller. In: 7th international workshop on biosignal interpretation
Mollakazemi MJ, Atyabi SA, Ghaffari A (2015) Heart beat detection using a multimodal data coupling method. J Physiol Meas 36(8):1729–1742
MATLAB software help. http://www.mathworks.com/help/bioinfo/ref/knnimpute.html
Ghaffari A, Atyabi SA, Mollakazemi MJ, Niknazar M, Niknami M, Soleimani A (2013) PhysioNet/CinC Challenge 2013: a novel noninvasive technique to recognize the fetal QRS complexes from noninvasive fetal electrocardiogram signals. Comput Cardiol 40:293–296
Mooney DM, Grooome LJ, Bentz LS, Wilson JD (1995) Computer algorithm for adaptive extraction of fetal cardiac electrical signal. In: Proceedings of the 1995 ACM symposium on applied computing pp 113–117
Azad KAK (2000) Fetal QRS complex detection from abdominal ECG: a fuzzy approach. IEEE Nordic signal processing symposium (NORSIG). Kolmarden, Sweden, pp 275–278
Piéri JF, Crowe JA, Hayes-Gill BR, Spencer CJ, Bhogal K, James DK (2001) Compact longterm recorder for the transabdominal foetal and maternal electrocardiogram. J Med Biol Eng Comput 39(1):118–125
Ibrahimy MI, Ahmed F, Ali MAM, Zahedi E (2003) Real-time signal processing for fetal heart rate monitoring. J IEEE Trans Biomed Eng 50(2):258–261
Karvounis EC, Papaloukas C, Fotiadis DI, Michalis LK (2004) Fetal heart rate extraction from composite maternal ECG using complex continuous wavelet transform. Comput in Cardiol 2004:737–740
Martens SMM, Rabotti C, Massimo M, Sluijte RJ (2007) A robust fetal ECG detection method for abdominal recordings. J Physiol Meas 28(4):373–388
Karvounis EC, Tsipouras MG, Fotiadis DI (2009) Detection of fetal heart rate through 3-D phase space analysis from multivariate abdominal recordings. J IEEE Trans Biomed Eng 56(5):1394–1406
Hasan M, Reaz M (2012) Hardware prototyping of neural network based fetal electrocardiogram extraction. Meas Sci Rev 12(2):52–55
Hasan MA, Reaz MBI, Ibrahimy MI, Hussain MS, Uddin J (2009) Detection and processing techniques of FECG signal for fetal monitoring. J Biol Proced Online 11(1):263–295
Sameni R (2008) Extraction of fetal cardiac signals from an array of maternal abdominal recordings. Ph.D. Thesis, Sharif University of Technology, Institut National Polytechnique de Grenoble, 2008. http://www.sameni.info/Publications/Thesis/PhDThesis.pdf
Acknowledgments
The authors would like to thank Obstetrician Sasan Heshmat, Pars General Hospital, Tehran, Iran for helping us to produce reliable annotations, checking the final results, and providing necessary medical information.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors report no declarations of interest.
Rights and permissions
About this article
Cite this article
Ghaffari, A., Mollakazemi, M.J., Atyabi, S.A. et al. Robust fetal QRS detection from noninvasive abdominal electrocardiogram based on channel selection and simultaneous multichannel processing. Australas Phys Eng Sci Med 38, 581–592 (2015). https://doi.org/10.1007/s13246-015-0381-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13246-015-0381-2