Abstract
Plant-specific WUSCHEL-related homeobox (WOX) transcription factors are known to be involved in plant developmental processes, especially in embryogenesis. In this study, a total of thirteen WOX members were identified in the banana (Musa acuminata) genome (MaWOX) and characterized for in-silico analysis. Phylogenetic analysis revealed that these genes were divided into three clades (ancient, intermediate and modern) which reflected the evolutionary history of WOX families. Furthermore, modern clade members have shown higher variations in gene structural features and carried unique conserved motifs (motif 3 and motif 4) when compared to the members of other clades. The differential expression of all 13 MaWOX was observed in early (embryogenic cell suspension (ECS), multiplying ECS, germinating embryos, young leaflet and node of germinated plantlets) and late (unripe fruit peel and pulp, ripe fruit peel and pulp) developmental stages of banana cultivar Grand Naine. The maximum expression of MaWOX6 (18 fold) and MaWOX13 (120 fold) was found during somatic embryogenesis and in unripe fruit pulp, respectively. Moreover, numerous cis-elements responsive to drought, cold, ethylene, methyl jasmonate (MeJA), abscisic acid (ABA) and gibberellic acid (GA) were observed in all MaWOX promoter regions. The subsequent expression analysis under various abiotic stresses (cold, drought and salt) revealed maximum expression of the MaWOX3 (830 fold), MaWOX8a (30 fold) and MaWOX11b (105 fold) in salt stress. It gives evidence about their possible role in salt stress tolerance in banana. Hence, the present study provides precise information on the MaWOX gene family and their expression in various tissues and stressful environmental conditions that may help to develop climate-resilient banana plants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Wuschel related homeobox (WOX) gene family belongs to the homeodomain (HD) superfamily which encodes one of the largest groups of transcription factors containing 60–66 amino acids conserved DNA binding domain (Alvarez et al. 2018). The homeobox (HB) transcription factors have been classified into 14 classes in plants which are HD-ZIP I-IV, KNOTTED-1 like homeobox (KNOX), plant homeodomain (PHD), DDT, nodulin homeobox gene (NDX), luminidependens (LD), BEL, wuschel related homeobox (WOX) and plant interactor homeobox (PINTOX), SAWADEE and plant zinc finger (PLINC) (Mukherjee et al. 2009; Alvarez et al. 2018). Evolutionary studies indicate that the WOX gene family of HB transcription factors have diverged into three clades. A total of 15 WOX proteins of the dicot model plant Arabidopsis thaliana have been classified into modern or Wuschel (WOX 1–7 and WUS proteins), intermediate (WOX 8, 9, 11 and 12 proteins), and ancient (WOX 10, 13 and 14 proteins) clades according to amino acid sequences of HD (Lian et al. 2014; Zhang et al. 2014).
Recent studies elucidate that the WOX genes have been shown to play a pivotal role in key developmental processes such as stem cell maintenance, embryo patterning and organ development in plants (Schoof et al. 2000; Ueda et al. 2011; Lin et al. 2013b). In A. thaliana, WUS (AtWUS) plays an important role in the maintenance of root and shoot apical meristem as well as vegetative to embryonic transition (Mayer et al. 1995; He et al. 2019). It also acts as a repressor in stem cell regulation and an activator in floral patterning (Ikeda et al. 2009). The overexpression of AtWOX1 was reported to develop abnormal meristem which subsequently formed small leaves, low fertility and dwarf A. thaliana plants (Graaff et al. 2009; Zhang et al. 2011). AtWOX3 plays a vital role in lateral organ formation by recruiting organ founder cells (Shimizu et al. 2009; Alvarez et al. 2018). AtWOX2, 8 and 9 are reported to be expressed during early embryonic development stages in plants (Alvarez et al. 2018; Wang et al. 2019). AtWOX4 and AtWOX14 are involved in cambial meristem differentiation and maintenance (Hirakawa et al. 2010; Liu et al. 2014). AtWOX5 and Populus trichocarpa WOX5a (PtWOX5a) are known for their role in auxin-responsive regulation of stem cell differentiation in root apical meristem (Lopez-Moya et al. 2017; He et al. 2019). Four PtoWOX genes, when ectopically expressed in transgenic poplars; were reported to promote adventitious root formation (Liu et al. 2014). In rice, OsWOX11 has been shown to modulate root development imparting enhanced drought resistance and is also involved in crown root emergence activation by cytokinin-auxin signaling integration (Zhao et al. 2009; Cheng et al. 2016). Therefore, the functional diversification of WOX genes makes them a research hotspot in plant developmental biology.
Bananas are perennial monocot herbs belonging to the Musaceae family and known as a staple food/fruit crop worldwide. Banana is considered the fourth most important global food crop while India is ranked first in the production of bananas with an average output of about 32 million tonnes per year (FAOSTAT 2020, http://faostat.fao.org). Total banana production in the world is reported as about 108.7 million tonnes in 2020 (FAOSTAT 2020, http://faostat.fao.org). Bananas are cultivated in the provinces of humid subtropics, humid tropics, and semiarid tropics, and some banana genotypes can grow up to an elevation of 2000 mean sea level (MSL). The selected cultivar in the present study i.e., Grand Naine is the most commonly grown commercially cultivated variety of banana worldwide. It is considered a member of the Cavendish banana cultivar group (AAA genome) and contributes not only to household food and nutritional security but also for income generation as a cash crop. Grand Naine is a high yielding variety amongst others in India and occupies the first position in production share with 63% (Devarajan et al. 2021). The major constraints for banana production are largely dominated by biotic and abiotic stresses (Tushemereirwe et al. 2004; Tripathi et al. 2008). Abiotic stresses such as salinity, extreme temperature, drought, flooding, low temperature, ultraviolet rays, nutritional imbalances, and oxidative stress results in yield losses. The productivity of the banana crop was significantly impacted by the drought and has been estimated in the range from 20 to 65%, particularly for bananas from east Africa with the AAA genome (van Asten et al. 2011). In case of biotic stresses, various viral, bacterial and fungal pathogens like banana streak virus (BSV), banana bunchy top virus (BBTV), Ralstonia solanacearum (bugtok and moko disease), Fusarium oxysporum f. sp. cubense (Fusarium wilt), and Xanthomonas campestris pv. musacearum (Xcm) (Xanthomonas wilt) etc. significantly affects the overall yield of banana cultivation (Tripathi et al. 2019).
The availability of the whole-genome sequence of banana enabled researchers to study the genome-wide evolution and divergence among the different members of various gene families (D’Hont et al. 2012; Davey et al. 2013). In the present study, we have identified 13 WOX (MaWOX) genes in the banana genome and characterized them using various bioinformatics tools. The basic local alignment search tool (BLAST) was used to categorize MaWOX genes in different groups. The structural features, conserved motifs and physicochemical properties were predicted using online tools like HMMER (http://www.ebi.ac.uk/Tools/hmmer), MEME (http://meme-suite.org/) and ExPASy (https://www.expasy.org/), respectively. The phylogenetic analysis was carried out to understand the evolutionary relationship of MaWOX with other plant species. Further, the differential expression analysis of all 13 MaWOX genes during somatic embryo’s developmental stages, vegetative and fruit tissues have revealed their potential role in the growth and development of banana plants. The expression response of these genes was also performed in tissue culture raised banana plants under different abiotic stress conditions (cold, salt and drought). Further, promoter analysis was performed as the promoter’s architecture and the presence of different cis-regulatory elements have also emerged as the critical factor that influences the tissue-specific expression level of a gene in terms of its transcript abundance in response to developmental phase or stress condition (Wang et al. 2019). WOX gene family has been explored in many plant species including arabidopsis, rice, poplar, tea, watermelon, wheat, Brassica and cotton (Zhang et al. 2015; Yang et al. 2017; Wang et al. 2019). In the present study, potential role of cis-elements in the promoter regions of predicted WOX genes in banana was analysed. However, studies highlighting the role of WOX gene family in banana are still lacking. Hence, intending to investigate the crucial role of this gene family, we have conducted genome-wide studies of WOX gene family in banana.
Materials and methods
Plant materials, growth conditions and stress treatments
To examine the tissue-specific expression of MaWOX genes, tissues from different development stages of the banana cultivar (cv.) Grand Naine were collected from the experimental field of National Agri-Food Biotechnology Institute (NABI), Mohali, Punjab, India (310 m above sea level; Latitude 30° 47´ North; Longitude 76° 41´ East). The early development stages included in-vitro cultures viz., embryogenic cell suspension (ECS), multiplying ECS, germinating embryos, young leaf and nodal portion of germinated plantlet. The ECS was developed following the previously established protocol in our lab (Shivani et al. 2017; Shivani and Tiwari 2019). In short, immature male flowers were inoculated on callus induction medium containing 2, 4-dichlorophenoxyacetic acid (2, 4-D) under the dark condition at 27 °C. The embryogenic callus was isolated and inoculated in 2, 4-D (18 μM) and zeatin (0.2 μM) containing suspension medium and kept in dark at 27 °C with 90 rotations per minute agitation to generate ECS. The ECS was cultured on different semi-solid media for embryos multiplication, regeneration, germination and plantlets development. The fruit tissues (unripe peel, unripe pulp, ripe peel and ripe pulp) collected from the research field were considered as the late development stages.
For the gene expression analysis, tissue culture raised banana plants were treated under different abiotic stress conditions (cold, salt and drought). For cold stress, plants were shifted at 10 °C in the plant tissue culture chamber (Percival, USA). For the drought and salt stress treatments, plants were transferred to 10% (w/v) PEG-4000 and 150 μM NaCl solutions, respectively. The young leaves from control (untreated) and different treatments were harvested at 6 h (hour), 12 h, 24 h and 48 h. All treatments were performed with three independent biological replicates, and the samples were frozen in liquid nitrogen and stored at − 80 °C.
Identification of banana WOX (MaWOX) genes
The complete genome sequence of Musa acuminata (DH-Pahang v2) was downloaded from the banana genome hub (Droc et al. 2013; http://banana-genome.cirad.fr/). The WOX protein sequences of A. thaliana, Oryza sativa and Zea mays were downloaded from The Arabidopsis Information Resource (TAIR) (Swarbreck et al. 2007; http://www.arabidopsis.org), Oryzabase (Kurata and Yamazaki 2006; https://shigen.nig.ac.jp/rice/oryzabase/) and MaizeGDB (Lawrence et al. 2004; https://www.maizegdb.org/) databases. The BLASTP search was performed in the banana genome using WOX protein sequences from the selected plant species (Arabidopsis, rice and maize) as queries (E-value < 10–10). The conserved domain database search (Marchler-Bauer et al. 2010) was used to confirm the presence of the homeobox domain (PF00046) in the extracted genes from the banana genome. Expasy program (Gasteiger et al. 2003) was used to calculate parameters like molecular weight, peptide length and isoelectric point in the identified protein sequences. Each MaWOX gene has been given a unique name based on the BLAST results and their similarity with AtWOX genes (Table 1).
Phylogenetic analysis of MaWOX proteins
The WOX proteins from monocots (Oryza sativa and Zea mays) as well as from dicots (A. thaliana, Citrullus lanatus, and Cucumis sativus) were considered to analyse the evolutionary relationship with MaWOX proteins. Sequences of WOX proteins were extracted from respective genome databases Oryzabase (https://shigen.nig.ac.jp/rice/oryzabase/) for rice, MaizeGDB (https://www.maizegdb.org/) for Z. mays, TAIR (https://www.arabidopsis.org/) for A. thaliana, Dicots PLAZA 3.0 (Proost et al. 2015; https://bioinformatics.psb.ugent.be/plaza/versions/plaza_v3_dicots/) for C. lanatus and Cucumis sativus v1.0. (https://genome.jgi.doe.gov/portal/cucumber/cucumber.home.html) for C. sativus. Multiple sequence alignment was done using Clustal X (Larkin et al. 2007; http://www.clustal.org/) and a neighbor-joining tree with bootstrap value 1000 was built using MEGA X software (Kumar et al. 2018; https://www.megasoftware.net/). The final tree was improved manually using iTOL tool (Letunic and Bork 2021; https://itol.embl.de/).
Gene structure, conserved motif and domain analyses
The coding and genomic sequences of the MaWOX genes were extracted from the banana genome hub (http://banana-genome.cirad.fr/) and uploaded on Gene Structure Display Server 2.0 (Hu et al. 2015; http://gsds.gao-lab.org/) to understand the exon–intron patterns and position of homeodomain in each MaWOX gene. The location of the homeodomain was extracted using the HMMER web server (Finn et al. 2011; http://www.ebi.ac.uk/Tools/hmmer). Further, MEME (http://meme-suite.org/) software was used to analyze the motifs present in WOX genes (Bailey et al. 2015). Motifs with E-value < 10–10 were retained for analysis.
Subcellular localization and chromosomal distribution of MaWOX genes
The genomic coordinates of all MaWOX genes were extracted by running BLASTN against the banana genome database (http://banana-genome.cirad.fr/). The chromosomal localization of the genes was displayed using the MapGene2Chrom (http://mg2c.iask.in) tool (Jiangtao et al. 2015). The syntenic relationship of WOX genes in A. thaliana and M. acuminata was drawn using circa (http://circos.ca/) software (Musliner et al. 1993). The subcellular localization of MaWOX proteins is predicted using Plant-mPLOC software (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/) (Chou and Shen 2010).
Cis-acting element analysis of MaWOX promoters
The 2000 bp regions upstream of the MaWOXs coding region were downloaded from the NCBI database. Subsequently, cis-acting regulatory elements in the promoter sequences of MaWOX gene family were analyzed using Plant-CARE software (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Lescot et al. 2002; Wang et al. 2019).
RNA isolation and cDNA synthesis
The total RNA was isolated from different tissues using the Spectrum™ Plant Total RNA Kit (Sigma–Aldrich, USA) and given DNase (Thermo Scientific, USA) treatment to remove DNA contamination. The integrity and size of RNA were analyzed by agarose gel electrophoresis. DNase treated RNA was quantified and cDNA was synthesized as per the user manual of cDNA synthesis kit (Thermo Scientific, USA).
Expression pattern analysis of MaWOX genes by quantitative real-time PCR (qRT-PCR)
The qRT-PCR was executed using ABI 7500 sequence detector fast real-time PCR machine (Applied Biosystems, USA). Actin1 (GenBank Accession No. AF246288) was used as a housekeeping gene to normalize the variable expression of MaWOX genes in different tissues (Shivani et al. 2017). Primers were checked by amplification with conventional end-point PCR. A melting curve study was carried out using conventional qRT-PCR. The total volume of each reaction was kept 10 μl and components were as follows: (1) 1X SYBR Green Master mix (Applied Biosystems, USA), (2) 5 pmol of primers (forward and reverse), (3) 0.5 μl template (cDNA), and (4) sterile distilled H2O. Further, real-time PCR experiment was performed following the given parameters step (1) 95 °C 3 min, step (2) (95 °C 10 s, 55 °C 10 s, 72 °C 30 s) × 40 cycles, step (3) 95 °C 10 s, step (4) 65 °C 5 s and step (5) 95 °C 5 s following the thermal dissociation curve. The data were calculated in the form of fold change in expression with respect to control plants using the 2−ΔΔCt method (Livak and Schmittgen 2001). While ΔΔCt is defined as Ct target − Ct actin at timex − (Ct target − Ct actin) at time 0. Data was expressed in the form of a heat map using TBtools software (Chen et al. 2020). All the experiments were executed taking three biological replicates and were comprised of three technical replicates of each sample. For statistical significance, Statistical analysis was performed using Student’s unpaired t-test and statistical significance was checked at *P ≤ 0.01; **P ≤ 0.001; ***P ≤ 0.0001.
Results
Genome-wide identification, chromosomal location and physicochemical property analysis of MaWOX genes
Total 13 MaWOX candidate genes were found in the banana genome by considering WOX proteins from A. thaliana, Oryza sativa and Zea mays as query sequences in BLASTP search. The putative MaWOX genes which were common from all the species were considered for downstream analysis. The homeobox domain (PS50071) was identified in all 13 retrieved sequences using PROSITE, HMMER and NCBI-CDD based analyses (Table 1). All the identified genes have the complete open reading frame and are encoding WOX protein-like features. Hence, they have been designated as putative members of the WOX gene family in banana. Physicochemical properties of the predicted MaWOX proteins were depicted in Table 2. The length of the proteins varied between 173 amino acids (MaWOX9a) to 404 amino acids (MaWOX8b). The molecular weight of proteins ranged from 19.86 kDa (MaWOX4) to 43.56 kDa (MaWOX8b), while the isoelectric point from 5.22 (MaWOX13) to 9.54 (MaWOX4). It has been observed that all the predicted MaWOX proteins were localized in the nucleus highlighting their role as transcription factors. Furthermore, the chromosomal distribution of MaWOX genes has shown that all MaWOX genes are scattered on 8 chromosomes, except chromosome numbers 3, 5 and 11. The highest numbers of genes were positioned on chromosome 8 (3 genes), followed by chromosomes 7, 9 and 10 with two genes, while the rest of the chromosomes harbor one gene only. Fine mapping of MaWOX genes on the respective chromosome is given in Fig. 1.
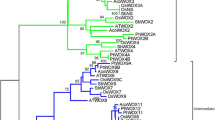
Phylogenetic analysis of MaWOX proteins
Phylogenetic analysis was performed to establish the evolutionary correlation of MaWOX genes with other plant species. We created neighbor-joining phylogenetic tree using a total of 83 WOX proteins from A. thaliana (15), Z. mays (20), O. sativa (13), C. lanatus (11), C. melo (12), C. sativus (11) and M. acuminata (13). The phylogenetic evaluation shows that all the banana WOX proteins are divided into three clades i.e., ancient, intermediate and modern clades (Fig. 2a). The total number of WOX sequences in ancient and intermediate clades were 14 and 24, respectively. Modern clade occupied 45 members, which is the highest as compared to others. The ancient clade harbors WOX13 and WOX14, while the intermediate clade has WOX8, WOX9a, WOX11a and WOX12 proteins. Further, the modern clade disseminated into subclades based on the presence of specific WOX proteins such as WOX2, WOX3, WOX4, WOX5 and Wuschel. Each subclade in modern clade has at least one member of each of the seven plants (A. thaliana, Z. mays, O. sativa, C. lanatus, C. melo, C. sativus and M. acuminata), indicating their potential role throughout evolution. Furthermore, it was observed that each clade was comprised of both monocots and dicots. Moreover, synteny analysis using circa software showed that the orthologues of all the 13 MaWOX genes were present in A. thaliana. For synteny analysis, 13 WOX gene sequences from M. acuminata genome and 15 WOX gene sequences from A. thaliana have been studied. Among these 13 orthologous pairs of M. acuminata—A. thaliana gene sequences mapped on A. thaliana genome, where one on each Machr2, Machr4, Machr6; two on each Machr7, Machr9, Machr10 and four on Machr8 (Fig. 2b).
Phylogenetic tree and synteny analysis. a Phylogenetic analysis of WOX proteins in A. thaliana (AT), Z. mays (ZM), O. sativa (OS), C. lanatus (CL), C. melo (CM), C. sativus (CS) and M. acuminata (MA). b Synteny analysis of Arabidopsis thaliana and Musa acuminata by CIRCOS Plot. Lines are ending with orthologs in M. acuminata and A. thaliana. Machr represents M. acuminata chromosomes while Atchr represents A. thaliana chromosomes
Gene structure and conserved motif analysis
Exon-introns and conserved motifs have been analyzed to understand the evolution of the MaWOX gene family. The intron–exon association of MaWOX genes is represented in Fig. 3. All the MaWOX genes in banana contain exons as well as introns mainly in their coding regions. A maximum of three exons and two introns are present in MaWOX genes. Total four conserved motifs were identified in each sequence and those were extracted using the MEME tool. Motifs 1 and 2 encoding for homeodomain are highly conserved in all MaWOXs protein sequences. A comprehensive list of predicted motifs in each MaWOX putative protein along with the sequence logo is represented in Fig. 4a, b.
MaWOX protein sequence analysis. a Representation of motif present in all MaWOX in banana. b Sequence logos of HD domains generated from all MaWOX. The x-axis represents the relative positions of the HD motifs and the y-axis represents the information content as measured in bits. 1,2,3 and 4 depict the different conserved motifs present in MaWOX protein sequences
Prediction of cis‑regulatory elements
The cis-elements in the promoter regions of 13 MaWOX genes were analyzed to further understand the regulatory network control of the MaWOX family genes. In addition to the core cis-elements such as TATA-box and CAAT-box, several other cis-elements were distributed into three categories viz., plant growth and development, stress responsiveness and hormone responsiveness (Fig. 5). In the first category (plant growth and development), cis-elements like light responsive (Box 4 and G-box), meristem specific expression (CAT-box), zein metabolism regulation (O2-site) and endosperm specific expression (GCN4-motif) were identified. In the second category (stress responsiveness), five types of cis-elements such as ARE for anaerobic induction, MBS and MYB for drought inducibility, MYC for low-temperature responsiveness and GC-motif for anoxic specific inducibility were identified. In the third category (hormone responsive), eight types of cis-elements involved in MeJA responsiveness (CGTCA and TGACG motifs), abscisic acid responsiveness (ABRE), salicylic acid responsiveness (TCA element), auxin responsiveness (AuxRR-core and TGA element) and gibberellin responsiveness (P-box and GARE motif) were observed. Therefore, considering the presence of the large number of cis-elements involved in different abiotic stresses, subsequent gene expression analysis was validated following cold, drought and salt stress treatments in banana plantlets.
Expression of the MaWOX genes in different tissues
The MaWOX genes were found to be differentially expressed in various tissues of banana. The expression of MaWOX genes was normalized with the expression of Actin1 gene (GenBank Accession No. AF246288). The expression in the early developing tissues viz., ECS, multiplying ECS, germinating embryos, young leaflet and node of germinated plantlets revealed that MaWOX6, MaWUS, MaWOX9a, MaWOX11a and MaWOX13 were highly expressed, while MaWOX6 has shown maximum (18 fold) expression (Figs. 6a and S1). The expression in late-stage developing tissues (unripe fruit peel and pulp, ripe fruit peel and pulp) showed that four MaWOX genes (MaWOX3, MaWOX6, MaWOX8 and MaWOX13) were highly expressed whereas MaWOX13 showed 120 fold higher expression in unripe pulp tissue (Figs. 6b and S1). The WOX4 and WOX1 genes were not expressed or expressed at a low level in banana plant tissues. The differential expression of most of the WOX genes belonging to modern, intermediate and ancient clades have been noticed while comparing their expression with WOX1 and WOX4 genes of the modern clade. These results suggest that the different WOX genes might be playing a crucial role at various stages of plant growth and development.
The MaWOX genes expression patterns in a early and b late developmental stages of tissues. Early developmental stages viz., ECS (Embryogenic cell suspension), EM (multiplying ECS), EG (germinating embryos), EGL (Embryo germinated young leaflet), EGN (Embryo germinated young node). Late developmental stages viz., UPL (Unripe pulp), UPE (Unripe peel), RPL (Ripe peel) and RPE (Ripe pulp). The heat map was generated by TBtools software. Columns and rows in the heat map represent tissues and MaWOX genes, respectively. Color scale indicates relative fold change in gene expression calculated by qRT-PCR analysis using three independent replicates of each tissue. The colour towards the red side shows the higher expression of MaWOX genes
Expression analysis of MaWOX genes under abiotic stress conditions
To investigate the effects of drought, salt and cold stresses, we analyzed the expression profiles of the MaWOX genes at four-time points (0 h, 6 h, 12 h, 24 h and 48 h) after stress exposure. The expression of MaWOX at different time points was compared with the untreated (0 h) condition. Under drought stress, the expression of MaWOX9a was upregulated 35 fold at 6 h; then expression declined gradually to ~ sevenfold at 48 h (Figs. 7a and S2). Other MaWOX genes viz., MaWOX1 and MaWOX3 were also expressed ≥ fivefold at 6 h and 12 h. In response to cold stress, only MaWOX3 and MaWOX8a have shown expression 2 and fourfold, respectively at 6 h (Figs. 7b and S2). In salt stress treatment, expression of MaWOX1, MaWOX4, MaWUS, MaWOX13 was significantly induced (≥ 11 fold) at 12 h compared to the untreated control. We noted expression of the MaWOX3, MaWOX11b and MaWOX8a upregulated by 830, 105 and 30 folds, respectively at 12 h as compared to control (Figs. 7c and S2). These observations have suggested that some of the MaWOX genes might have an important role in the regulation of abiotic stresses, particularly in the salt stress condition at the early stage of treatment.
Expression analysis of MaWOX genes during the exposure of various stress treatments in banana plants. a Drought stress (DS), b cold stress (CS) and c salt stress (SS) treatments. The heat map was generated by TBtools software. Columns and rows in the heat map represent stress treatments with time-points of tissue collection and MaWOX genes, respectively. Color scale indicates relative fold change in gene expression calculated by qRT-PCR analysis using three independent replicates of each treatment. The colour towards the red side shows the higher expression of MaWOX genes
Discussion
The WOX gene family is crucial for plants as its members control cell division and differentiation, which in turn affects plant growth and development (Breuninger et al. 2008; Costanzo et al. 2014; Dolzblasz et al. 2016). Many plants have been the subject of prior research on the WOX gene family, including cotton, tobacco and maize (Zhang et al., 2010; Yang et al. 2017; Zhou et al. 2018). However, there were no reports on the identification of this gene family in banana. Our study is focused on studying the role of MaWOX genes in development processes as well as in abiotic stress condition (drought, salt, and cold) in Musa acuminata cv. Grand Naine. The expression of these genes in different tissues as well as under different stress conditions revealed their promising role in the growth, development and abiotic stress management in banana plants (Alvarez et al. 2018). The 13 WOX genes found in the banana genome are concomitant with previous reports in different plant species. For instance, 15, 12, 13, 21 and 11 WOX genes were reported in the genome of A. thaliana, P. trichocarpa, O. sativa, Z. mays and Sorghum bicolor, respectively (Nardmann and Werr 2006; Zhang et al. 2010). The chromosomal localization suggested that all the MaWOX genes are distributed on 8 chromosomes, except chromosome numbers 3, 5 and 11. In the watermelon (C. lanatus), CtWOX2 gene which showed similarity with MaWOX2 found to be presented on chromosome 3.
The phylogenetic analysis with a total of 83 WOX proteins from A. thaliana, Z. mays, O. sativa, C. lunatus and C. sativa indicated that MaWOX proteins can be divided into three separate clades namely ancient, intermediate and modern clades. This pattern showed similarity to other plant species (Graaff et al. 2009). The banana derived WOX protein sequences presented in ancient (MaWOX13 and MaWOX8a), intermediate (MaWOX8b, MaWOX9b, MaWOX11a, and MaWOX11b) and modern (MaWOX6, MaWOX3, MaWOX1, MaWOX2, MaWOX4, MaWOX9a and MaWUS) clades showed consistent patterns of distributions as found previously in other plants (Lian et al. 2014). For example, A. thaliana derived AtWOX13, AtWOX10 and AtWOX14 (ancient clade), AtWOX11, AtWOX12, (intermediate clade) and AtWOX1, AtWOX2, AtWUS, AtWOX4, AtWOX5, AtWOX6 and AtWOX7 (WUS clade) proteins also showed the similar patterns of distribution in three clades.
The exon–intron distribution analysis predicted that there were 1–2 introns present in each MaWOX gene, that is similar to the arabidopsis, maize and rice (Zhang et al. 2010). Both the ancient and intermediate clades of MaWOX family genes have similar numbers of intron, as also reported previously in the C. sinensis plant, however, variations were noted in the genes belonging to the modern clade (Liu and Xu, 2018; Wang et al. 2019). The differences between the sequence of MaWOXs in the modern and other clades were further supported by the conserved motif analysis. Out of four (1–4), motifs 1 and 2 were found to be common in all MaWOX proteins. These two motifs are responsible for encoding the WOX homeodomain that contained helix-loop-helix-turn-helix structure consisting of fourteen amino acid residues i.e., Leu in helix 1, Pro, Ile, Ile and Leu in helix 2, Asn, Val, Trp, Phe, Gln, Asn and Arg in helix 3, Gly in loop and turn, were found to be presented in each MaWOX protein with high-level conservation. The third conserved motif (motif 3) was observed in the protein sequences belonging to the ancient and intermediate clades. This observation is also similar to WOX family members in the intermediate clade of walnut which is responsible for root development (Liu et al. 2014; Chang et al. 2020). All the members of the modern clade are having motif 4 (WUS-box motif), except in MaWOX4 which might be responsible for transcriptional inhibition and play a crucial role in flower and leaf development (Lin et al. 2013a).
The cis-regulatory elements analysis in MaWOXs promoter regions found to be responsive for hormonal (AuxRR-core, ABA, GA, Auxin Me-JA responsive), plant growth development (GCN4-motif, O2 site, CAT-box) and stress (ARE, MBS and GC-motif) responses in banana. The presence of a higher number of stress-responsive elements might be responsible for higher expression of MaWOX8a, MaWOX11b and MaWOX3 during abiotic stress treatments. Similar observations have been reported previously in other plants such as C. sinensis and G. hirsutum (Wang et al. 2019).
Previous reports were mainly focused on characterizing role of WOX genes in plant growth, especially during embryogenesis (Zuo et al. 2002; Breuninger et al. 2008; Zhang et al. 2014). Previous study has shown that the expression of WOX genes in cotton was low under different developmental stages emphasising that they are mainly involved in embryogenesis and hence their expression was restricted (Yang et al. 2017). However, the present study demonstrated that MaWOXs are not only involved in growth and development but also responded to various abiotic stress conditions. For instance, MaWOX13, MaWOX6, MaWOX3 and MaWOX8 showed higher expression in at least four tissues i.e., ECS, regenerated cells, unripe pulp and unripe peel. The higher expression of four MaWOX genes i.e., MaWOX9a, MaWOX13, MaWOX6 and MaWOX3 in unripe peel, ripe peel and ripe pulp tissues suggested their expression is regulated during fruit development stages in banana. Other MaWOX genes (MaWOX1, MaWOX4, MaWOX11a) showed either low (≤ twofold) or no expression in any of tested tissues. These observations indicated that the various MaWOX genes may have diverse role at different growth and development stages of banana. A previous study of MaWOX genes indicated that MaWOX11 in transgenic rice plants enhances drought resistance by regulating root hair development (Cheng et al. 2016). GhWOX13 gene is specifically expressed at a high level in cotton fiber as they influence cotton fiber development. Expression analysis showed that GhWOX13a and GhWOX1b genes were induced by multiple stresses, indicating that these genes might be involved during multiple stress (cold, heat and salt) conditions (Yang et al. 2017). In the present study, different time points of abiotic stress (drought, salt and cold) showed significant variation in MaWOX genes expression. Under drought stress, MaWOX1, MaWOX9a and MaWOX3 upregulated their expression at 6 h and 12 h, while in salinity stress all the MaWOX genes upregulated their expression at 12 h in which MaWOX8a, MaWOX11b and MaWOX3 depicted the highest expression. Banana is a highly salt sensitive crop and salt stress negatively impacts each physiological process that harmed its growth and development. As high soluble salt contents in soil causes accelerated breakdown of banana root system and ultimately compromise in the overall productivity (Turner et al. 2013; Sreedharan et al. 2015). However, most of the MaWOX genes were showing low expression in case of cold stress conditions. Henceforth, this study paved the way for functional validation of MaWOX genes that have shown promising expression responses at different developmental stages and under abiotic stress conditions.
Conclusion
Previous research has shown that the WOX gene family members play important roles in the regulation of plant growth and development. The results of the present study provide a genomic framework of MaWOX genes and contribute in understanding its evolutionary aspects as well as expression studies at different tissues developmental stages in banana. A total of 13 MaWOXs genes were identified in banana genome (DH-Pahang v2). Cis-acting element analysis of promoter region of MaWOX genes revealed their role in the regulation and synthesis of various plant hormones that are responsible for plant growth and stress response at different stages. The expression studies demonstrated in different banana tissues provided possible dimensions of MaWOX genes acting as conserved factors during the banana embryogenecity and various stress responses. Further, MaWOX13, MaWOX6, MaWOX3, MaWOX8 and MaWOX9a showed higher expression (≥ 20 fold) in developing tissues suggesting their role during growth and fruit development. While in salinity stress, MaWOX8a, MaWOX11b and MaWOX3 depicted the highest expression (≥ 100 fold). The results have provided useful information for further studies on functional aspects of MaWOXs, particularly for the MaWOX8a, MaWOX11b and MaWOX3 that may have the potential ability to respond under the salt stress conditions. Therefore the functional characterization of selected MaWOX genes (MaWOX8a, MaWOX11b and MaWOX3) could expand our understanding of their role in salt stress regulation resulting in amelioration of the incidence and consequences of salt stress in banana plants.
References
Alvarez JM, Bueno N, Cañas RA, Avila C, Cánovas FM, Ordás RJ (2018) Analysis of the WUSCHEL-RELATED HOMEOBOX gene family in Pinus pinaster: new insights into the gene family evolution. Plant Physiol Biochem 123:304–318. https://doi.org/10.1016/j.plaphy.2017.12.031
Bailey TL, Johnson J, Grant CE, Noble WS (2015) The MEME suite. Nucleic Acids Res 43(1):39–49. https://doi.org/10.1093/nar/gkv416
Breuninger H, Rikirsch E, Hermann M, Ueda M, Laux T (2008) Differential expression of WOX genes mediates apical-basal axis formation in the arabidopsis embryo. Dev Cell 14(6):867–876. https://doi.org/10.1016/j.devcel.2008.03.008
Chang Y, Song X, Zhang Q, Liu H, Bai Y, Lei X, Pei D (2020) Genome-wide identification of WOX gene family and expression analysis during rejuvenational rhizogenesis in walnut (Juglans regia L.). Forests 11(1):16. https://doi.org/10.3390/f11010016
Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R (2020) TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant 13(8):1194–1202. https://doi.org/10.1016/j.molp.2020.06.009
Cheng S, Zhou DX, Zhao Y (2016) WUSCHEL-related homeobox gene WOX11 increases rice drought resistance by controlling root hair formation and root system development. Plant Signal Behav 11(2):11130198. https://doi.org/10.1080/15592324.2015.1130198
Chou KC, Shen HB (2010) Plant-mPLoc: a top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS ONE 5(6):113–135. https://doi.org/10.1371/journal.pone.0011335
Costanzo E, Trehin C, Vandenbussche M (2014) The role of WOX genes in flower development. Ann Bot 114(7):1545–1553. https://doi.org/10.1093/aob/mcu123
D’hont A, Denoeud F, Aury JM, Baurens FC, Carreel F, Garsmeur O, Noel B, Bocs S, Droc G, Rouard M, Da Silva C (2012) The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature 488(7410):213–217. https://doi.org/10.1038/nature11241
Davey MW, Gudimella R, Harikrishna JA, Sin LW, Khalid N, Keulemans J (2013) A draft Musa balbisiana genome sequence for molecular genetics in polyploid, inter-and intra-specific Musa hybrids. BMC Genom 14(1):1–20. https://doi.org/10.1186/1471-2164-14-683
Devarajan R, Jayaraman JK, Somasundaram SM, Ragupathy S, Raman P, Sathiamoorthy K, Subbaraya U (2021) Genetic diversity in fresh fruit pulp mineral profile of 100 Indian Musa accessions. Food Chem 361:130080. https://doi.org/10.1016/j.foodchem.2021.130080
Dolzblasz A, Nardmann J, Clerici E, Causier B, van der Graaff E, Chen J, Davies B, Werr W, Laux T (2016) Stem cell regulation by arabidopsis WOX genes. Mol Plant 9(7):1028–1039. https://doi.org/10.1016/j.molp.2016.04.007
Droc G, Lariviere D, Guignon V, Yahiaoui N, This D, Garsmeur O, Dereeper A, Hamelin C, Argout X, Dufayard JF, Lengelle J (2013) The banana genome hub. Database. https://doi.org/10.1093/database/bat035
FAOSTAT (2020). http://www.fao.org/faostat/en/#data/TP. Accessed 3 Aug 2022
Finn RD, Clements J, Eddy SR (2011) HMMER web server: interactive sequence similarity searching. Nucleic Acids Res 39(2):29–37. https://doi.org/10.1093/nar/gkr367
Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A (2003) ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res 31(13):3784–3788. https://doi.org/10.1093/nar/gkg563
Graaff EVD, Laux T, Rensing SA (2009) The WUS homeobox-containing (WOX) protein family. Genome Biol 10(12):1–9. https://doi.org/10.1186/gb-2009-10-12-248
He P, Zhang Y, Liu H, Yuan Y, Wang C, Yu J, Xiao G (2019) Comprehensive analysis of WOX genes uncovers that WOX13 is involved in phytohormone-mediated fiber development in cotton. BMC Plant Biol 19(1):1–12. https://doi.org/10.1186/s12870-019-1892-x
Hirakawa Y, Kondo Y, Fukuda H (2010) TDIF peptide signaling regulates vascular stem cell proliferation via the WOX4 homeobox gene in arabidopsis. Plant Cell 22(8):2618–2629. https://doi.org/10.1105/tpc.110.076083
Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G (2015) GSDS 20: an upgraded gene feature visualization server. Bioinformatics 31(8):1296–1297. https://doi.org/10.1093/bioinformatics/btu817
Ikeda M, Mitsuda N, Ohme-Takagi M (2009) Arabidopsis WUSCHEL is a bifunctional transcription factor that acts as a repressor in stem cell regulation and as an activator in floral patterning. Plant Cell 21(11):3493–3505. https://doi.org/10.1105/tpc.109.069997
Jiangtao C, Yingzhen K, Qian W, Yuhe S, Daping G, Jing LV, Guanshan L (2015) MapGene2Chrom, a tool to draw gene physical map based on Perl and SVG languages. Yi Chuan 37(1):91–97. https://doi.org/10.16288/j.yczz.2015.01.013
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35(6):1547. https://doi.org/10.1093/molbev/msy096
Kurata N, Yamazaki Y (2006) Oryzabase. An integrated biological and genome information database for rice. Plant Physiol 140(1):12–17. https://doi.org/10.1104/pp.105.063008
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23(21):2947–2948. https://doi.org/10.1093/bioinformatics/btm404
Lawrence CJ, Dong Q, Polacco ML, Seigfried TE, Brendel V (2004) MaizeGDB, the community database for maize genetics and genomics. Nucleic Acids Res 32(1):393–397. https://doi.org/10.1093/nar/gkh011
Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30(1):325–327. https://doi.org/10.1093/nar/30.1.325
Letunic I, Bork P (2021) Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res 49(1):293–296. https://doi.org/10.1093/nar/gkab301
Lian G, Ding Z, Wang Q, Zhang D, Xu J (2014) Origins and evolution of WUSCHEL-related homeobox protein family in plant kingdom. Sci World J. https://doi.org/10.1155/2014/534140
Lin H, Niu L, McHale NA, Ohme-Takagi M, Mysore KS, Tadege M (2013a) Evolutionarily conserved repressive activity of WOX proteins mediates leaf blade outgrowth and floral organ development in plants. Proc Natl Acad Sci USA 110(1):366–371. https://doi.org/10.1073/pnas.1215376110
Lin H, Niu L, Tadege M (2013b) STENOFOLIA acts as a repressor in regulating leaf blade outgrowth. Plant Signal Behav 8(6):366–371. https://doi.org/10.4161/psb.24464
Liu W, Xu L (2018) Recruitment of IC-WOX genes in root evolution. Trends Plant Sci 23(6):490–496. https://doi.org/10.1016/j.tplants.2018.03.011
Liu J, Sheng L, Xu Y, Li J, Yang Z, Huang H, Xu L (2014) WOX11 and 12 are involved in the first-step cell fate transition during de novo root organogenesis in arabidopsis. Plant Cell 26(3):1081–1093. https://doi.org/10.1105/tpc.114.122887
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 25(4):402–408. https://doi.org/10.1006/meth.2001.1262
Lopez-Moya F, Escudero N, Zavala-Gonzalez EA, Esteve-Bruna D, Blázquez MA, Alabadí D, Lopez-Llorca LV (2017) Induction of auxin biosynthesis and WOX5 repression mediate changes in root development in arabidopsis exposed to chitosan. Sci Rep 7(1):1–14. https://doi.org/10.1038/s41598-017-16874-5
Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M (2010) CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res 39(1):225–229. https://doi.org/10.1093/nar/gkq1189
Mayer KF, Schoof H, Haecker A, Lenhard M, Jürgens G, Laux T (1995) Role of WUSCHEL in regulating stem cell fate in the arabidopsis shoot meristem. Cell 95(6):805–815. https://doi.org/10.1016/s0092-8674(00)81703-1
Mukherjee K, Brocchieri L, Bürglin TR (2009) A comprehensive classification and evolutionary analysis of plant homeobox genes. Mol Biol Evol 26(12):2775–2794. https://doi.org/10.1093/molbev/msp201
Musliner DJ, Durfee EH, Shin KG (1993) CIRCA: A cooperative intelligent real-time control architecture. IEEE Trans Syst Man Cybern Syst 23(6):1561–1574
Nardmann J, Werr W (2006) The shoot stem cell niche in angiosperms: expression patterns of WUS orthologues in rice and maize imply major modifications in the course of mono-and dicot evolution. Mol Biol Evol 23(12):2492–2504. https://doi.org/10.1093/molbev/msl125
Proost S, Van Bel M, Vaneechoutte D, Van de Peer Y, Inzé D, Mueller-Roeber B, Vandepoele K (2015) PLAZA 30: an access point for plant comparative genomics. Nucleic Acids Res 43(1):974–981. https://doi.org/10.1093/nar/gku986
Schoof H, Lenhard M, Haecker A, Mayer KF, Jürgens G, Laux T (2000) The stem cell population of arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100(6):635–644. https://doi.org/10.1016/s0092-8674(00)80700-x
Shimizu R, Ji J, Kelsey E, Ohtsu K, Schnable PS, Scanlon MJ (2009) Tissue specificity and evolution of meristematic WOX3 function. Plant Physiol 149(2):841–850. https://doi.org/10.1104/pp.108.130765
Shivani AP, Tiwari S (2019) Enhanced Agrobacterium-mediated transformation efficiency of banana cultivar Grand Naine by reducing oxidative stress. Sci Hortic 246:675–685. https://doi.org/10.1016/j.scienta.2018.11.024
Shivani AP, Sharma V, Kaur N, Kaur N, Pandey P, Tiwari S (2017) Genome-wide analysis of transcription factors during somatic embryogenesis in banana (Musa spp.) cv. Grand Naine. PLoS ONE 12(8):e0182242. https://doi.org/10.1371/journal.pone.0182242
Sreedharan S, Shekhawat UK, Ganapathi TR (2015) Constitutive and stress-inducible overexpression of a native aquaporin gene (MusaPIP2; 6) in transgenic banana plants signals its pivotal role in salt tolerance. Plant Mol Biol 88(1):41–52. https://doi.org/10.1007/s11103-015-0305-2
Swarbreck D, Wilks C, Lamesch P, Berardini TZ, Garcia-Hernandez M, Foerster H, Li D, Meyer T, Muller R, Ploetz L, Radenbaugh A (2007) The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Res 36(1):1009–1014. https://doi.org/10.1093/nar/gkm965
Tripathi L, Tripathi JN, Tushemereirwe WK (2008) Rapid and efficient production of transgenic East African highland banana (Musa spp.) using intercalary meristematic tissues. Afr J Biotechnol 7(10):1438–1445
Tripathi L, Ntui VO, Tripathi JN (2019) Application of genetic modification and genome editing for developing climate-smart banana. Food Energy Secur 8(4):00168. https://doi.org/10.1002/fes3.168
Turner NC, Colmer TD, Quealy J, Pushpavalli R, Krishnamurthy L, Kaur J, Singh G, Siddique KH, Vadez V (2013) Salinity tolerance and ion accumulation in chickpea (Cicer arietinum L.) subjected to salt stress. Plant Soil 365(1):347–361. https://doi.org/10.1007/s11104-012-1387-0
Tushemereirwe W, Kangire A, Ssekiwoko F, Offord LC, Crozier J, Boa E, Rutherford M, Smith JJ (2004) First report of Xanthomonas campestris pv. musacearum on banana in Ugan. Plant Pathol 53(6):802. https://doi.org/10.1111/j.1365-3059.2004.01090.x
Ueda M, Zhang Z, Laux T (2011) Transcriptional activation of arabidopsis axis patterning genes WOX8/9 links zygote polarity to embryo development. Dev Cell 20(2):264–270. https://doi.org/10.1016/j.devcel.2011.01.009
van Asten PJ, Fermont AM, Taulya G (2011) Drought is a major yield loss factor for rainfed East African highland banana. Agric Water Manag 98(4):541–552. https://doi.org/10.1016/j.agwat.2010.10.005
Wang P, Guo Y, Chen X, Zheng Y, Sun Y, Yang J, Ye N (2019) Genome-wide identification of WOX genes and their expression patterns under different hormone and abiotic stress treatments in tea plant (Camellia sinensis). Trees 33(4):1129–1142. https://doi.org/10.1007/s00468-019-01847-0
Yang Z, Gong Q, Qin W, Yang Z, Cheng Y, Lu L, Ge X, Zhang C, Wu Z, Li F (2017) Genome-wide analysis of WOX genes in upland cotton and their expression pattern under different stresses. BMC Plant Biol 17(1):1–7. https://doi.org/10.1186/s12870-017-1065-8
Zhang X, Zong J, Liu J, Yin J, Zhang D (2010) Genome-wide analysis of WOX gene family in rice, sorghum, maize, arabidopsis and poplar. J Integr Plant Biol 52(11):1016–1026. https://doi.org/10.1111/j.1744-7909.2010.00982.x
Zhang Y, Wu R, Qin G, Chen Z, Gu H, Qu LJ (2011) Over-expression of WOX1 leads to defects in meristem development and polyamine homeostasis in arabidopsis. F J Integr Plant Biol 53(6):493–506. https://doi.org/10.1111/j.1744-7909.2011.01054.x
Zhang F, Wang Y, Li G, Tang Y, Kramer EM, Tadege M (2014) STENOFOLIA recruits TOPLESS to repress ASYMMETRIC LEAVES2 at the leaf margin and promote leaf blade outgrowth in Medicago truncatula. Plant Cell 26(2):650–664. https://doi.org/10.1105/tpc.113.121947
Zhang N, Huang X, Bao Y, Wang B, Liu L, Dai L, Chen J, An X, Sun Y, Peng D (2015) Genome-wide identification and expression profiling of WUSCHEL-related homeobox (WOX) genes during adventitious shoot regeneration of watermelon (Citrullus lanatus). Acta Physiol Plant 37(11):1–2. https://doi.org/10.1007/s11738-015-1964-y
Zhao Y, Hu Y, Dai M, Huang L, Zhou DX (2009) The WUSCHEL-related homeobox gene WOX11 is required to activate shoot-borne crown root development in rice. Plant Cell 21(3):736–748. https://doi.org/10.1105/tpc.108.061655
Zhou X, Guo Y, Zhao P, Sun MX (2018) Comparative analysis of WUSCHEL-related homeobox genes revealed their parent-of-origin and cell type-specific expression pattern during early embryogenesis in tobacco. Front Plant Sci 8(9):311. https://doi.org/10.3389/fpls.2018.00311
Zuo J, Niu QW, Frugis G, Chua NH (2002) The WUSCHEL gene promotes vegetative-to-embryonic transition in Arabidopsis. Plant J 30(3):349–359. https://doi.org/10.1046/j.1365-313x.2002.01289.x
Acknowledgements
The authors express their gratitude to the National Agri-Food Biotechnology Institute (NABI), Department of Biotechnology (DBT), Government of India for research facilities and support. The present research was also supported by the Biotechnology Industry Research Assistance Council (BIRAC) for a banana biofortification project grant to NABI. RC and SS are thankful to the Regional Center of Biotechnology, Faridabad, for Ph.D. registration. Authors acknowledge to DBT-eLibrary Consortium (Del-CON) for providing access to online journals.
Funding
This work was supported by Core Research Grant of National Agri-Food Biotechnology Institute (NABI) and partially supported by Biotechnology Industry Research Assistance Council (BIRAC) through banana biofortification project grant (BIRAC/TechTransfer/08/I2/QUT-BBF). We greatly acknowledge research fellowship support to RC and SS from the Council of Scientific and Industrial Research (CSIR) and the Department of Biotechnology (DBT), Government of India, respectively. The funders had no role in the design of the study; in the collection, analyses and interpretation of data; in the writing of the manuscript; or in the decision to publish the result.
Author information
Authors and Affiliations
Contributions
ST conceived the idea, designed the experiment, analyzed the data; KK, RC and SS prepared the materials and performed bioinformatics analysis drafted the manuscript; RC and SS did qRT-PCR analysis; KK and ST contributed to the refining and writing of the manuscript. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Research involving human participants and/or animals
The authors declare that the present research does not involve any human participants and/or animals.
Supplementary Information
Below is the link to the electronic supplementary material.
13205_2022_3387_MOESM1_ESM.pptx
Fig. S1 Expression profiling of MaWOX genes during different stages of developmental stages of banana. Early developing tissues including ECS (Embryogenic cell suspension), EM (multiplying ECS), EG (germinating embryos), EGL (embryo germinated young leaflet), EGN (embryo germinated young node) and late-developing tissues including UPL (Unripe pulp), UPE (Unripe peel), RPL (Ripe peel) and RPE (Ripe pulp). Bars in graphs show the relative expression in term of fold change calculated by qRT-PCR analysis using three independent replicates. The error bars indicates standard error of mean. Statistical analysis was performed using Student’s unpaired t-test and statistical significance was checked at *P ≤ 0.01; **P ≤ 0.001; ***P ≤ 0.0001. Fig. S2 Expression profiling of MaWOX genes during drought stress, cold stress and salt stress treatments. Bars in graphs show the relative expression of genes at different time-points in terms of fold change calculated by qRT-PCR analysis using three independent replicates. The error bars indicate standard error of mean. Student’s unpaired t-test was performed for statistical analysis. Asterisk * (*P ≤ 0.01; **P ≤ 0.001; ***P ≤ 0.0001) are level of significance
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chaudhary, R., Singh, S., Kaur, K. et al. Genome-wide identification and expression profiling of WUSCHEL-related homeobox (WOX) genes confer their roles in somatic embryogenesis, growth and abiotic stresses in banana. 3 Biotech 12, 321 (2022). https://doi.org/10.1007/s13205-022-03387-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-022-03387-w