Abstract
Phragmites australis (common reed) is a widespread perennial grass of wetland habitats, with cryptic native and introduced subspecies in North America. We determined the relative abundance of the subspecies and the distributions of plastid DNA haplotypes throughout British Columbia, Canada, at the northwestern distribution limit of common reed in North America. Of 203 specimens assigned to subspecies using molecular markers, we identified only 9 plants as the introduced ssp. australis; all remaining samples were the native ssp. americanus. The two subspecies co-occurred at only one locality. We identified four native haplotypes (one widespread in British Columbia and three others more localized) and two introduced haplotypes. Using plants of known haplotype, we assessed the utility of different morphological traits and trait combinations for distinguishing native and introduced subspecies in this geographic region. No single morphological trait was diagnostic, but principal components analysis and identification indices based on combinations of traits consistently separated the native and introduced subspecies in our sample. Two- or three-trait combinations of ligule length, lemma length and stem anthocyanic coloration gave the best separation. These indices could reduce the need for confirmation of the introduced subspecies using molecular tools, facilitating efforts to monitor and control this invasive plant.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cryptic invasive species pose a particularly difficult problem for the understanding and control of biological invasions (Pyšek et al. 2013). Phragmites australis (Cav.) Trin. ex Steud., the common reed, is a cosmopolitan perennial grass occurring in a range of wetland habitats. In North America these include pond and lake margins, estuaries, salt marshes, roadside ditches, road and rail right-of-ways, and wetter microsites in agricultural fields. The species spreads vegetatively by rhizomes and can form extensive swards, but also establishes readily by seed (Brisson et al. 2008; McCormick et al. 2010; Kettenring et al. 2012) and may be favored by anthropogenic disturbance.
The species has been present in North America for thousands of years, as indicated by fossil evidence (Orson 1999). However, it has greatly increased in abundance during the last 150 years, especially in the eastern part of the continent. Saltonstall (2002) showed that P. australis in North America consisted of both native and introduced races. The introduced form was first documented in North America in the 1870s (Meyerson et al. 2009), and subsequently became widespread and abundant; it is now considered an important invasive grass in many North American wetlands.
Phragmites australis is highly variable worldwide with multiple races, some of which have been formally described as varieties or subspecies. Although currently there is no general agreement on the disposition of infraspecific taxa over much of the geographic range, commonly recognized segregates of P. australis in North America include ssp. americanus Saltonst., P.M.Peterson & Soreng (Saltonstall et al. 2004) and ssp. berlandieri (E.Fourn.) Saltonst. & Hauber (Saltonstall and Hauber 2007). The remaining North American material not included under the above names, though often referred to only as introduced or European P. australis, is generally treated in North American floras and checklists as ssp. australis. In the absence of taxonomic consensus, we use these three names for convenience in this paper. Two of the forms, the native ssp. americanus and the introduced ssp. australis, are widespread in North America. The third, ssp. berlandieri, is an introduced race of possibly hybrid origins (Lambertini et al. 2012b) that is restricted to the southern USA, occurring primarily along the Gulf Coast (Lambertini et al. 2006, 2012a).
The two widespread subspecies in North America have slight morphological differences, but overlap to some degree in most or all of their traits and are therefore difficult to tell apart in the field. To address the problem of identification, Saltonstall (2002, 2003a, 2003c) surveyed molecular variation in two plastid DNA noncoding regions and demonstrated that the two subspecies are readily distinguished using these markers. She identified 27 distinct haplotypes worldwide using these DNA regions, of which 11 were found to be native to North America (ssp. americanus). In contrast, all samples of ssp. australis in North America were of a single haplotype, M (Saltonstall 2002, 2003a). Comparisons of DNA samples from herbarium specimens with samples from extant populations (Saltonstall 2002) indicated that before 1910 the introduced haplotype M (and thus ssp. australis) was rare in North America, and that over the last 150 years it has dramatically increased in abundance, displacing the native subspecies in many areas and spreading into habitats where the species was previously uncommon or absent. This introduced lineage initially became established along the eastern seaboard of the USA (Saltonstall 2002), most likely originating from Europe (Plut et al. 2011; Lambertini et al. 2012b). From there it has spread widely in eastern and southern North America. It has also increased in the west, especially in disturbed areas (Lambert et al. 2016).
Genetic surveys to date show that although the introduced haplotype M is prevalent across North America, other introduced haplotypes are locally abundant. At least three distinct introduced lineages have been found along the Gulf Coast (Saltonstall 2003a; Hauber et al. 2011; Lambertini et al. 2012a). In southwestern North America, introduced P. australis is abundant in some areas (Kulmatiski et al. 2010; Lambert et al. 2016) but remains less common than the native subspecies. Lambert et al. (2016) reported two previously unknown Asian haplotypes from the southwest, as well as the widespread haplotype M. Microsatellite DNA markers are variable among introduced populations, suggesting that even haplotype M populations may have originated from several sources; current evidence (Meyerson and Cronin 2013) suggests that new introductions of P. australis are continuing to arrive and become established in North America.
Because interfertility might be expected between the native and introduced subspecies of P. australis, co-occurrence and hybridization are of possible concern in the control of the introduced subspecies, as this offers a potential pathway for invasive characteristics to enter native populations. Hybrids have rarely been reported, but do occur where the native and introduced subspecies are found together (Saltonstall et al. 2014; Wu et al. 2015). Hybridization may be especially likely in anthropogenically disturbed wetlands, and some hybrids appear to be of very recent origin (Lambert et al. 2016; Saltonstall et al. 2016).
Although P. australis has been well studied in southern and eastern USA and in eastern Canada, the distributions of the introduced and native subspecies are less well documented in the northwestern part of the continent. Here we evaluate molecular and morphological variation in P. australis at its northwestern extent in North America. Our objectives were to: 1) determine the locations and abundance of introduced forms of P. australis in western Canada, as well as the distributions of native DNA haplotypes; and 2) analyze morphological differences between native and introduced subspecies using plants of known DNA haplotype, in order to devise indices that could be useful for identifying the subspecies in the field.
Methods and Materials
Plant Sampling
Samples for this study were collected from populations throughout British Columbia in 2012 and 2014, in association with ongoing invasive species monitoring. Sampling was carried out in all accessible localities where common reed was known to occur, and as far as possible all distinct clones in an area were sampled. We obtained DNA sequence data from 189 leaf samples taken from these populations (listed in Table S1). To examine historical collections of P. australis (1975–2012; Table S1), we also sequenced leaf material from 14 herbarium specimens at the Royal British Columbia Museum (V).
DNA Analyses
DNA was extracted from silica-dried leaves or pressed specimens using a modified CTAB method (Doyle and Doyle 1987) or the Phire Plant Direct PCR kit (Thermo Fisher Scientific, Waltham, MA, USA). We amplified and sequenced the two plastid DNA (pDNA) regions, the trnT-trnL and rbcL-psaI spacers, identified as diagnostic by Saltonstall (2002, 2003a). For the trnT-trnL region, we used forward primer a with reverse primer b (Taberlet et al. 1991); for the rbcL-psaI spacer, we used forward primer rbcL with reverse primer psaI (Saltonstall 2002). For all DNA regions, PCR amplification was performed in 50 μL volumes with the following reagents: 5 μL template DNA (1:10 dilution), 5 μL 10X PCR Buffer, 5 μL 2 mM dNTPs, 0.25 μL Taq DNA polymerase (Invitrogen, Carlsbad, California, USA), 2.5 μL of 5 μM of each primer and 1.5 μL 100X BSA (New England Biolabs, Ipswich, Massachusetts, USA). We carried out all PCR using Techne TC-312 thermal cyclers. The standard PCR protocol for amplification of the two chloroplast DNA regions was 3 min at 94 °C, 30 cycles of 94 °C for 30 s, 50 °C for 1 min, and 72 °C for 1 min, with a final 5 min at 72 °C. For samples analyzed using the Phire Plant Direct PCR kit, the PCR amplification protocol for the trnT-trnL region was 5 min at 98 °C, 40 cycles of 98 °C for 5 s, 60 °C for 5 s, and 72 °C for 20 s, with a final 1 min at 72 °C; for the rbcL-psaI region, the PCR amplification protocol was 5 min at 98 °C, 40 cycles of 98 °C for 5 s, 56 °C for 5 s, and 72 °C for 20 s, with a final 1 min at 72 °C. All samples were sequenced in both forward and reverse directions using the amplifying primers. PCR products were purified before sequencing using the QIAquick PCR Purification Kit (Qiagen, Valencia, California, USA). DNA sequencing was carried out by Macrogen Inc. (Seoul, Korea). Sequences representing the distinct genetic variants found in this study are deposited in GenBank, with accession numbers of KY765930-KY765935 for the trnT-trnL region and KY765924-KY765929 for the rbcL-psaI region. Sequences were aligned using ClustalX (Thompson et al. 1997) and Jalview (Waterhouse et al. 2009). The two DNA regions were concatenated into a single combined sequence for each sample. We constructed unrooted haplotype networks using the statistical parsimony software TCS v. 1.21 (Clement et al. 2000) for all complete sequences in our data set, together with accessions downloaded from GenBank for other haplotypes for which complete sequences were available. All indels were coded as single-gap traits for the analyses. Separate runs were carried out with repetitive DNA (microsatellite) sequence variation (Saltonstall 2016) either included or excluded. The distributions of individual native and introduced haplotypes were mapped using Mappad 2.0 (http://www.ncdc.noaa.gov/paleo/softlib.html).
Morphological Measurements
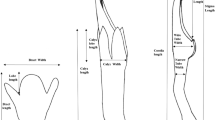
Morphological measurements were taken from plant specimens collected in late summer (during flowering or soon after). Only specimens in good condition were measured. All plants used for morphological measurement were voucher specimens of known molecular haplotype; each voucher consisted of a complete single culm with associated leaves and inflorescence. We obtained measurements from 42 specimens of ssp. americanus and 6 specimens of ssp. australis. The traits that were used (Table 1) included features of the inflorescence, spikelets, florets, leaves and culm. Traits were selected for ease of measurement with minimum damage to voucher specimens, and included features previously used to separate the introduced and native subspecies (Saltonstall et al. 2004; Catling et al. 2007; Swearingen and Saltonstall 2010). We excluded some traits that have been suggested as useful but were difficult to evaluate in the available voucher specimens (e.g., attachment of leaf sheath, leaf color, presence/absence of fungal spots on culms). Measurement details for individual traits are as follows.
Spikelets (Glumes and Lemmas)
Five spikelets were selected at random from the inflorescence of each voucher. Lengths (in mm) of the first (lower) glume, second (upper) glume, and a lemma (the lowest lemma except where damaged or absent, in which case another lemma was used) were measured for each spikelet.
Inflorescences
Inflorescence length was measured from the lowest branch attachment point to the inflorescence tip. Inflorescence branch length was measured on a branch selected at random from the second-lowest branch cluster of each inflorescence; the branch was measured from its attachment point to the tip.
Leaves (Blades and Ligules)
Leaf blade length (cm) and maximum width (cm) were measured on the uppermost five consecutive leaves of each specimen (omitting leaves with damaged tips). Ligule lengths (measured to the nearest 0.1 mm), including the membranous portion but excluding divergent hairs of the hairy fringe, were measured at the midpoint and margin on the same five leaves.
Culm Surface and Coloration
Culm surfaces varied from shiny and smooth to dull and rough with longitudinal ridges. Culm surface texture was ranked on a scale of 1–5 (1 = smooth; 5 = rough/ridged). Culm coloration (presence of reddish to purple anthocyanins), which occurred on exposed stem internodes not covered by leaf sheaths, varied from absent to extensive. Coloration was scored on a scale of 1–4 (1 = pigmentation absent; 2 = pigmentation slight and limited to less than 1/3 of the internode; 3 = pigmentation moderate over 1/3 or more of the internode; 4 = pigmentation very dark over most of the internode).
Morphological Analyses
Means and standard deviations of all traits were calculated separately for specimens of introduced and native DNA haplotypes. Traits measured on multiple structures per plant (all spikelet and leaf traits) were averaged to give a single value per plant for purposes of comparisons among individual plants. Trait distributions were assessed for their degree of overlap and their utility in distinguishing the introduced from the native subspecies.
Principal components analyses (PCA) were carried out based on various combinations of morphological traits. Trait correlations were assessed using Pearson’s correlation coefficient; some highly correlated traits were subsequently omitted from analysis. The final PCA was based on 11 traits (Table 1), including all those commonly used to distinguish the subspecies. All analyses were carried out using Statistix v. 9 (Analytical Software, Tallahassee, Florida, USA).
Combinations of the traits that showed greatest divergence between subspecies were assessed to determine whether a multiple-trait index could be developed to help in distinguishing the subspecies in the field. For each plant, ID indices were calculated as follows: (1) each individual trait value was scaled between 0 and 1, using the maximum and minimum trait values in the data set (scaled trait value = the difference between the observed individual value and the minimum value, divided by the difference between maximum and minimum values); (2) the scaled values were averaged for the included traits, then multiplied by 10 to give an index on a scale of 0 to 10. Maximum and minimum values used in these calculations were taken directly from the entire range of values over all specimens (native and introduced) for each trait in our morphological data set. We calculated several different indices based on various combinations of the following traits: lower and upper glume length, lemma length, ligule length at the centre, length of uppermost leaf, and culm color score. The distributions of the resulting indices were then examined for overlap between the two subspecies.
Results
Occurrence of Introduced and Native Haplotypes
On the basis of sequence data from 203 plants, we identified 6 plastid DNA haplotypes in British Columbia (Fig. 1). Four of these haplotypes (A, B, D, E) were native and two (L, M) were introduced. The sequences for all haplotypes matched previously published sequences in GenBank.
Plastid DNA haplotype network for Phragmites australis, including all haplotypes known from North America as well as a majority of the haplotypes reported from other regions. Colored circles indicate haplotypes found in British Columbia; black circles = inferred intermediate haplotypes not known from P. australis
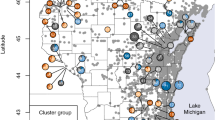
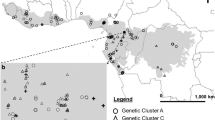
The native subspecies of P. australis (ssp. americanus) was found to be much more common in British Columbia than the introduced subspecies (Fig. 2a, b). Haplotype E, which is widespread elsewhere in Canada and in the northern USA, was the most abundant native haplotype. The remaining three native haplotypes were more localized in British Columbia (Fig. 2a). Haplotype A was found only on southern Vancouver Island in southwestern British Columbia. Haplotype B was found only in the Rocky Mountain Trench of southeastern British Columbia, and haplotype D was found only in the south Okanagan Valley of south-central British Columbia. Haplotypes B and D both co-occurred with the common haplotype E.
Introduced haplotypes were found in only 9 plants from 7 localities (Fig. 2b; Table S1). All of these belonged to the common introduced haplotype M, except for one Vancouver Island sample with variegated leaves which was identified as haplotype L (Fig. 2b). Only one herbarium specimen (collected near Vernon, B.C in 2006) was of a non-native haplotype (M); resampling at this locality in 2012 yielded the same haplotype. Eight of nine plants of introduced haplotype were from small populations and did not co-occur with native haplotypes. The exception was a single sample from the south Okanagan Valley, which was found in association with extensive populations of the native subspecies.
Morphological Differences between Subspecies of P. australis
Means, standard deviations and ranges of variation for the 11 individual morphological traits used in the PCA (Table 1) indicate overlap between introduced and native groups in most traits. Nevertheless, several traits were sufficiently well differentiated between the subspecies to be of value in separating them (Fig. 3a-e). Introduced Phragmites australis ssp. australis had smaller flower parts on average, including smaller lower and upper glumes (Figs. 3a,b) and smaller lemmas (Fig. 3c). It also had shorter leaf ligules (Fig. 3d). Culms of the introduced subspecies had little or no purple pigmentation, whereas those of the native subspecies tended to be moderately to strongly pigmented (Fig. 3e). Inflorescence and leaf size traits were less diagnostic, and broadly overlapped. Leaf length and width both averaged somewhat larger for the introduced subspecies, but varied with leaf position and were highly variable overall (Table 1; supplementary Figs. S1, S2).
Principal components analysis (PCA) using the 11 morphological features showed a clear overall differentiation of the native and introduced subspecies based on specimens identified by molecular haplotype, especially along the first axis (Fig. 4). However, the two subspecies were not strongly separated in the PCA (Fig. 4), a reflection of the high degree of overlap in some of the measured traits. The first PCA axis (PCA1) captured 41% and the second PCA axis (PCA2) 23% of the total variation in the data set (64% for the two axes combined). Both axes included multiple variables with high loadings (absolute value >0.3). The highest-loading variables were ligule length at midpoint and inflorescence branch length on PCA1, and first glume length on PCA2. Individual variable loadings on each axis are given in supplementary Table S2.
We derived two ID indices (Fig. 5) using the traits that showed greatest divergence and least overlap between the two P. australis subspecies (lemma length, ligule length and culm color). The best separation of the subspecies was obtained using vegetative or a combination of vegetative and floral traits. Index 1 (Fig. 5a), based only on the two vegetative traits (ligule length, culm color), varied from 0.6–3.1 for ssp. australis and 4.3–9.7 for ssp. americanus. Index 2 (Fig. 5b), based on these two vegetative traits and one floret trait (lemma length), ranged from 0 to 2.5 for ssp. australis and 3.9–9.9 for ssp. americanus. These two indices clearly differentiated the specimens in our sample.
Separation of native and introduced subspecies of Phragmites australis using two identification indices based on combined scaled values of selected morphological traits. Introduced = ssp. australis; native = ssp. americanus. a) Index 1 (= ligule length + culm color) for non-flowering specimens; b) Index 2 (= ligule length + lemma length + culm color) for flowering specimens. See text for details of trait measurements and index calculations
Discussion
Native and Introduced Phragmites australis in British Columbia
Molecular analyses confirm that almost all populations of P. australis in British Columbia are the native subspecies, a pattern previously reported by Catling and Mitrow (2011) based on morphological determination of Canadian Phragmites populations. Of the four native haplotypes of Phragmites australis we identified, the most common (haplotype E) was reported from British Columbia in previous work (Saltonstall 2003) and is widespread across the northern USA and in central Canada. The other three, though not previously reported from British Columbia, have been documented from adjacent regions (Saltonstall 2003). Haplotype A, which we found on southern Vancouver Island, is also known from across the western USA (northern Washington to the Rocky Mountains). Haplotype B, reported here from southeastern British Columbia, occurs also in the western USA, extending to the Great Plains. Haplotype D, which we found in the southern Okanagan Valley, also occurs in Oregon and Montana.
The introduced subspecies is still uncommon in British Columbia, in contrast with its abundance in parts of eastern North America where it has often displaced the native subspecies. With the exception of a single occurrence of haplotype L, all samples of introduced P. australis were of the previously reported haplotype M, with sequences identical to those previously reported by Saltonstall (2002). This suggests that they are likely the result of westward spread from existing populations and are not novel introductions. The single specimen of haplotype L was a variegated specimen collected from the margin of a landscaped pond on private property, and was probably planted. This haplotype corresponds to microsatellite variant L1 reported by Lambertini et al. (2012b). It is widespread in Europe, but in North America it has only been reported previously from a locality in Quebec (Meyerson and Cronin 2013).
Differentiation of the Subspecies
Our analyses of morphological variation in specimens from British Columbia extend earlier work (Saltonstall et al. 2004) that was based on 21 specimens identified to molecular haplotype. We show that measurements from selected combinations of vegetative and floral traits can be used to identify the subspecies of Phragmites australis with considerable confidence, even if individual traits are not diagnostic. Although the rarity of the introduced subspecies in British Columbia limited our sample size in this study, and larger sample sizes will likely show greater morphological overlap between the subspecies, the ranges of variation reported here for these traits are similar to those described previously (Saltonstall et al. 2004; Swearingen and Saltonstall 2010). Catling and colleagues (Catling 2007; Catling et al. 2007; Catling and Mitrow 2011) have discussed which traits are most useful for separating the subspecies; they identified ligule length, stem anthocyanin coloration and glume length as particularly helpful. Our findings generally agree with their conclusions. We found (confirming the results of Saltonstall et al. 2004) that lemma length (included here in Index 2) contributed to better separation of subspecies than did glume length, but since the glumes persist after flowering, the latter trait is useful for identifying late-season specimens that have shed their florets.
The two subspecies of P. australis remain sometimes difficult to identify with complete certainty. All of the traits used in this study vary within plants and perhaps geographically, and are subject to possible measurement error. Some traits (e.g., culm color) can be difficult to assess objectively, especially in winter material. For greater accuracy in assigning specimens to subspecies using the indices described here, it is preferable to use specimens in good condition and to average multiple measurements from a specimen where possible. For size determination of small structures (glumes, lemmas, ligules), a hand lens or microscope may be needed. However, we show that the morphological traits we used here can be combined quantitatively to provide more accurate identification of the introduced and native subspecies. A small fraction of specimens may be morphologically intermediate and require molecular analysis for positive identification; this could especially be the case where hybridization is suspected. Nevertheless, the use of a quantitative index can greatly reduce the necessity for confirming identification with molecular methods, thus saving time and resources.
Prospects for Spread of P. australis ssp. australis in Northwestern North America
The phylogeographic history of P. australis is complex (Lambertini et al. 2006, 2012b), with recent evidence indicating multiple introductions of non-native haplotypes in North America (Hauber et al. 2011; Lambertini et al. 2012a; Meyerson and Cronin 2013; Lambert et al. 2016). Although the chloroplast haplotype M is the most widespread introduced form in North America, it is associated with multiple nuclear genotypes (Saltonstall 2003b). We identified all non-native samples in British Columbia as haplotype M, except for one sample of haplotype L (variant L1) which is probably an escaped cultivated form. This population has not increased over several years and appears not to be strongly invasive (K.L. Marr, personal observation).
Recent evidence confirms that the native and introduced races of Phragmites australis can hybridize (Meyerson et al. 2010; Paul et al. 2010; Meyerson and Cronin 2013; Saltonstall et al. 2014; Wu et al. 2015). Because Phragmites propagates within populations extensively by rhizomatous spread and seedlings may not be common in established stands, the two subspecies have been considered not to hybridize to any great extent. However, this may vary by region. Saltonstall et al. (2016) report extensive occurrence of hybrids in disturbed wetlands of southern Nevada. The occurrence of hybrids (even rarely) is a potentially important consideration in managing invasive populations. The possibility exists that invasive characteristics could enter native populations through hybridization, although in most parts of the continent this is not yet a serious concern. In our study, co-occurring specimens of the introduced and native subspecies were identified from only a single locality (at this site the native subspecies was abundant but the introduced haplotype was represented by only a single clump).
The introduced subspecies is now the most common form in much of the eastern US (Saltonstall 2003a), implying that native haplotypes have been displaced. Non-native Phragmites australis has had major adverse effects on wetlands in eastern North America, and is a threat to various other wetland species (Vasquez et al. 2005, 2006; Meyerson et al. 2009) as well as to the native subspecies. Its initial introduction along the Atlantic coast has been followed by continued gradual expansion over many decades (Saltonstall 2002), suggesting that it will eventually become abundant across the continent (Catling and Mitrow 2011). The non-native form may have benefitted from “ecological escape” from herbivores and pathogens on its continents of origin (enemy-release hypothesis; Cronin et al. 2015). However, recent genetic and ecological work also indicates that the subspecies differ ecologically (Kirk et al. 2011; Wu et al. 2015). Trends toward global warming may favor the introduced subspecies (Guo et al. 2013), but under some circumstances the native subspecies may have greater potential for vegetative spread than the introduced subspecies, especially further north (Douhovnikoff and Hazelton 2014; Cronin et al. 2015). Wu et al. (2015) showed that ssp. americanus occupies lower-salinity habitats than ssp. australis, and remnant populations of the native subspecies appear to retain considerable genetic variability (Saltonstall 2011). Climate warming and the direct effects of human activities (e.g., clearing of wetland habitats or eutrophication of marshlands) may increase the success of introduced Phragmites australis (Kirk et al. 2011; Wu et al. 2015), which appears to require disturbance for seedling establishment (Kettenring et al. 2015). However, P. australis is a wetland species and tends to occur more sporadically in drier climates, particularly in inland habitats. This may limit spread of the introduced form in summer-dry climatic regimes of western North America, providing time for the implementation of control measures.
Conclusions
The introduced form of the wetlands grass Phragmites australis is still uncommon in the northwest part of the species range in North America, in contrast to other regions of the continent. This may be a transient phenomenon, or may in part reflect ecological differences between native and introduced subspecies of this grass. We show that quantitative indices based on combinations of morphological traits can be used to distinguish the two subspecies with substantial confidence, reducing the need to confirm ID using molecular methods.
References
Brisson J, Paradis E, Bellavance M-E (2008) Evidence of sexual reproduction in the invasive common reed (Phragmites australis subsp. australis; Poaceae) in eastern Canada: a possible consequence of global warming? Rhodora 110:225–230
Catling PM (2007) Additional notes on the identification of alien Phragmites in Canada. Botanical Electronic News No 370. http://www.ou.edu/cas/botany-micro/ben/ben366.html
Catling PM, Mitrow G (2011) The recent spread and potential distribution of Phragmites australis subsp. australis in Canada. Canadian Field-Naturalist 125(2):95–104
Catling PM, Mitrow G, Black L (2007) Analysis of stem color and correlated morphological characters for grouping Phragmites (Poaceae) taxa in eastern Ontario. Rhodora 109:125–136
Clement M, Posada D, Crandall K (2000) TCS: a computer program to estimate gene genealogies. Molecular Ecology 9:1657–1660
Cronin JT, Bhattarai GP, Allen WJ, Meyerson LA (2015) Biogeography of a plant invasion: plant-herbivore interactions. Ecology 96:1115–1127
Douhovnikoff V, Hazelton ELG (2014) Clonal growth:invasion or stability? A comparative study of clonal architecture and diversity in native and introduced lineages of Phragmites australis (Poaceae). American Journal of Botany 101:1577–1584
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin 9:11–15
Guo W-Y, Lambertini C, Li X-Z, Meyerson LA, Brix H (2013) Invasion of old world Phragmites australis in the new world: precipitation and temperature patterns combined with human influences redesign the invasive niche. Global Change Biology 19:3406–3422
Hauber DP, Saltonstall K, White DA, Hood CS (2011) Genetic variation in the common reed, Phragmites australis, in the Mississippi River Delta marshes: evidence for multiple introductions. Estuaries and Coasts 34:851–862
Kettenring KM, de Blois S, Hauber DP (2012) Moving from a regional to a continental perspective of Phragmites australis invasion in North America. AoB PLANTS 2012: pls040. doi:10.1093/aobpla/pls040
Kettenring KM, Whigham DF, Hazelton ELG, Gallagher SK, Weiner HM (2015) Biotic resistance, disturbance, and mode of colonization impact the invasion of a widespread, introduced wetland grass. Ecological Applications 25:466–480
Kirk H, Paul J, Straka J, Freeland JR (2011) Long-distance dispersal and high genetic diversity are implicated in the invasive spread of the common reed, Phragmites australis (Poaceae), in northeastern North America. American Journal of Botany 98:1180–1190
Kulmatiski A, Beard KH, Meyerson LA, Gibson JR, Mock KE (2010) Nonnative Phragmites australis invasion into Utah wetlands. Western North American Naturalist 70:541–552
Lambert AM, Saltonstall K, Long R, Dudley TL (2016) Biogeography of Phragmites australis lineages in the southwestern United States. Biological Invasions 18:2597–2617
Lambertini C, Gustafsson MHG, Frydenberg J, Lissner J, Speranza M, Brix H (2006) A phylogeographic study of the cosmopolitan genus Phragmites (Poaceae) based on AFLPs. Plant Systematics and Evolution 258:161–182
Lambertini C, Mendelssohn IA, Gustafsson MHG, Oleson B, Riis T, Sorrell BK, Brix H (2012a) Tracing the origin of Gulf Coast Phragmites (Poaceae): a story of long-distance dispersal and hybridization. American Journal of Botany 99:538–551
Lambertini C, Sorrell BK, Riis T, Olesen B, Brix H (2012b) Exploring the borders of European Phragmites within a cosmopolitan genus. AoB plants 2012: pls020. doi:10.1093/aobpla/pls020
McCormick MK, Kettenring KM, Baron HM, Whigham DF (2010) Extent and reproductive mechanisms of Phragmites australis spread in brackish wetlands in Chesapeake Bay, Maryland (USA). Wetlands 30:67–74
Meyerson LA, Cronin JT (2013) Evidence for multiple introductions of Phragmites australis to North America: detection of a new non-native haplotype. Biological Invasions 15:2605–2608
Meyerson LA, Saltonstall K, Chambers RM (2009) Phragmites australis In eastern North America: a historical and ecological perspective. In: Silliman BR, Grosholz E, Bertness MD (eds) Human impacts on salt marshes: a global perspective. University of California Press, Berkeley and Los Angeles, pp 57–81
Meyerson LA, Viola DV, Brown RN (2010) Hybridization of invasive Phragmites australis with a native subspecies in North America. Biological Invasions 12:103–111
Orson RA (1999) A paleoecological assessment of Phragmites australis in New England tidal marshes: changes in plant community structure during the last few millennia. Biological Invasions 1:149–158
Paul J, Vachon N, Garroway CJ, Freeland JR (2010) Molecular data provide strong evidence of natural hybridization between native and introduced lineages of Phragmites australis in North America. Biological Invasions 12:2967–2973
Plut K, Paul J, Ciotir C, Major M, Freeland JR (2011) Origin of non-native Phragmites australis in North America, a common wetland invader. Fundamentals of Applied Limnology 179:121–129
Pyšek P, Hulme PE, Meyerson LA, Smith GF, Boatwright JS, Crouch NR, Figueiredo E, Foxcroft LC, Jarošík V, Richardson DM, Suda J, Wilson JRU (2013) Hitting the right target: taxonomic challenges for, and of, plant invasions. AoB PLANTS 5: plt042. doi:10.1093/aobpla/plt042
Saltonstall K (2002) Cryptic invasions by a non-native genotype of the common reed, Phragmites australis, into North America. Proceedings of the National Academy of Sciences (USA) 99:2445–2449
Saltonstall K (2003a) Genetic variation among north American populations of Phragmites australis: implications for management. Estuaries 26:444–451
Saltonstall K (2003b) Microsatellite variation within and among north American lineages of Phragmites australis. Molecular Ecology 12:1689–1702
Saltonstall K (2003c) A rapid method for identifying the origin of north American Phragmites populations using RFLP analysis. Wetlands 23:1043–1047
Saltonstall K (2011) Remnant native Phragmites australis maintains genetic diversity despite multiple threats. Conservation Genetics 12:1027–1033
Saltonstall K (2016) The naming of Phragmites haplotypes. Biological Invasions 18:2433–2441
Saltonstall K, Hauber D (2007) Notes on Phragmites australis (Poaceae: Arundinoideae) in North America. Journal of the Botanical Research Institute of Texas 1(1):385–388
Saltonstall K, Peterson P, Soreng R (2004) Recognition of Phragmites australis subsp. americanus (Poaceae: Arundinoideae) in North America: evidence from morphological and genetic analysis. SIDA, Contributions to Botany 21(2):683–692
Saltonstall K, Castillo HE, Blossey B (2014) Confirmed field hybridization of native and introduced Phragmites australis (Poaceae) in North America. American Journal of Botany 101:211–215
Saltonstall K, Lambert AM, Rice N (2016) What happens in Vegas, better stay in Vegas: Phragmites australis hybrids in the Las Vegas wash. Biological Invasions 18:2463–2474
Swearingen J, Saltonstall K (2010) Phragmites field guide: distinguishing native and exotic forms of common reed (Phragmites australis) in the United States. Plant Conservation Alliance, Weeds Gone Wild http://www.nps.gov/plants/alien/pubs/index.htm
Taberlet P, Gielly L, Pautou G, Bouvet J (1991) Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Molecular Biology 17:1105–1109
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research 24:4876–4882
Vasquez EA, Glenn EP, Brown JJ, Guntenspergen GL, Nelson SG (2005) Salt tolerance underlies the cryptic invasion of north American salt marshes by an introduced haplotype of the common reed Phragmites australis (Poaceae). Marine Ecology Progress Series 298:1–8
Vasquez EA, Glenn EP, Guntenspergen GL, Brown JJ, Nelson SG (2006) Salt tolerance and osmotic adjustment of Spartina alterniflora (Poaceae) and the invasive M haplotype of Phragmites australis (Poaceae) along a salinity gradient. American Journal of Botany 93:1784–1790
Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ (2009) Jalview version 2- a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191
Wu CA, Murray LA, Hefferman KE (2015) Evidence for natural hybridization between native and introduced lineages of Phragmites australis in the Chesapeake Bay watershed. American Journal of Botany 102:805–812
Acknowledgements
This project was carried out under a contract funded by the Ministry of Forests, Lands and Natural Resource Operations, Government of British Columbia. We acknowledge the help of all those who collected specimens for this study, including S. Cesselli, M. DeWolf, R. Haegedorn, C. MacRae, K. May, D. McLean, J. E. Portelance, R. Rudland, B. Smith and E. Sonntag. We also thank the Royal British Columbia Museum (V) for permission to analyze leaf tissue from herbarium specimens of P. australis.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Allen, G.A., McCormick, L.J., Jantzen, J.R. et al. Distributional and Morphological Differences between Native and Introduced Common Reed (Phragmites australis, Poaceae) in Western Canada. Wetlands 37, 819–827 (2017). https://doi.org/10.1007/s13157-017-0914-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13157-017-0914-4