Abstract
Chemotherapy-induced cognitive impairment (CICI) is widely recognized as a frequent adverse side effect following the administration of chemotherapeutic agents. This study aimed to explore the neuroprotective functions and mechanisms of microRNAs (miRNAs) mediated by dexmedetomidine (Dex) on cisplatin-induced CICI. The model rats received 5 mg/kg cisplatin injections once per week for 4 weeks. Dex (30 μg/kg) was administered before cisplatin treatment. The protective effects of Dex were evaluated using Morris water maze, Nissl staining, and transmission electron microscopy. Dex-mediated miRNAs were screened via miRNA sequencing. The effects of Dex and key miRNAs on mitochondrial DNA gene mt-ND1 and caspase-9 expression were tested. Dex exhibited a protective effect against decreased learning memory ability, hippocampal neuronal damage, and mitochondrial damage in CICI rats. Thirty-nine differentially expressed (DE) miRNAs were screened, 13 of which responded positively to Dex treatment. Gene Ontology annotation identified that DE miRNAs were mainly involved in transcription, DNA-templated. Kyoto Encyclopedia of Genes and Genomes pathway analysis showed that DE miRNAs were mainly involved in neuronal function and brain development-related pathways, such as axon guidance and calcium signaling pathways. Compared to cisplatin treatment, the expression of miR-429-3p responded more strongly to Dex treatment. In cisplatin-treated cultured hippocampal neurons, Dex treatment and miR-429-3p overexpression significantly increased mitochondrial DNA gene mt-ND1expression and decreased caspase-9 expression. Our study suggests that Dex alleviates CICI by modulating miR-429-3p expression in rats. Thus, Dex may be effective in preventing the side effects of cisplatin.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Chemotherapy-induced cognitive impairment (CICI) refers to impairment in cognitive function that often occurs in patients with tumors outside the central nervous system during diagnosis, treatment, or after treatment (Zhou et al. 2016). CICI is an ever-increasing problem of great clinical concern. Longitudinal studies have identified that nearly 40% of cancer patients develop CICI before treatment, nearly 75% during treatment, and 60% after treatment. Most recent studies regarding CICI have focused on breast cancer patients (Wefel et al. 2015). However, the underlying mechanisms involved in CICI are not well understood, and few drugs have been proven effective in reducing or preventing CICI.
There is emerging evidence that CICI also frequently develops in cancer patients treated with platinum-based compounds, such as cisplatin. Cis-diamineplatinum (II) dichloride (cisplatin), which is widely used in the treatment of ovarian, lung, and esophageal cancer, and other malignancies (Dasari and Tchounwou 2014; Ho et al. 2016), and recent studies have shown that cisplatin can cross the blood–brain barrier, elevate mitochondrial DNA (mtDNA) content, and decrease SOD activity in the hippocampus (Lomeli et al. 2017).
Dexmedetomidine (Dex) is a commonly used sedative and is notable for its ability to provide sedation without risk of respiratory depression. In recent years, Dex has attracted wide attention due to its potential neuroprotective effects. Dex has been shown to prevent brain injury caused by subarachnoid hemorrhage (Cosar et al. 2009). In addition, several studies have shown that Dex can significantly reduce hypoxia/re-oxygenation injury to hippocampal neurons in rats, which may be related to inhibition of excessive mitochondrial fission and mitochondrial apoptosis (Li et al. 2016).
MicroRNAs (miRNAs), as a family of small non-coding RNAs, play important roles in cell proliferation and apoptosis, tumorigenesis, and active immunity. There are many studies concerning miRNAs in cognitive impairment; for example, the up-regulation of miR-192-5p expression can restore neural function and attenuate cognitive impairment in depressed mice via the TGF-β1 signaling pathway (Tang et al. 2019). Studies have shown that miRNAs may be involved in the functional regulation of mitochondria, and thus in the regulation of cognitive damage (Ling et al. 2013; Reddy et al. 2017). Therefore, in this study, differential expression of miRNAs was identified in rats with cognitive damage to clarify the roles of miRNAs in cisplatin-induced CICI.
This study aimed to explore neuroprotective functions and mechanisms involving Dex and to provide new concepts for the therapeutic treatment of CICI. To characterize CICI in rats, we examined learning and memory capacity, hippocampal neuron survival, and mitochondrial functions. We further screened differentially expressed (DE) miRNAs by miRNA sequencing, and evaluated the effects of Dex in mediating key DE miRNAs on mitochondrial DNA gene and apoptosis of hippocampal neurons.
Materials and methods
Ethics statement
The animal experiments reported in this study, and all procedures involving the handling and treatment of rats during this study, were approved by the Institutional Animal Care and Use Committee of the Second Affiliated Hospital of Nanchang University. All experiments were performed according to the National Institutes of Health Guide for the Care and Use of Laboratory Animals. At the planned or humane endpoint, rats were euthanized by intraperitoneally injection of pentobarbital sodium (150 mg/kg). All efforts were made as much as possible to reduce animal suffering.
Animals and Dex treatments
Thirty male Sprague–Dawley rats (200–250 g) were housed in inlet valve-closed cages on a 12 h light/dark cycle (lights on 6:00 am). Water and food were available ad libitum. Cisplatin and Dex dosage and injection times followed previous reports (Zhou et al. 2016). Thirty rats were randomized into three groups and treated as follows: (1) intraperitoneally injected with physiologic (0.9%) saline per week for 4 weeks (control group); (2) injected with cisplatin (Sigma-Aldrich, Saint Louis, MO, USA) at 5 mg/kg per week for 4 weeks (model group); c) pretreated with Dex (BD123881, Bidepharm, Shanghai, China) at 30 μg/kg for 30 min prior to injection of cisplatin at 5 mg/kg per week for 4 weeks (Dex group). Each group contained ten rats.
Morris water maze (MWM)
Morris water maze equipment (Cat: XR-XM101) purchased from Shanghai Xinruan Information Technology Co., Ltd. MWM testing was performed in a black circular pool 160 cm in diameter and 50 cm in depth, and filled to a depth of 30 cm with warm water. The pool temperature was maintained at 25 ± 1 °C. An escape platform with 10 cm in diameter is placed 2 cm below the water surface. A camera connected to a computer was placed 2 m above the tank to record the rats’ routes, which were captured by digital tracking system and analyzed by SuperMaze Version 3.3.0.0 software.
The water tank was divided into nominal southeast, northeast, northwest, and southwest quadrants. A place navigation test was first performed. Rats were permitted to swim freely for 1 min to adapt to the water environment before training. A place navigation test was repeated for 4 days, four times each day. Rats were then allowed to stay on the platform for 20 s. If the underwater platform was not found by rats within 120 s, the experimenter guided them to find the platform and allowed them to stay on the platform for 20 s, recording the escape latency as 120 s. The interval of the four consecutive experiments was 20 s, and the escape latencies of each rat on the test day were calculated as the average escape latency of these four repeated escape experiments. After experiments, rats were thoroughly dried with towels and a hair drier, and then returned to their home cages.
Spatial probe testing was conducted on the sixth day. The escape platform was removed from the pool during spatial probe testing, which was performed to assess the rats’ ability to remember the location of the platform. During the probe test, rats were allowed to swim freely for 120 s with the platform removed from the pool. The swimming speed, swimming path and the numbers of the rats crossing the platform were recorded.
Nissl staining
A minimum of three slices of hippocampal tissues from each group of rats was dewaxed. The slices were stained with preheated 1% toluidine blue (60 °C) for 40 min, rinsed with distilled water three times, and then decolorized with an alcohol gradient (95%, 80%, 70%) for 3 min each to identify whether the Nissl granules were clear under the microscope. A light blue to colorless background was considered to be appropriate. Then, the slices were dehydrated with an ethanol gradient, cleared with xylene, and sealed with neutral balsam after drying. Nissl bodies in nerve cells were stained bluish violet as viewed under light microscopy. Image-J software was used to analyze images and to calculate nerve cell numbers in the hippocampal CA1 area (Zheng et al. 2019).
Transmission electron microscope (TEM) observations
Rat hippocampal tissues were fixed with 2.5% glutaraldehyde and 1% osmium acid. Hippocampal tissues were then dehydrated with an acetone gradient, embedded in Epon 812 epoxy resin, cut into ultra-thin sections using a microtome, and stained with lead citrate. Hippocampal neurons were observed under a JEM-1230 electron microscope (JEOL, Japan).
miRNA extraction and expression profiling
After behavioral analysis, hippocampal tissues were dissected from rats, and total RNA was extracted using a high-purity miRNA isolation kit (Takara Bio USA, Inc., USA) following the manufacturer’s instructions. The yield of RNA was quantified using a NanoDrop instrument (Invitrogen, Carlsbad, CA, USA). Complementary DNA (cDNA) libraries of miRNAs from rats hippocampal tissue were constructed according to published miRNA cloning protocols (Pandey et al. 2015; Boese et al. 2016). Approximately 20 μg of the extracted, purified RNA was sequenced on a NextSeq 500 (Illumina) platform. The remaining RNA was subjected to reverse transcription for subsequent miQPCR analysis. Fast-QC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) software was used to evaluate the overall quality of sequencing data, including the distribution of the mass value of the bases, distribution of the position of the mass values, and GC content.
DE miRNAs analysis
Clean NextSeq reads were mapped to the human genome using the miRBase 21.0 database and RfamBase, and sequences matching the above database were considered as known miRNA sequences. Reads per million miRNAs mapped (RPM) values were used to represent miRNA expression levels. DE miRNAs were analyzed using DESeq 2.0 (v1.8.3). P < 0.05 and |Log2Fold Change (Log2FC) |≥ 1 were used to screen DE miRNAs. The miRNA target prediction software Miranda (Score ≥ 150 and Energy < − 20) and RNAhybrid (Energy < − 25) was used to predict binding sites of the different miRNAs. The intersection of the two algorithms was taken as the final result of target gene prediction. Functional annotation and ontology analysis of candidate target genes predicted to be regulated by different miRNAs were performed using Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) functional classifications.
Validation of miRNA expression by miQPCR
To validate the reliability of Illumina sequence analysis, the expression of five miRNAs expressed in mouse hippocampal tissue was detected by miQPCR (a quantitative real-time polymerase chain reaction (qPCR) method for miRNA quantification). miQPCR was performed in 96-well plates using a Bio-Rad Real-time PCR Detection System (Bio-Rad, Hercules, CA, USA), according to the manufacturer’s protocols. All reactions were performed as three technical replicates, including a template-free (water) negative control. U6 served as an internal reference gene, and relative gene expression was standardized by the 2−ΔΔCt method. All primer sequences used in this study are listed in Supplementary Table 1.
Cell culture and treatments
Neonatal hippocampal neurons were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum in an incubator containing 5% CO2 at 37 °C. Experiments using hippocampal neurons were carried out at passages 3–8. When cells were approximately 80–90% confluent, new medium was added before cisplatin or Dex treatment. Addtionally, cisplatin and Dex dosage followed the reports (Lomeli et al. 2017; Hwang et al. 2013). In cisplatin experiments, cells were treated with 12.5 mg/L cisplatin for 24 h. In Dex experiments, cells were treated with 12.5 mg/L cisplatin and Dex (1 μM) for 24 h.
MiRNAs transient transfection
60 nM miR-429-3p mimic (Genepharma, Shanghai, China) was transfected into neonatal hippocampal neurons using Lipofectamine RNAiMAX (Invitrogen, Carlsbad, CA) compared to a scrambled control miRNA. Similarly, 60 nM antisense inhibitor miR-429-3p, or scrambled control antisense inhibitor, was transfected into neonatal hippocampal neuron cells using Lipofectamine RNAiMAX. Cells were incubated for 48 h and harvested with Trizol reagent (LifesTechnologies, Tokyo, Japan).
Immunofluorescence staining
Cultured neonatal hippocampal neurons were collected and fixed in 4% paraformaldehyde at 4 °C for 24 h. Cells were washed with PBS and sealed with 0.2% Triton X-100 PBS at room temperature for 1 h. Rabbit anti-caspase-9 polyclonal antibody (1:200, Abcam, Cambridge, MA, USA) was added and incubated overnight at 4 °C. Next, the cells were washed three times with PBS and incubated with DyLight 488 conjugated donkey anti-rabbit IgG mixture for 1 h at room temperature. Then, DAPI (1:500, Beyotime, Shanghai, China) was used to stain cells for 10 min at room temperature. After dyeing, the greenish Dylight 488 or the reddish Cy3 was observed using an Olympus confocal laser scanning microscope (Olympus Corporation, Tokyo, Japan). Japanese Olympus FV10 asw-3.1 software was used for imaging.
Statistical analysis
All data were analyzed using Graphpad Prism 6.0 software. Results are expressed as means ± SEM. Student’s t test was used for comparisons of two groups, and one-way ANOVA was used for comparisons involving multiple groups. P values ≤ 0.05 were considered statistically significant.
Results
Dex protects learning and memory ability of rats
The MWM experiments were conducted to determine the effects of cisplatin on the learning and memory abilities of rats, and to explore whether Dex exerted protective effect on such abilities. The results showed that swimming paths of Dex group animals were significantly shorter than that of the control and model groups (Fig. 1a). The swimming speed of the model group was significantly less than that of the control and model groups by the third and fourth days (P < 0.05; Fig. 1b). In addition, the escape latency time of the model group rats was always the longest over the three stages (days 1, 3, and 4), and showed significant differences on days 3 and 4. However, the escape latency time was significantly reduced to that of the control group level after Dex treatment (P < 0.05; Fig. 1c). These results indicated that Dex had a protective effect on cisplatin-induced impairment of learning and memory in rats.
Results of Morris Water Maze (MWM) test in rats from control, chemotherapy-induced cognitive impairment (CICI) model, and dexmedetomidine (Dex) groups. a Swimming paths of rats in each group in the MWM assay. b Average swimming speed of rats in each group. The swimming speed of rats treated with Dex was increased compared to those of the model group. ANOVA results of repeated measurements showed that there was a significant difference in the average swimming speed with increasing duration of treatment in each group (*indicates P < 0.05). c Post pairwise comparisons using Bonferroni method showed escape latency time (the time from launch to the first landing on the platform) of rats in each group. The escape latency of rats in the Dex group was significantly lower than those of model group animals on Days 3 and 4. (n = 5 per group, *indicates P < 0.05; **indicates P < 0.01)
Dex promotes survival of hippocampal neurons and maintains normal hippocampus morphology in rats
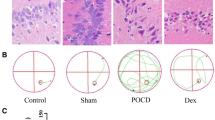
The survival of hippocampal neurons in each group was detected by Nissl staining. Results showed that compared with the control group, the survival of neurons in hippocampus tissues in the model group was significantly diminished (P < 0.05); However, Dex treatment increased the survival of neurons in hippocampus tissues compared with the model group (P < 0.05; Fig. 2a–c). TEM was utilized to observe the ultrastructure of hippocampal tissues, and indicated as the clear and complete nuclear membrane and the complete mitochondrial in the control group (Fig. 2d). Hippocampal neurons in the model group exhibited swollen neuronal cells, partial organelle damage, and destruction to the structural continuity of the cristae (Fig. 2e). Hippocampus neuronal morphology in the Dex group was intermediate between the two groups (Fig. 2f). Altogether, compared with the control group, cisplatin induced mitochondrial damage in hippocampal neurons, while Dex significantly reduced such damage.
Dex protected the survival of rat hippocampal neurons and hippocampal morphology. Representative Nissl-stained images of surviving hippocampal neurons of a control group, b model group, and c Dex group (×200 magnification); Representative transmission electron microscope (TEM) ultrastructural images of hippocampus tissues of d control group, e model group, and f Dex group (×20,000 magnification)
miRNA expression profiling of hippocampal tissue
We carried out miRNA sequencing to reveal miRNA expression profiles of rats in control, model, and Dex groups. The filtered results of sequence quality control data for all samples are shown in Supplementary Table 2. The quality of the reads was assessed using quality scores, which showed that 99.9% of reads were < 30, indicating very high-quality sequencing data (Supplement Figure 1).
DE miRNAs in model and Dex group hippocampus
We employed DESeq2.0 (P < 0.05 and |Log2Fold Change (Log2FC) |≥ 1) DE analyses to identify miRNAs with altered expression in the model and Dex groups. After probe screening and data normalization, DE fold-changes of 1.5 or higher were observed. We identified 39 significantly DE miRNAs between the Dex and model groups. These DE miRNAs included 24 upregulated miRNAs and 15 downregulated miRNAs (Fig. 3a). Meanwhile, we found 166 significantly DE miRNAs between the model and control groups. These 166 involved 17 upregulated miRNAs and 149 downregulated miRNAs (Fig. 3b). Interestingly, after further analysis, we found that compared with those in the healthy control group, 13 miRNAs were downregulated in the model group, whereas these levels were restored after Dex treatment (Fig. 3c, d). Thus, we selected these 13 candidate genes for further miQPCR validation.
Differentially expressed (DE) miRNAs in hippocampus tissues from rats administered CICI and Dex treatment. a Cluster map of miRNAs with significant differences between the control and model groups. b Cluster map of miRNAs with significant differences between the model and Dex groups. c, d revealed 13 DE miRNAs which were downregulated in the model group but upregulated in the Dex group
Prediction of DE miRNA targets and pathway analysis
The target genes of DE miRNAs though taking intersection of Miranda (Score ≥ 150 and Energy < − 20) and RNAhybrid (Energy < − 25) were 80,070 (Supplement Figure 2A), and the target genes of DE miRNAs were 17,482 between the model and Dex groups (Supplement Figure 2B).
To better understand the functions of Dex-mediated miRNAs, we mapped all DE miRNAs between the model and Dex groups to the GO and KEGG databases. The GO results indicated that these highly correlated target genes are involved in “transcription, DNA-templated”, “regulation of transcription, DNA-templated” and “positive regulation of transcription from RNA polymerase II promoter” (Fig. 4a). KEGG pathway analysis revealed crucial target gene functions, and the results indicated that these DE miRNAs mainly participate in axon guidance and calcium signaling pathways. In addition, several pathways (WNT and Hippo signaling pathways in cancer, the neurotrophin signaling pathway, and mitogen-activated protein kinase (MAPK) signaling pathway) are also involved, and these signaling pathways are all associated with neuronal function or brain development (Fig. 4b).
Validation of candidate DE miRNAs
To verify the concordance of DE miRNA expression with our miRNA sequencing data, we next confirmed the expression of 13 candidate miRNAs in these nine samples by miQPCR. Surprisingly, eight candidate miRNAs did not exhibit the same expression trend in samples from the same group, and thus the remaining five candidate miRNAs (mmu-miR-429-3p, mmu-miR-200a-5p, mmu-miR-6537-3p, mmu-miR-3100-3p, and mmu-miR-6965-5p) were confirmed at the next validation stage. To determine the expression pattern of all miRNAs used for technical validation, we then expanded the samples size to ten (control group, n = 10; model group, n = 10; Dex group, n = 10), and conducted miQPCR to further confirm expression of the five candidate miRNAs in the three groups.
As a result (shown in Fig. 5) only mmu-miR-429-3p and mmu-miR-6965-5p of the five genes had the expected expression patterns. Compared with the control group, the expression levels of mmu-miR-429-3p and mmu-miR-6965-5p were significantly decreased in the model group, and were significantly increased after Dex treatment. Moreover, the expression of mmu-miR-429-3p in the Dex group was higher than that of the control group, while the response of mmu-miR-6965-5p to Dex was not as profound as that of mmu-miR-429-3p. Therefore, we will conduct further exploratory experiments involving miR-429-3p.
Effects of Dex on expression of mtDNA and caspase-9 in hippocampal neurons
Immunofluorescence results showed that in hippocampal neurons, Dex treatment significantly decreased the expression of caspase-9, compared to the cisplatin-induced model group (Fig. 6a). In addition, the relative expression of mt-ND1, one of the major mtDNA genes, also increased after Dex treatment in cultured hippocampal neurons, suggesting Dex improved mtDNA damage derived from cisplatin induction (Fig. 6b; P < 0.05).
Cisplatin-induced caspase-9 expression and mitochondrial DNA loss in hippocampal neurons can be mitigated by Dex treatment. a Representative immunofluorescence images of caspase-9 expression in hippocampal neurons treated with cisplatin and Dex. b Expression of mitochondrial DNA gene mt-ND1 in hippocampal neurons exposed to Dex or cisplatin. Data are representative of three independent experiments, and expressed as means ± SD. # indicates P < 0.05 vs. the model group
Effects of miR-429-3p on expression of caspase-9 and mtDNA in hippocampal neurons
Previous studies have shown that cisplatin induces mitochondrially-mediated apoptosis in cultured hippocampal neurons and miRNA sequencing through caspase-9 activation (Lomeli et al. 2017). However, whether Dex-mediated miR-429-3p can protect against caspase-9 activation is unclear. Caspase-9 is a member of the cysteine aspartic acid protease (caspase) family. Under apoptotic stimulation, cytochrome c is released from mitochondria and binds with procaspase/Apaf-1, which activates caspase-9 by cleaving it at Asp315. Cleaved caspase-9 further activates other caspase proteins to initiate a caspase cascade that culminates in apoptosis (Kuwahara et al. 2000; Jiang et al. 2000). We used miR-429-3p mimics for transient transfection, and antisense RNAs for miR-429-3p inhibition, so as to alter the abundance of miR-429-3p. The effectiveness of the mimic and inhibitor manipulations was very high, as assessed by qPCR (Fig. 7a, b; P < 0.05). Immunofluorescence results showed that transfection of miR-429-3p mimics significantly decreased the expression of caspase-9 and increased the relative expression of mt-ND1 in hippocampal neurons (Fig. 7c, d; P < 0.05). However, miR-429-3p inhibitor transfection activated the expression of caspase-9 and reduced the expression of mt-ND1 in cultured hippocampal neurons (Fig. 7c, e; P < 0.05), suggesting that miR-429-3p might play important roles in regulating mtDNA damage and apoptosis in hippocampal neurons.
mir-429-3p mediated by Dex attenuated cisplatin-induced caspase-9 expression and mitochondrial DNA loss in hippocampal neurons. a, b Transfection efficiency of miR-429-3p mimics or inhibitor in hippocampal neurons. c–e miR-429-3p gain- and loss-of-expression regulated caspase-9 expression and mitochondrial DNA mt-ND1 levels in hippocampal neurons. Data are presented as means ± SD. *indicates P < 0.05 vs. scrambled miRNA
Discussion
Growing evidence shows that CICI frequently occurs in cancer patients treated with platinum-based compounds, such as cisplatin. However, therapeutic drugs and their underlying mechanisms remain elusive. Recently, Dex has attracted wide attention due to its potential neuroprotective effects. In this study, we first demonstrated that Dex treatment improved cognitive function in cisplatin-induced CICI rats. Then, we employed miRNA sequencing to investigate the miRNAs involved in the process of Dex-attenuated, cisplatin-induced CICI. We identified 13 DE miRNAs that were downregulated in the model group. Such downregulation was restored after Dex treatment, and KEGG pathway analysis revealed that DE miRNAs were mainly enriched in axon guidance and calcium signaling pathways. Subsequently, miQPCR validation indicated that the expression of miR-429-3p was significantly increased after treatment with Dex compared to that in the model group, which was consistent with miRNA sequencing data. Thus, we suspected that miR-429-3p might play a key role in improving cognitive function deficits in CICI rats by administration of Dex. Lastly, we validated our observation that Dex might attenuate CICI through regulation of mtDNA damage (mt-ND1) and apoptosis (caspase-9) via miR-429-3p.
The phenomenon of host neurological dysfunction caused by chemotherapy has been observed in several studies. A multilevel meta-analysis of cognitive function in survivors receiving chemotherapy (Ch+) showed that compared with healthy and chemotherapy-negative (Ch−) control groups, cognitive impairment was greater in patients receiving chemotherapy (Ch+) (Bernstein et al. 2017). Magnetic resonance imaging (MRI) and diffusion tensor imaging (DTI) in breast cancer patients showed decreased hippocampal volume, abnormal structure involving gray and white matter, and decreased ability of episodic memory (Filippi and Agosta 2016; Chanraud et al. 2010). Compared with the control group, the ability to engage in learning and memory was impaired in rats undergoing chemotherapy (Christie et al. 2012). Moreover, cisplatin, a chemotherapy drug, is widely used in the treatment of several malignancies, including ovarian, lung, and esophageal cancers, as well as other tumors. Recent studies have shown that cisplatin can accumulate in the hippocampus after crossing the blood–brain barrier and result in cognitive impairment. Mechanistically, cisplatin could interact with mtDNA causing DNA-Pt adducts which would result in the inhibition of mtDNA replication and transcription, mitochondrial function, and even apoptosis of neurocytes (Lomeli et al. 2017; Podratz et al. 2017). Similarly, in our research, we found that a 4-week duration of cisplatin injections could significantly impair the spatial learning and memory ability of rats, and significantly reduced the survival of neurons in hippocampus tissues, and that altered hippocampal neurons were abnormally shaped in cisplatin-treated rats.
Currently, however, few drugs have proven effective in attenuating or preventing CICI. In recent years, Dex has been shown to exert a neuroprotective effect by alleviating neuronal damage in the hippocampal CA1 region (Hwang et al. 2013). Dex is reported to prevent brain injury caused by subarachnoid hemorrhage through inducing neuroprotection by activating the MAPK pathway (Cosar et al. 2009). In rats with cerebral hemorrhage, Dex has been reported to enhance the expression of brain-derived neurotrophic factor to inhibit cell apoptosis, attenuate short-term and spatial learning, and memory damage (Hwang et al. 2013). Additionally, Dex contributes to cognitive improvement of rats with cognitive dysfunction. For example, Dex attenuates isoflurane-induced injury in the developing brain, providing neurocognitive protection in neonatal rats (Hwang et al. 2013). Dex may exert preventive and protective effects against anxiety-like behaviors and cognitive impairments in rats with post-traumatic stress disorder after repeated administration (Hwang et al. 2013). In our study, Dex administration improved memory ability in rats and effectively protected them from cognitive impairment induced by cisplatin. Furthermore, Dex promoted the survival of hippocampal neurons and maintained the normal morphology of hippocampus in rat, suggesting that Dex may be a potential drug in treatment and prevention of CICI.
Recently, miRNAs have been reported to play important roles in disease therapy. miRNA-based RNA interference (RNAi) therapy has the advantage of lower toxicity than other gene silencing methods (Lam et al. 2015; Anthiya et al. 2018). It has been reported that in the absence of induced toxicity, the delivery of miRNAs in vivo successfully inhibits tumor activity (Chen et al. 2015). Intravenous injection of anti-miRNAs could reduce cell proliferation and tumor size of glioblastomas in the brain and surrounding organs (Chen et al. 2015). However, so far, most studies have focused on the roles of miRNAs in tumor inhibition, with poor development of miRNA therapy for CICI. Therefore, we performed NGS and obtained miRNA profiles after CICI treatment with Dex. We identified 166 DE miRNAs (17 upregulated and 149 downregulated) involving control and model group animals, and 39 DE miRNAs (24 upregulated and 15 downregulated) in the model and Dex groups. Further analysis revealed that 13 DE miRNAs were downregulated in the model group compared with the control group, but were upregulated after Dex treatment. Further validation by miQPCR analysis revealed that mmu-miR-429-3p, mmu-miR-6537-3p, and mmu-miR-6965-5p exhibited the same expression patterns as revealed by miRNA sequencing.
miR-429 belongs to the miRNA-200 family and is derived from chromosome 1p33.36 (Feng et al. 2014; Peng et al. 2018). Numerous studies have shown that miR-200 family members, as epigenetic regulatory molecules, can control the proliferation and invasion of glioma cells, and modulate therapeutic responses and prognosis in glioma patients by targeting multiple downstream genes to regulate physiological and pathological processes (Qin et al. 2017; Zhang et al. 2012). Additionally, increased expression of miR-200 family members promotes neuronal differentiation (Zhao et al. 2017), while decreased expression of the miRNA-200 family promotes neuronal proliferation by targeting SOX2 and KLF4 expression (Dong et al. 2017; Pandey et al. 2015). Expression of miR-200 family members is strongly decreased in West Nile virus infection, ischemic and psychiatric states, and is usually accompanied by a reduction in dendritic spine density (Kumar and Nerurkar 2014). Because the miR-200 family includes host factors that can influence dendritic spine density, synaptic morphology, and function, their expression is particularly sensitive to environmental stress, injury or disease (Boese et al. 2016). Moreover, studies have shown that miR-429 plays a neuroprotective role in brain ischemia. It has been demonstrated that downregulated of miR-429 expression by lncRNA MALAT1 sponge action, can promote apoptosis of hippocampal neurons in hypoxic-ischemic brain damage (HIBD) mice via activation of WNT1 signaling. In addition, silencing of WNT1 signaling by miR-429 enhances the cerebral protective effects of Dex against HIBD mice (Fang et al. 2019). In our study, we also identified miR-200 family members, including the DE miRNA, miR-429-3p, which might contribute to the protective effects against CICI in rat. Further analyses are evidently required.
Functional analysis revealed that DE miRNAs are mainly involved in “transcription, DNA-templated” and “positive regulation of transcription from RNA polymerase II promoter”. Pathway analysis revealed that DE miRNAs are mainly involved in axon guidance, calcium, WNT, and MAPK signaling pathways. To date, these pathways have been demonstrated to be closely associated with neuronal function or brain development. For instance, activation of axon guidance factors and calcium signaling pathways could improve memory ability and attenuate cognitive impairment (Rosso and Inestrosa 2013; Yuan et al. 2016), and the Hippo signaling pathway is associated with memory performance and cognitive ability in mammals (Zhang et al. 2014; Xiao et al. 2011). Moreover, most of the above-mentioned signaling pathways are involved in the regulation of transcription by RNA polymerase II, suggesting that Dex might improve cognitive function via regulation of epigenetic blockade; this proposal needs further study.
Conclusions
This study is the first to show that Dex treatment can attenuate memory deficits and cognitive impairment caused by chronic cisplatin injection in rats, indicating that Dex might be a candidate drug to treat and prevent CICI. Additionally, we identified miRNA expression profiles and revealed that miR-429-3p might play key roles in protection against CICI by Dex in rats.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Anthiya S, Griveau A, Loussouarn C, Baril P, Garnett M, Issartel JP, Garcion E (2018) MicroRNA-based drugs for brain tumors. Trends Cancer 4(3):222–238. https://doi.org/10.1016/j.trecan.2017.12.008
Bernstein LJ, McCreath GA, Komeylian Z, Rich JB (2017) Cognitive impairment in breast cancer survivors treated with chemotherapy depends on control group type and cognitive domains assessed: a multilevel meta-analysis. Neurosci Biobehav Rev 83:417–428. https://doi.org/10.1016/j.neubiorev.2017.10.028
Boese AS, Saba R, Campbell K, Majer A, Medina S, Burton L, Booth TF, Chong P, Westmacott G, Dutta SM, Saba JA, Booth SA (2016) MicroRNA abundance is altered in synaptoneurosomes during prion disease. Mol Cell Neurosci 71:13–24. https://doi.org/10.1016/j.mcn.2015.12.001
Chanraud S, Zahr N, Sullivan EV, Pfefferbaum A (2010) MR diffusion tensor imaging: a window into white matter integrity of the working brain. Neuropsychol Rev 20(2):209–225. https://doi.org/10.1007/s11065-010-9129-7
Chen Y, Gao DY, Huang L (2015) In vivo delivery of miRNAs for cancer therapy: challenges and strategies. Adv Drug Deliv Rev 81:128–141. https://doi.org/10.1016/j.addr.2014.05.009
Christie LA, Acharya MM, Parihar VK, Nguyen A, Martirosian V, Limoli CL (2012) Impaired cognitive function and hippocampal neurogenesis following cancer chemotherapy. Clin Cancer Res 18(7):1954–1965. https://doi.org/10.1158/1078-0432.CCR-11-2000
Cosar M, Eser O, Fidan H, Sahin O, Buyukbas S, Ela Y, Yagmurca M, Ozen OA (2009) The neuroprotective effect of dexmedetomidine in the hippocampus of rabbits after subarachnoid hemorrhage. Surg Neurol 71(1):54–59. https://doi.org/10.1016/j.surneu.2007.08.020(discussion 59)
Dasari S, Tchounwou PB (2014) Cisplatin in cancer therapy: molecular mechanisms of action. Eur J Pharmacol 740:364–378. https://doi.org/10.1016/j.ejphar.2014.07.025
Dong H, Hao X, Cui B, Guo M (2017) MiR-429 suppresses glioblastoma multiforme by targeting SOX2. Cell Biochem Funct 35(5):260–268. https://doi.org/10.1002/cbf.3271
Fang H, Li HF, He MH, Yan JY, Yang M, Zhang FX, Wang RR, Wang QY, Zhang JP (2019) Long non-coding RNA MALAT1 sponges microRNA-429 to regulate apoptosis of hippocampal neurons in hypoxic-ischemic brain damage by regulating WNT1. Brain Res Bull 152:1–10. https://doi.org/10.1016/j.brainresbull.2019.06.004
Feng X, Wang Z, Fillmore R, Xi Y (2014) MiR-200, a new star miRNA in human cancer. Cancer Lett 344(2):166–173. https://doi.org/10.1016/j.canlet.2013.11.004
Filippi M, Agosta F (2016) Diffusion tensor imaging and functional MRI. Handb Clin Neurol 136:1065–1087. https://doi.org/10.1016/B978-0-444-53486-6.00056-9
Ho GY, Woodward N, Coward JI (2016) Cisplatin versus carboplatin: comparative review of therapeutic management in solid malignancies. Crit Rev Oncol Hematol 102:37–46. https://doi.org/10.1016/j.critrevonc.2016.03.014
Hwang L, Choi IY, Kim SE, Ko IG, Shin MS, Kim CJ, Kim SH, Jin JJ, Chung JY, Yi JW (2013) Dexmedetomidine ameliorates intracerebral hemorrhage-induced memory impairment by inhibiting apoptosis and enhancing brain-derived neurotrophic factor expression in the rat hippocampus. Int J Mol Med 31(5):1047–1056. https://doi.org/10.3892/ijmm.2013.1301
Kumar M, Nerurkar VR (2014) Integrated analysis of microRNAs and their disease related targets in the brain of mice infected with West Nile virus. Virology 452–453:143–151. https://doi.org/10.1016/j.virol.2014.01.004
Lam JK, Chow MY, Zhang Y, Leung SW (2015) siRNA versus miRNA as therapeutics for gene silencing. Mol Ther Nucleic Acids 4:e252. https://doi.org/10.1038/mtna.2015.23
Li J, Xiong M, Nadavaluru PR, Zuo W, Ye JH, Eloy JD, Bekker A (2016) Dexmedetomidine attenuates neurotoxicity induced by prenatal propofol exposure. J Neurosurg Anesthesiol 28(1):51–64. https://doi.org/10.1097/ANA.0000000000000181
Ling H, Fabbri M, Calin GA (2013) MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov 12(11):847–865. https://doi.org/10.1038/nrd4140
Lomeli N, Di K, Czerniawski J, Guzowski JF, Bota DA (2017) Cisplatin-induced mitochondrial dysfunction is associated with impaired cognitive function in rats. Free Radic Biol Med 102:274–286. https://doi.org/10.1016/j.freeradbiomed.2016.11.046
Pandey A, Singh P, Jauhari A, Singh T, Khan F, Pant AB, Parmar D, Yadav S (2015) Critical role of the miR-200 family in regulating differentiation and proliferation of neurons. J Neurochem 133(5):640–652. https://doi.org/10.1111/jnc.13089
Peng L, Fu J, Ming Y (2018) The miR-200 family: multiple effects on gliomas. Cancer Manag Res 10:1987–1992. https://doi.org/10.2147/CMAR.S160945
Podratz JL, Lee H, Knorr P, Koehler S, Forsythe S, Lambrecht K, Arias S, Schmidt K, Steinhoff G, Yudintsev G, Yang A, Trushina E, Windebank A (2017) Cisplatin induces mitochondrial deficits in Drosophila larval segmental nerve. Neurobiol Dis 97(Pt A):60–69. https://doi.org/10.1016/j.nbd.2016.10.003
Qin Y, Chen W, Liu B, Zhou L, Deng L, Niu W, Bao D, Cheng C, Li D, Liu S, Niu C (2017) MiR-200c inhibits the tumor progression of glioma via targeting moesin. Theranostics 7(6):1663–1673. https://doi.org/10.7150/thno.17886
Reddy PH, Williams J, Smith F, Bhatti JS, Kumar S, Vijayan M, Kandimalla R, Kuruva CS, Wang R, Manczak M, Yin X, Reddy AP (2017) MicroRNAs, aging, cellular senescence, and Alzheimer’s disease. Prog Mol Biol Transl Sci 146:127–171. https://doi.org/10.1016/bs.pmbts.2016.12.009
Rosso SB, Inestrosa NC (2013) WNT signaling in neuronal maturation and synaptogenesis. Front Cell Neurosci 7:103. https://doi.org/10.3389/fncel.2013.00103
Tang CZ, Yang JT, Liu QH, Wang YR, Wang WS (2019) Up-regulated miR-192-5p expression rescues cognitive impairment and restores neural function in mice with depression via the Fbln2-mediated TGF-beta1 signaling pathway. FASEB J 33(1):606–618. https://doi.org/10.1096/fj.201800210RR
Wefel JS, Kesler SR, Noll KR, Schagen SB (2015) Clinical characteristics, pathophysiology, and management of noncentral nervous system cancer-related cognitive impairment in adults. CA Cancer J Clin 65(2):123–138. https://doi.org/10.3322/caac.21258
Xiao L, Chen Y, Ji M, Dong J (2011) KIBRA regulates Hippo signaling activity via interactions with large tumor suppressor kinases. J Biol Chem 286(10):7788–7796. https://doi.org/10.1074/jbc.M110.173468
Yuan L, Hu S, Okray Z, Ren X, De Geest N, Claeys A, Yan J, Bellefroid E, Hassan BA, Quan XJ (2016) The Drosophila neurogenin tap functionally interacts with the Wnt-PCP pathway to regulate neuronal extension and guidance. Development 143(15):2760–2766. https://doi.org/10.1242/dev.134155
Zhang C, Zhang J, Hao J, Shi Z, Wang Y, Han L, Yu S, You Y, Jiang T, Wang J, Liu M, Pu P, Kang C (2012) High level of miR-221/222 confers increased cell invasion and poor prognosis in glioma. J Transl Med 10:119. https://doi.org/10.1186/1479-5876-10-119
Zhang L, Yang S, Wennmann DO, Chen Y, Kremerskothen J, Dong J (2014) KIBRA: in the brain and beyond. Cell Signal 26(7):1392–1399. https://doi.org/10.1016/j.cellsig.2014.02.023
Zhao C, Ma ZG, Mou SL, Yang YX, Zhang YH, Yao WC (2017) Targeting effect of microRNA on CD133 and its impact analysis on proliferation and invasion of glioma cells. Genet Mol Res 16(1):gmr16019281. https://doi.org/10.4238/gmr16019281
Zheng P, Bin H, Chen W (2019) Inhibition of microRNA-103a inhibits the activation of astrocytes in hippocampus tissues and improves the pathological injury of neurons of epilepsy rats by regulating BDNF. Cancer Cell Int 19:109. https://doi.org/10.1186/s12935-019-0821-2
Zhou W, Kavelaars A, Heijnen CJ (2016) Metformin prevents cisplatin-induced cognitive impairment and brain damage in mice. PLoS ONE 11(3):e0151890. https://doi.org/10.1371/journal.pone.0151890
Funding
This study was provided by Science and technology project of education department of Jiangxi province (project no. GJJ190035).
Author information
Authors and Affiliations
Contributions
Conceptualization and Funding acquisition: GX. Data curation: CL and FD. Formal analysis: JN and BZ. Investigation: CL, JN and WD. Methodology: ZZ and BZ. Writing—original draft: CL, JN and WD. Writing—review and editing: BZ, FD and ZZ. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declared that they have no conflict of interest.
Ethics approval and consent to participate
The animal experiments in this study and all procedures involving the handling and treatment of rat during this study were approved by the Institutional Animal Care and Use Committee of the Second Affiliated Hospital of Nanchang University. All the experiments were performed according to the guidelines of the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Electronic supplementary material
Below is the link to the electronic supplementary material.
13205_2020_2217_MOESM3_ESM.tif
Supplementary file3 (TIF 120 kb) Quality control results of a representative sample sequence. The horizontal axis indicates the number of bases or the range of bases, the vertical axis displays quality score values; quality scores > 30 revealed the accuracy of mapping to be greater than 99.9%
13205_2020_2217_MOESM4_ESM.tif
Supplementary file4 (TIF 697 kb) Wayne diagrams illustrating predicted DE miRNAs target genes between control and model groups (A), as well as DE miRNA target genes involving the Dex and model groups (B)
Rights and permissions
About this article
Cite this article
Li, C., Niu, J., Zhou, B. et al. Dexmedetomidine attenuates cisplatin-induced cognitive impairment by modulating miR-429-3p expression in rats. 3 Biotech 10, 244 (2020). https://doi.org/10.1007/s13205-020-02217-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-020-02217-1