Abstract
WUSCHEL-related homeobox (WOX) gene family has important role in plant developmental process and represents a class of plant specific transcription factors that are involved in early phase of embryogenesis by coordinating gene transcription. Recent available pineapple whole genome sequences provide a base for detail comprehensive phylogenetic analysis of the WOX genes in pineapple. Ten WOX genes were found in pineapple genome and were located on 6 chromosomes. On the basis of phylogenetic analysis, WOX gene family was divided into three clades and structural analysis revealed that WOX gene family is highly conserved in pineapple. Gene structure analysis revealed that five genes had two exons with only one intron. Expression profiles based on RNA-Seq of different tissues revealed seven genes showed very low expression in almost all tissues (leaf, flower, root, fruit, stamen, sepal, petal and ovule) at all developmental stages. AcoWOX13 exhibited relatively higher expression in all tissues while the highest expression was shown by AcoWUS at ovule stage 1. Expression analysis under abiotic stresses analyzed by qRT-PCR, showed a dynamic response of WOX genes to abiotic stresses. Interestingly qRT-PCR had result pattern similar to RNA-Seq data as AcoWOX13 had high expression for all abiotic stresses while other WOX genes were down-regulated. This is the first genome-wide study of the WOX genes in pineapple coupled with high throughput RNA-seq for the identification, structural and functional analysis of 10 WOX genes in pineapple. Our study provides more insight for future studies regarding the functions of WOX genes in pineapple.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The WUSCHEL-related homeobox (WOX) genes are specifically expressed in plants and WOX proteins, form a large family, which is a subgroup of the homodomain (HD)-containing transcription factors. Typically the homodomain contains 60 amino acid residues with the helix-loop-helix-turn-helix structure (Kamiya et al. 2003). First studied homeobox gene was in Drosophila (Gehring et al. 1994; van der Graaff et al. 2009). After that in most eukaryotes a number of homeobox members have been identified (Lian et al. 2014). WOX (WUSCHEL-related homeobox) is the member of ZIP superfamily belonging to homeobox proteins family (Bharathan et al. 1997). Phylogenetic analyses have divided the WOX gene family into three major clades: WUX, intermediate, and ancient clade. The WUX and intermediate clades with representatives found in ferns and seed plants, whereas the ancient clade with representatives found in all lineages of green algae and land plants (Nardmann et al. 2009; van der Graaff et al. 2009).
Homeobox transcription factors play an important role in the developmental process of eukaryotes, as illustrated by the animal homeobox transcription factors HOX proteins (van der Graaff et al. 2009). In Drosophila melanogaster HOX genes play a role in homeotic mutations in which one body part transform into another which suggest that HOX proteins are involved in body patterning along the main body axis (Gehring 1993). A characteristic DNA segment of Homeotic genes, the homeobox encodes homeotic proteins domain. The homeo domain binds to specific DNA sequences, whereby the homeotic proteins play a role in gene regulatory function. Fusing the protein-coding sequences of the normal Antennapedia gene to an inducible promoter and reintroduce to the germline of flies, it has been possible to alter the body plan in a predicted way by transforming head structures into thoracic structures (Gehring 1987). Homeobox transcription factors also play a wide variety of roles in plants. The WUSCHEL (WUS) homeobox transcription factor is the significant member of the one of plant homeobox transcription factor families, the WUS homeobox (WOX) protein family (van der Graaff et al. 2009). Members of WOX family in plants play vital role in physiological and developmental processes. The genome of model plant Arabidopsis thaliana consists of at least 15 members of WOX gene family, and certain members of this family are essential for the vital processes like stem cell maintenance, organ formation and embryonic patterning (Laux et al. 1996; Haecker et al. 2004; van der Graaff et al. 2009). Arabidopsis WUSCHEL (AtWUS) is able to maintain stem cell homeostasis in the shoot apical meristem (SAM) at all development stages and in wus mutant the SAM maintenance is disrupted both embryonically and postembryonically (Laux et al. 1996; Mayer et al. 1998). WOX5 is required for the same function in the apical meristem of root (Sarkar et al. 2007). WOX3 play its role in lateral stamens and stamens development in flower and also the development of lateral stipules of the leaf (Matsumoto and Okada 2001; Nardmann et al. 2004), While WOX4 is required for cambium tissue activity in Arabidopsis main stem (Suer et al. 2011). WOX6 acts as a key regulator in ovule development (Park et al. 2005). WOX9 is involved in cell division maintenance and also control center zone premature differentiation in SAM (Wu et al. 2005). WOX2 and WOX8 is required for embryo formation and differentiation and both are co-expressed in egg cells and zygotes (Haecker et al. 2004). WOX gene family was studied in several plants including Arabidopsis, rice and maize (Kamiya et al. 2003; Nardmann et al. 2007), however, the role of this family in pineapple is not yet determined.
Pineapple (Ananas comosus L.) is an important fruit crop famous for its aroma and taste and a high valued fruit crop in horticultural industries in many countries. It is a tropical plant belongs to family Bromeliaceae, subfamily Bromelioideae, in the order Bromeliales. It’s a native crop of South America and in terms of its production it is third most important tropical fruit crop after banana and mango all over the world. In term of its consumption pineapple is used as fresh fruit, canned slices and juice, it is also the source of a meat-tenderizing enzyme bromelain, a valuable pharmaceutical agent. Based on traditional morphology, pineapple is a diploid (2n) perennial monocotyledonous plant consist of two genera, Ananas and Pseudananas having nine species (Smith 1979). MD-2 has gained popularity over other pineapple varieties because of its production, taste, quality, aroma, internal browning resistance and resilience to chilled storage. All these qualities have made MD2 a significant fruit crop in pineapple industry (Redwan et al. 2016).
Abiotic stresses such as cold, heat, salinity and drought have a devastating effect on plant growth and yield production (Suzuki et al. 2014). As its sessile lifestyle, plants are constantly exposed to a broad range of environmental stresses as high salt, drought and extremes of temperature. Due to these abiotic stress factors growth of plant is retarded and result in significant loss of plant productivity (Lata et al. 2011). More than 10% of arable land is affected by salinity and drought and more than 50% of important yield crop production is reduced worldwide (Bray et al. 2000). These abiotic stresses can also affect different stages of plant development and several stresses often concurrently affect the plants (Chinnusamy et al. 2004).
In this study, we identified 10 genes in WOX homeobox gene family in pineapple. These WOX family genes were grouped into three clades based on their phylogenetic relationship and were located specific chromosomes. Gene structure, protein structure, protein motifs and RNA Seq data for different tissues were also investigated for further study. Expression profiles under four stress treatments (cold, heat, salt, and drought) were evaluated to determine the responses of WOX family genes to abiotic stresses in the pineapple variety MD2. Our results will provide novel insights into the stress responses of WOX family genes and promote a better perceptive to understand about the utility and functions of WOX family genes in pineapple.
Results
Identification of WUSCHEL-Related Homeobox (WOX) Gene Family in Pineapple
To identify the WOX gene family in pineapple (Ananas comosus), the Hidden Markov Model (HMM) profile of the WOX domain (PF00046) was downloaded from the Pfam database. For the identification of WOX genes of pineapple, the HMM profile of the WOX domain was used to query pineapple genome database as well as non-redundant protein database with the help of the BLAST program. Ten WOX gene sequences were identified from the HMMER database in pineapple genome.
Phylogenetic Analysis of WOX Genes
To study the evolutionary history of pineapple WOX genes, phylogenetic analysis was carried out using neighbor joining (NJ) and maximum likelihood (ML) methods with the aligned WOX protein family sequences by MEGA 6.0. 100 iterations bootstrap test was performed based on the alignment of the WOX domain sequences. To demonstrate the possible evolutionary history of the WOX family in pineapple, we aligned the homeo-domains of pineapple WOX proteins against the WOX homeo-domains from Arabidopsis, rice, sorghum and poplar. Based on phylogenetic tree, there were 16 WOX genes in Arabidopsis, 14 WOX genes in rice, 11 WOX genes in sorghum, 18 WOX genes in poplar against 10 pineapple WOX genes. All of these 70 WOX genes from all the five plants were divided into 3 clades, such as WUS clade, intermediate clade and ancient clade. WUS clade contained 8 Arabidopsis, 7 rice, 11 popular, 5 sorghum and 5 pineapple WOX genes, intermediate clade included 5 WOX genes of Arabidopsis, 6 WOX genes of rice, 4 WOX genes of popular, 5 WOX genes of sorghum and 3 WOX genes of pineapple while ancient clade consist of 3 Arabidopsis, 1 rice, 3 popular, 1 sorghum against 2 WOX genes of pineapple (Fig. 1).
Phylogenetic tree of Arabidopsis, Rice and pineapple WOX gene family. The un-rooted phylogenetic tree was constructed using MEGA 6.0 and the Neighbour-Joining method. The full length amino acid sequences were aligned by using ClustalX. The bootstrap test was performed with 100 iterations. AT (Arabidopsis), Os (Rice), Pt (Poplar), Sb (Sorghum) and Aco (Pineapple)
Exon/Intron Structure Analysis and Identification of Conserved Motifs and their Structure
Number of exons and introns were calculated from the gene structures of WOX genes of pineapple. AcoNS, AcoWOX3, AcoWOX5 genes of WUS clade and AcoWOX14 and AcoWOX13 genes of ancient clade had 2 exons and one intron. AcoWOX11 gene from WUS clade and AcoWOX16 gene of intermediate clade had 3 exons and 2 introns. AcoWUS gene of WUS clade and AcoWOX2 gene from intermediate clade had only one exon without intron. AcoWOX9 gene from intermediate clade was the largest gene as it had 4 exons and 3 introns. (Figure 2) as listed in Table 1. The phylogenetic relationship and classification of WOX gene in pineapple were further supported by motif analysis. Four conserved motifs for pineapple WOX genes were sensitized by motif analysis using MEME software (Fig. 3). Moreover the structural information of theses motifs gives further insight to understand WOX genes in pineapple (Fig. 4).
Characteristics of Pineapple WOX Genes
Information about all these 10 WOX genes such as gene ID, locus, isoionic point (IP), protein molecular weight (MW), ORF lengths, number of amino acid and number of introns and exons. The value of isoionic point (IP) varied from 5.64 (AcoWOX13) to 10.02 (AcoWOX14). The corresponding molecular weight varied from 17.21 KDa to 45.71 KDa for AcoWOX2 and AcoWOX9 respectively. ORF length and number of amino acids ranged from 474 to 1272 and 158 to 424 respectively (Table 1).
Gene Location of WOX Genes on Pineapple Chromosomes
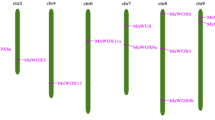
Mapinspect software was used to locate pineapple WOX genes on chromosome which will provide an insight into the organization of WOX genes in pineapple. Based on the available pineapple genome sequence, the pineapple WOX gene mapping revealed that 9 WOX genes out of 10 pineapple WOX genes were distributed among six chromosomes out of total 25 pineapple chromosomes, while gene AcoWOX9 was located on scaffold_1266 which has not still been mapped onto a chromosome. Three WOX genes were located on chromosome 23. Chromosomes 4, 8, 24 and 25 had only one WOX genes each, showing the fewest WOX genes among all of the chromosomes studied (Fig. 5).
Expression Profiles of WOX Genes
RNA-Seq is emerging cost effective, high-throughput and powerful tool for transcriptome profiling (Wang et al. 2009). It is an attractive tool to detect and quantify gene expression and especially for low-expressed genes (Mortazavi et al. 2008). RNA-seq data represents a high level of reproducibility in technical as will in biological replicates (Marioni et al. 2008).
RNA-seq analysis for different tissues at various developmental stages were reported from recently sequenced pineapple genome (Ming et al. 2015). Hierarchical clustering analyses for expression of WOX gene family in pineapple revealed that gene AcoWOX13 expressed in all of 8 different tissue at every stage while other genes of this family had either no expression or very low expression but in ovule, gene AcoWUS had more high expression than gene AcoWOX13 (Fig. 6). Moreover in stage 1of ovule, AcoWUS had the highest expression. Gene AcoWOX13 had higher expression in root while moderate expression in all stages of fruit and in all other tissue at all stages (Fig. 6). RNA-Seq was confirmed and validated by using qRT-PCR. Three different genes were selected and were tested by qRT-PCR for five various tissues i.e. root, fruit, flower, leaf and stamen. The results obtained were accordant with RNA-Seq expression data of these genes (Fig. 7).
Expression Profiles of Pineapple WOX Genes under Abiotic Stresses
Production of many plants crops are adversely affected by different abiotic stresses like cold, heat, salt or drought (Bhattacharjee et al. 2015). WOX genes play an important role in plant development (Lian et al. 2014). We investigate the functions of WOX genes correlated with their expression, 10 WOX genes were subjected to the qRT-PCR to analyze the expression profiles of WOX genes under cold, heat, salt, and drought treatments in ‘MD2’ variety of pineapple. To investigate the pineapple WOX genes in response to cold stress, plants were subjected to 4 °C as a cold stress. In cold treatment the expression level of most of WOX genes were down-regulated. WOX gene AcoNS had moderate expression at 24 h treatment while AcoWOX13 had moderate expression at 48 h treatment but high significant expression at 24 h treatment in response to cold. (Figure 8). Heat stress is another important abiotic stress that also has drastic effects on plant growth and development and causes major loss to crop yield. (Cai et al. 2015). To examine whether WOX genes were involved in the response of pineapple to heat stress plants were kept at 45 °C. All genes except AcoWOX13 were down-regulated while AcoWOX13 had highly significant expression both at 24 and 48 h with 24 h was the highest. Salt stress is also a major stress which adversely affects the plant and cause major loss in crop yield. To test whether WOX genes in the pineapple genome are involved in the response of pineapple to salt stress, we examined the expression patterns of all the WOX genes in pineapple under salt stress treatment by qRT-PCR (Fig. 8). Among 10 WOX genes in pineapple only AcoWOX13 was highly expressed at both 24 and 48 h with maximal at 48 h. Drought is another major problem and drought stress can also affect crop yield. To observe if WOX genes are involved in the response of pineapple to drought stress, 400 mMol/ml manitol was used to perform drought stress trial. Total of 10 WOX genes, AcoNS at 24 h had moderate expression and AcoWOX13 had highly significant expression at both 24 and 48 h with the highest expression at 48 h while all other genes were down-regulated. (Figure 8).
Heat map of expression profiles of WOX genes under abiotic stresses [cold (4 °C), heat (45 °C), Salt (NaCl) and drought (Manitol)]. qRT-PCR was used to analyze the relative expression level of each WOX gene. The expression level of pineapple Actin was used as the internal control to standardize the RNA samples for each reaction, and the expression at 0 h was set as 1 (data not shown). Expression level can be understood using the given scale
Discussion
The concept of genome-wide analysis has become a major approach for studying gene functional analysis, in reference to their evolution and structure. In this study, comparative analysis of the WOX family across species allows us to inquire various functions of the pineapple WOX family members and helps to assist further gene function analysis. Recently genome wide analysis has become very useful for gene structure and to understand its phylogeny. Evolution created high degree of variation in the shape of organs in plants (Lian et al. 2014). WOX genes are considered to play vital role in various biological, physiological and developmental processes in many plant species. The WOX genes encode WOX homeodomain-containing proteins which act as transcription factors that are involved in various biological processes and regulate the expression of genes in a spatial, temporal and tissue-specific pattern (Gehring 1987; Wolberger 1996). Genome wide study in many plant species about WOX gene family indicates that there are 15 WOX genes in Arabidopsis, 13 in rice, 11 in sorghum, 12 in poplar and 21 WOX genes in maize (Zhang et al. 2010). Vitis vinifera has 12 VvWOX genes (Gambino et al. 2011). Selaginella kraussiana has 8 SkWOX genes (Ge et al. 2016). In the present study, we identified 10 WOX genes in pineapple genome, through a combination of bioinformatics approaches and analyzed their genomic locations and phylogenetic relationship in pineapple, compared their expression profile in different tissues at different stages through RNA-seq and exposed these genes to various abiotic stresses.
Phylogenetic, Exon/Intron Structure Analysis and Identification of Conserved Motifs
Phylogenetic analysis reveals the resemblance of genes on basis of their protein structure. WOX gene family was divided into 3 clades on the basis of their structural similarity. The close evolutionary relationship among WOX genes in Arabidopsis, rice and pineapple orthologs (Fig. 1). Using the alignment of the amino acid homodomain sequences of WOX genes of pineapple, Arabidopsis and rice, we constructed phylogenetic tree. The phylogenetic analyses of WOX gene family in pineapple showed the typical subdivision into three major evolutionary lineages, with a major clade containing the WUS and most WOX proteins, confirming previous studies in Arabidopsis (Haecker et al. 2004; Deveaux et al. 2008; Vandenbussche et al. 2009).
Three orthologs of Arabidopsis ATWOX6 were found during phylogenetic analysis of pineapple, rice, Arabidopsis, poplar and sorghum WOX genes i.e. ATWOX1, PtWOX1A, PtWOX1B.. This feature was also reported in grass genomes (Nardmann et al. 2007). Therefore, the functions of the members of WOX family are distinct in different species, which makes the phylogenetic studies crucial to investigate. To get further insight into WOX genes, the gene structure is important to understand. Introns length, position and phase can also represent the phylogenetic relationships of the gene family (Li et al. 2006; Guo et al. 2008) and we presumed the gene structure of WOX genes of pineapple. Similar gene structures of intron number and length in each clade were observed. For instance, 3 of 5 genes in the WUS clade had two exons and one intron except AcoWOX11 and AcoWUS. Intermediate clade had similar exon/intron pattern. Ancient clade had nearly same gene structure but had variations in number of introns and exons. All these results confirm the authenticity of the phylogenetic analysis. Interestingly, the length of the AcoWOX9 was 7 kb with third intron length was more than 3 kb (Fig. 2). Our results are in line with the phylogenetic assessment in rice, sorghum, maize, Arabidopsis and poplar, where there were very similar topologies with some exceptions (Zhang et al. 2010).
Protein structure and motif of WOX gene family in pineapple showed close resemblance to that of Arabidopsis and rice. The WOX protein motifs of all the three plants were highly conserved and also the protein structure of WOX of pineapple had higher similarity with rice. Ten motifs were identified for WOX protein for all the 3 plants with little variations. Variation among motifs holds some great significance in many biological phenomena. Pineapple genome had 25 chromosomes. Ten WOX genes identified in pineapple genome were unevenly distributed on 6 chromosomes. These WOX genes can be studied for different biological processes. Gene expression patterns provided important clues for gene function; thus, we conducted RNA-Seq analysis to study the expression profile of various tissue at different developmental stage for five stages of stamen, four stages of sepal, seven stages of ovule and three stages of petal development while RNA-Seq for flower, root, leaf, six stages for fruit was obtained by Ming et al. 2015. We saw some unusual expression pattern as most of the WOX genes were either not expressed or very low expression, except AcoWOX13, that had shown expression in every tissue at all developmental stages. Interestingly AcoWUS had the highest expression in ovule stage S1 (Fig. 6). No such results were reported by WOX gene family in other plant species.
Plants grow in intricate environments and bear many stresses, such as heat, cold, soil salinization and drought. These abiotic stresses cause loss in crop production and affect quality (Yamaguchi-Shinozaki et al. 1999; Singh et al. 2002). Plants evolve to resist damage by regulating plant-specific signals through gene expression by specific physiological and metabolic pathways (Rushton and Somssich 1998; Yamaguchi-Shinozaki and Shinozaki 2006). Transcription factors are a special type of regulators with highly conserved specific DNA-binding domains involved in stress resistance (Riechmann et al. 2000; Singh et al. 2002). Many evidences confirm that plant growth, development, and response to environmental stress are largely regulated at transcriptional level (Baena-González et al. 2007; Baena-González and Sheen 2008; Buscaill and Rivas 2014). In the present study, our data showed unique expression pattern as most of the genes had very low expression value and down-regulated except AcoWOX13, which showed highly significant expression in all of abiotic stresses with drought stress at 48 h was the maximum. The result obtained from both qRT-PCR analysis and RNA-Seq data suggested that both results had a strong correlation as only WOX gene AcoWOX13 was highly expressed. The expression data we described here will help to understand define conditions in which WOX functional defects may be expected. Our results will provide a novel insights into the stress responses of WOX genes and promote a better perceptive to understand about the utility and functions of WOX genes in pineapple.
Conclusions
The sequences of the 10 members belonging to the WOX gene family have been compiled from the pineapple genome database, and were divided into three clades in phylogenetic tree. Gene structure, protein structure and motif structures were designated. Also few basic characteristics of WOX genes (molecular weight, iso-electric point, ORF length) were identified. High-throughput sequencing RNA-Seq expression profile among different tissues revealed candidate genes with low expression profile except AcoWOX13 with high expression in almost all tissues followed by AcoWUS with high expression only in ovule, sepal and pistil. A vital role of WOX genes was observed in female gametophyte development in pineapple that can be utilized to study pineapple plant reproduction for its improvement and exposure of pineapple MD2 variety to four abiotic stresses (cold, heat, salt and drought) and genes response to these stresses was assessed and WOX genes expression was analyzed by using qRT-PCR.
Materials and Methods
Identification and Multiple Sequence Alignments and Phylogenetic Analysis of WOX Genes
The conserved domain of WOX based on a Hidden Markov Model (HMM) (PF00046) was downloaded from the Pfam protein family database (http://pfam.sanger.ac.uk/). As revealed by previous reports that members of WOX gene family are conserved particularly for homeo-domain at sequence level (Nardmann et al. 2009). We examine sequences homologous to AtWOX using TBLASTN, with default parameter settings and WOX homeo-domain sequences as queries at E-value cut-off of 1e-5. Following databases were used to retrieve WOX family proteins by TBLASTN: The Arabidopsis Information Resource (TAIR) database, the Rice Genome Annotation Project (RGAP) database, the Plant Transcription Factor Database (PlantTFDB), the National Center for Biotechnology Information (NCBI) database. To identify the WOX coding genes of Ananas comosus L, we used the HHMprofile of the WOX domain as a query to perform a HMMER search (http://hmmer.janelia.org/) against pineapple genome. All the redundant and non-WOX family sequences with different identification numbers were removed using the UniProt, the SMART, and Sanger database, respectively (Zhang et al. 2010). To determine the evolutionary relationship of WOX gene of pineapple with other plants, the predicted pineapple WOX genes (AcoWOX) were classified into subgroups based on sequence alignment with clearly classified WOX genes from Arabidopsis and rice. Multiple sequence alignments of amino acid sequences of pineapple and those of rice and Arabidopsis were conducted using MUSCLE 3.6 with the default parameter settings. To obtain a better alignment, results were adjusted manually based on corresponding amino acids location in the WOX motif using GeneDoc (version 2.6.002) software. MEGA (version 6.0) software was used to construct phylogenetic tree using the Neighbor-Joining (NJ) method with the following parameters: JTTmodel, poisson correction methods, pairwise deletion of gaps, and 100 bootstrap. Moreover maximum likelihood, minimal evolution and PhyML methods were used for the tree construction to validate the results of the NJ method.
Exon-Intron Structure and Motif Analysis
The DNA and cDNA sequences related to each predicted gene from the pineapple genome and the information about the distribution pattern of AcoWOX intron were obtained from the (http://www.phytozome. net/pineapple.php) UniPort (http://www.uniprot.org/). Comparisons to the Pfam database were used to identify motifs present in the AcoWOX protein sequences.
Characteristics of Pineapple WOX Genes
The gene length, open reading frame (ORF), number of amino acid, isoionic point (IP) of WOX genes and molecular weight for each WOX protein of pineapple was calculated using the ExPASy server (http://web.expasy.org/compute_pi/). Expression Heat map was constructed for 10 WOX genes form RNA Seq of reported tissues in pineapple.
Gene Location on Chromosomes
To obtain the location of genes on chromosomes, we drew a map of the distribution of AcoWOX genes throughout the pineapple genome. Gene start and stop points were calculated from pineapple genome for WOX genes and Mapinspect software was used to identify the position 10 WOX genes on 25 chromosomes on the bases of the start site of gene.
RNA-Seq Analysis
Healthy plants from MD2 variety were selected, samples collection was carried out at different developmental stages from different tissues. Prior to total RNA extraction samples were quickly stored in liquid nitrogen. Total RNA was extracted from specified tissues using RNA extraction kit (Omega Bio-Tek, Shanghai, China). mRNA was isolated from total RNA followed by fragmentation and priming. Then first strand and second strand cDNA was synthesized. After that purify the double stranded cDNA. Then construct End Repair cDNA library. Adopter was attached to cDNA followed by purification. 250-400 bp was set as insert site and 350-500 bp was set as final size of library. cDNA library was constructed and send for sequencing. (RNA library prep kit). RNA-Seq data was analyzed as (Trapnell et al. 2012). The RNA-Seq for different tissues i.e. flower, leaf, root, six developmental stages of fruit was obtained from Ming et al. 2015, six stages of stamen, four stages of sepal, three stages of petal and seven stages of ovule was conducted and WOX family genes expression were determined. Expression Heat map was constructed for expression profiles of 10 WOX genes of reported tissues in pineapple.
Quantitative Real-Time PCR Analysis
Gene specific primers were designed according the manufacturer’s instructions for the Bio-Rad Real-time PCR system (Foster City, CA, USA). Primers for 10 pineapple WOX genes and one Aco-actin were designed for qRT-PCR using Primer Quest Tool. To determine the expression analysis of selected WOX genes, RNA extraction Kit (Omega Bio-Tek, Shanghai, China) was used for isolation of total RNA from treated and untreated (control) fully grown pineapple leaves following manufacturer’s protocol. Subsequently First-strand cDNA was synthesized with gDNA remover using cDNA extraction kit (Trans Gen, Beijing, China) and 20-fold dilution was done, and 2-μL of cDNA was used in 20-μL of total for Bio-Rad Real-time PCR system with the following program: 95 °C for 30s; 95 °C for 5 s and 60 °C for 34 s; with 39 cycles (Cai et al. 2017).
Plant Material and Growth Conditions
Pineapple (Ananas comosus) variety MD2 was supplied by the Qin Lab (www.qinlab.net), Haixia Institute of Science and Technology, Fujian Agriculture and Forestry University, Fujian, China. Pineapple crowns were grown on soil mix [peat moss:perlite,2:1(v/v)] in plastic pots placed in greenhouse at 25 °C with light availability of 60–70 mMol photons m−2 s−1, under 65–70% humidity, with 16-hourslight/8-h dark photo-period (Su et al. 2017).
Abiotic Stress Treatment
Fully grown pineapple plants were exposed to cold, heat, drought and salt stresses. For cold stress plants were kept at 4 °C (Chen et al. 2016), heat stress at 45 °C, for drought and salt stress Manitol and NaCl in concentration of 400 mMol/ml were applied respectively. Samples were collected after 24 h and 48 h incubation. Untreated plant samples were also collected that were used as a control. All samples were immediately stored in liquid nitrogen prior to total RNA extraction.
Abbreviations
- WOX:
-
WUSCHEL-related homeobox
- RNA-Seq:
-
RNA sequencing
- qRT-PCR:
-
quantitative real-time PCR
- WUS:
-
Wuschel
- NaCl:
-
sodium chloride
References
Baena-González E, Sheen J (2008) Convergent energy and stress signaling. Trends Plant Sci 13:474–482
Baena-González E, Rolland F, Thevelein JM, Sheen J (2007) A central integrator of transcription networks in plant stress and energy signalling. Nature 448:938–942
Bharathan G, Janssen BJ, Kellogg EA, Sinha N (1997) Did homeodomain proteins duplicate before the origin of angiosperms, fungi, and metazoa? Proc Natl Acad Sci U S A 94:13749–13753
Bhattacharjee A, Ghangal R, Garg R, Jain M (2015) Genome-wide analysis of homeobox gene family in legumes: identification, gene duplication and expression profiling. PLoS One 10:e0119198
Bray EA, Bailey-serres J, Weretilnyk E (2000) Responses to abiotic stresses. In: Buchanan BB, Gruissem W, Jones RL (eds) Biochemistry and molecular biology of plants. American Society of Plant Biologists, Rockville, pp 1158–1203
Buscaill P, Rivas S (2014) Transcriptional control of plant defence responses. Curr Opin Plant Biol 20:35–46
Cai H, Cheng J, Yan Y, Xiao Z, Li J et al (2015) Genome-wide identification and expression analysis of calcium-dependent protein kinase and its closely related kinase genes in Capsicum Annuum. Front Plant Sci 6:737
Cai H, Zhao L, Wang L, Zhang M, Su Z, et al (2017) ERECTA signaling controls Arabidopsis inflorescence architecture through chromatin-mediated activation of PRE1 expression. New Phytol 214:1579–1596
Chen C, Zhang Y, Xu Z, Luan A, Mao Q, et al (2016) Transcriptome profiling of the pineapple under low temperature to facilitate its breeding for cold tolerance (J.-H. Liu, Ed.). PLoS One 11: e0163315
Chinnusamy V, Schumaker K, Zhu J-K (2004) Molecular genetic perspectives on cross-talk and specificity in abiotic stress signalling in plants. J Exp Bot 55:225–236
Deveaux Y, Toffano-Nioche C, Claisse G, Thareau V, Morin H et al (2008) Genes of the most conserved WOX clade in plants affect root and flower development in Arabidopsis. BMC Evol Biol 8:291
Gambino G, Minuto M, Boccacci P, Perrone I, Vallania R et al (2011) Characterization of expression dynamics of WOX homeodomain transcription factors during somatic embryogenesis in Vitis Vinifera. J Exp Bot 62:1089–1101
Ge Y, Liu J, Zeng M, He J, Qin P et al (2016) Identification of WOX family genes in Selaginella Kraussiana for studies on stem cells and regeneration in Lycophytes. Front Plant Sci 7:93
Gehring WJ (1987) Homeo boxes in the study of development. Science 236:1245–1252
Gehring WJ (1993) Exploring the homeobox. Gene 135:215–221
Gehring WJ, Affolter M, Burglin T (1994) Homeodomain Proteins. Annu Rev Biochem 63:487–526
Guo A-Y, Zhu Q-H, Gu X, Ge S, Yang J et al (2008) Genome-wide identification and evolutionary analysis of the plant specific SBP-box transcription factor family. Gene 418:1–8
Haecker A, Groß-Hardt R, Geiges B, Sarkar A, Breuninger H et al (2004) Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis Thaliana. Development 131:657–668
Kamiya N, Nagasaki H, Morikami A, Sato Y, Matsuoka M (2003) Isolation and characterization of a rice WUSCHEL-type homeobox gene that is specifically expressed in the central cells of a quiescent center in the root apical meristem. Plant J 35:429–441
Lata C, Yadav A, Pras M (2011) Role of plant transcription factors in abiotic stress tolerance. Ine: Abiotic stress response in plants - physiological, biochemical and genetic perspectives. InTech
Laux T, Mayer KF, Berger J, Jürgens G (1996) The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122:87–96
Li X, Duan X, Jiang H, Sun Y, Tang Y et al (2006) Genome-wide analysis of basic/helix-loop-helix transcription factor family in Rice and Arabidopsis. Plant Physiol 141:1167–1184
Lian G, Ding Z, Wang Q, Zhang D, Xu J (2014) Origins and evolution of WUSCHEL-related homeobox protein family in plant kingdom. ScientificWorldJournal 2014:534140
Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y (2008) RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res 18:1509–1517
Matsumoto N, Okada K (2001) A homeobox gene, PRESSED FLOWER, regulates lateral axis-dependent development of Arabidopsis flowers. Genes Dev 15:3355–3364
Mayer KF, Schoof H, Haecker A, Lenhard M, Jürgens G et al (1998) Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95:805–815
Ming R, VanBuren R, Wai CM, Tang H, Schatz MC et al (2015) The pineapple genome and the evolution of CAM photosynthesis. Nat Genet 47:1435–1442
Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5:621–628
Nardmann J, Ji J, Werr W, Scanlon MJ (2004) The maize duplicate genes narrow sheath1 and narrow sheath2 encode a conserved homeobox gene function in a lateral domain of shoot apical meristems. Development 131:2827–2839
Nardmann J, Zimmermann R, Durantini D, Kranz E, Werr W (2007) WOX gene phylogeny in Poaceae: a comparative approach addressing leaf and embryo development. Mol Biol Evol 24:2474–2484
Nardmann J, Reisewitz P, Werr W (2009) Discrete shoot and root stem cell-promoting WUS/WOX5 functions are an evolutionary innovation of angiosperms. Mol Biol Evol 26:1745–1755
Park SO, Zheng Z, Oppenheimer DG, Hauser BA (2005) The PRETTY FEW SEEDS2 gene encodes an Arabidopsis homeodomain protein that regulates ovule development. Development 132:841–849
Redwan RM, Saidin A, Kumar SV (2016) The draft genome of MD-2 pineapple using hybrid error correction of long reads. DNA Res 23:427–439
Riechmann JL, Heard J, Martin G, Reuber L, Jiang C et al (2000) Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290:2105–2110
Rushton PJ, Somssich IE (1998) Transcriptional control of plant genes responsive to pathogens. Curr Opin Plant Biol 1:311–315
Sarkar AK, Luijten M, Miyashima S, Lenhard M, Hashimoto T et al (2007) Conserved factors regulate signalling in Arabidopsis Thaliana shoot and root stem cell organizers. Nature 446:811–814
Singh K, Foley RC, Oñate-Sánchez L (2002) Transcription factors in plant defense and stress responses. Curr Opin Plant Biol 5:430–436
Smith LB (1979) (Bromeliaceae) 3. Bromelioideae. Hafner, New York
Su Z, Wang L, Li W, Zhao L, Huang X, et al (2017) Genome-wide identification of auxin response factor (ARF) genes family and its tissue-specific prominent expression in pineapple (Ananas comosus). Trop Plant Biol 10:86–96
Suer S, Agusti J, Sanchez P, Schwarz M, Greb T (2011) WOX4 imparts Auxin responsiveness to cambium cells in Arabidopsis. Plant Cell 23:3247–3259
Suzuki N, Rivero RM, Shulaev V, Blumwald E, Mittler R (2014) Abiotic and biotic stress combinations. New Phytol 203:32–43
Trapnell C, Roberts A, Goff L, Pertea G, Kim D et al (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and cufflinks. Nat Protoc 7:562–578
van der Graaff E, Laux T, Rensing SA (2009) The WUS homeobox-containing (WOX) protein family. Genome Biol 10:248
Vandenbussche M, Horstman A, Zethof J, Koes R, Rijpkema AS et al (2009) Differential recruitment of WOX transcription factors for lateral development and organ fusion in petunia and Arabidopsis. Plant Cell Online 21:2269–2283
Wang Z, Gerstein M, Snyder M (2009) RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10:57–63
Wolberger C (1996) Homeodomain interactions. Curr Opin Struct Biol 6:62–68
Wu X, Dabi T, Weigel D (2005) Requirement of Homeobox gene STIMPY/WOX9 for Arabidopsis meristem growth and maintenance. Curr Biol 15:436–440
Yamaguchi-Shinozaki K, Shinozaki K (2006) Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol 57:781–803
Yamaguchi-Shinozaki K, Kasuga M, Liu Q, Miura S, Shinozaki K (1999) Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat Biotechnol 17:287–291
Zhang X, Zong J, Liu J, Yin J, Zhang D (2010) Genome-wide analysis of WOX gene family in Rice, sorghum, maize, Arabidopsis and poplar. J Integr Plant Biol 52:1016–1026
Acknowledgments
This work was supported by NSFC (31522009; 31470284; U1605212 to Y.Q.), Fujian Innovative Center for Germplasm Resources and Innovation project of Characteristic Horticultural Crop Seed Industry (KLA15001D), FAFU international collaboration project and Natural Science Foundation of Fujian Province (2017 J01601).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by: Hongwei Cai
Syed Muhammad Azam and Yanhui Liu are co-author
Rights and permissions
About this article
Cite this article
Rahman, Z.u., Azam, S.M., Liu, Y. et al. Expression Profiles of Wuschel-Related Homeobox Gene Family in Pineapple (Ananas comosus L). Tropical Plant Biol. 10, 204–215 (2017). https://doi.org/10.1007/s12042-017-9192-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12042-017-9192-9