Abstract
Purpose
Although quantification of amyloid positron emission tomography (PET) is important for evaluating patients with cognitive impairment, its routine clinical use is hampered by complicated preprocessing steps and required MRI. Here, we suggested a one-step quantification based on deep learning using native-space amyloid PET images of different radiotracers acquired from multiple centers.
Methods
Amyloid PET data of the Alzheimer Disease Neuroimaging Initiative (ADNI) were used for this study. A training/validation consists of 850 florbetapir PET images. Three hundred sixty-six florbetapir and 89 florbetaben PET images were used as test sets to evaluate the model. Native-space amyloid PET images were used as inputs, and the outputs were standardized uptake value ratios (SUVRs) calculated by the conventional MR-based method.
Results
The mean absolute errors (MAEs) of the composite SUVR were 0.040, 0.060, and 0.050 of training/validation and test sets for florbetapir PET and a test set for florbetaben PET, respectively. The agreement of amyloid positivity measured by Cohen’s kappa for test sets of florbetapir and florbetaben PET were 0.87 and 0.89, respectively.
Conclusion
We suggest a one-step quantification method for amyloid PET via a deep learning model. The model is highly reliable to quantify the amyloid PET regardless of multicenter images and various radiotracers.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
A definitive diagnosis of Alzheimer’s disease (AD) requires both clinical features and histopathological confirmation by brain biopsy or autopsy [1,2,3]. The histopathology of AD brain is characterized by amyloid deposition, which can be currently noninvasively evaluated by amyloid positron emission tomography (PET) [1,2,3,4]. Therefore, amyloid imaging has become critical diagnostics to assess AD pathology for earlier diagnosis of AD and mild cognitive impairment (MCI), eventually affecting effective treatment [5,6,7,8]. The current routine clinical practice of amyloid imaging is dependent on the visual interpretation, which is limited in reliability because of interobserver variability, images acquired from multiple centers using variable radiotracers [3].
To overcome the visual interpretation and to use amyloid PET as a quantitative biomarker, there have been many efforts to reliably and accurately quantify the amyloid deposition using PET. Conventionally, both magnetic resonance imaging (MRI) and amyloid PET scans are employed to calculate amyloid deposition using standardized uptake value ratios (SUVRs) [9]. The MR-based processes to estimate SUVR have been variable according to studies; however, image co-registration, brain segmentation, and normalization processes to delineate cortex and the reference regions have been commonly included [9]. Even though these methods can accurately delineate the cortical and reference regions and show a high correlation with amyloid pathology [10, 11], there are hurdles in routine clinical use, including the requirement of structural MRI scan and complicated preprocessing steps [3]. Quantification results from multiple centers that can be affected by different PET machines and reconstruction algorithms make it difficult to standardized criteria for amyloid positivity [12]. Furthermore, due to recently developed various F-18 radiotracers for evaluating amyloid deposits, the quantification of amyloid deposition is considerably variable according to the types of radiotracers [12, 13].
In this study, we aimed to develop an automated amyloid aggregation quantification system based on end-to-end training of deep learning using multicenter amyloid PET. Recently, deep learning approaches have been widely used to evaluate brain imaging, particularly developing biomarkers for dementia [14,15,16,17]. These studies have attempted to predict future cognitive outcomes from multimodal images or to assess conventional biomarkers such as amyloid quantification. As the amyloid deposit is a key pathologic marker of AD, its accurate quantification is critical for understanding AD and its related disorders. Our approach of the end-to-end deep learning model has several advantages compared with previous conventional approaches: (1) amyloid PET quantification based on native-space PET without structural MRI, (2) quantification for multiple centers with different machines and reconstruction algorithms, and (3) a feasibility in the application of the same method for different F-18 radiotracers, florbetapir and florbetaben.
Material and Methods
Subjects
In this study, amyloid PET data of multiple centers collected from the Alzheimer disease neuroimaging initiative (ADNI) (http://adni.loni.usc.edu) database. ADNI total 1216 florbetapir PET and 89 florbetaben PET data were included for this study. Florbetapir PET images were divided into two datasets, a training/validation set and a test set. A training/validation consists of 850 baseline florbetapir PET images regardless of the diagnosis. Three hundred sixty-six florbetapir PET images as a 2-year follow-up were used as an independent test set to evaluate the accuracy of the model. Eighty-nine florbetaben PET images of ADNI-3 were also applied as an independent test set from the training data. The ADNI was launched in 2003 by a $60 million, 5-year public-private partnership. The initial purpose of ADNI has been to test whether serial MRI, PET, other biological markers, and clinical and neuropsychological data can access the progression of MCI and/or early AD. The principal investigator is Michael W. Weiner, MD, VA Medical Center and University of California San Francisco. Many co-investigators from a broad range of academic institutions and private corporations work to find participants then they have achieved to recruit over 50 sites across the USA and Canada. The institutional review boards of all participating institutions approved imaging studies and neuropsychological tests, and all participants signed a written informed consent form. For up-to-date information, see http://www.adni-info.org.
PET Images and Quantification of Amyloid Deposition
ADNI provides processed PET images of four different levels. Four different levels are different levels of preprocessed PET data that provide different initial points for the following analyses. Sets 1 and 2 of preprocessed PET data are native-space images and the original images. Set 1: the realigned dynamic images after co-registration, set 2: the single-frame averaged static images of set 1, set 3: transformation into a standardized orientation and intensity normalization of the images of set 2, and set 4: scanner-specific smoothing applying to image set 3. Four levels of preprocessed PET data can be downloaded from the ADNI, and we use the set 2 of preprocessed native-space PET images [11]. More specifically, to develop a deep learning model that can be applied to PET images with various centers of different machines and reconstruction parameters, we used native-space PET images without voxel size standardization or smoothing. Because PET images were reconstructed for multiple time frames for the ADNI database, we used averaged images across the time frames without further processing including a spatial transformation for developing deep learning model. SUVR calculation results based on structural MR were acquired from the ADNI database. The detailed SUVR calculation method is described in previous papers [18, 19]. Briefly, each amyloid PET scan was co-registered to the corresponding MRI which is segmented and parcellated with Freesurfer to define cortical subregions of interests (frontal, cingulate, parietal, and temporal), and reference regions [5, 9]. SUVRs were calculated by mean counts of the cortical subregions (frontal, cingulate, parietal, and temporal) divided by those of the whole cerebellum [5, 9]. The deep learning model was designed to predict normalized SUVRs of four regions calculated by the conventional MR-based method. The composite SUVR was calculated by the mean value of normalized SUVRs of four cortical subregions for both the conventional MR-based method and the deep learning-based predicted method.
End-to-End Training of Deep Learning
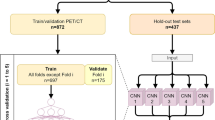
Florbetapir amyloid PET images were used as training/validation data. A total of 765 imaging data (90.0%) were used for training, and the remaining 85 imaging data (10.0%) were used for the internal validation. The validation data were used to determine architectures and parameters and were randomly selected among florbetapir training/validation data. The model was trained by 765 florbetapir PET dataset as a training set and 85 florbetapir PET datasets as an internal validation set to optimize the training parameters and architectures of the deep learning model (Fig. 1). The architecture of a 3D convolutional neural network (CNN) model we used in this study is shown in Fig. 2. We used supervised learning for the training. The input images were florbetapir PET, and the target outputs were conventional MR-based SUVRs. Thus, it was a regression problem instead of classification, and the label was a continuous variable.
Overall subjects for developing and validating the deep learning-based amyloid PET quantification method. A total of 1216 amyloid PET data were used for the training/validation and test datasets. Eight hundred fifty baseline florbetapir PET from ADNI data were used for developing the model and were randomly divided into two datasets, training (90%) and validation (10%) sets. Three hundred sixty-six baseline florbetapir PET images as a 2-year follow-up of ADNI data were used for an independent test set to validate the model. Eighty-nine florbetaben PET of ADNI data were used for another test set for validating the model
Convolutional neural network model and the architecture. The end-to-end deep learning model for SUVR prediction. Inputs are native-space PET images and outputs are conventional MR-based SUVRs. 3D CNN consists of multiple convolutional and deconvolutional layers to calculate amyloid PET to SUVR. The training of network was aimed at calculating SUVR, which cannot be distinguished from conventional MR-based SUVRs
The native-space PET images of multiple centers have different voxel size, and preprocessing steps were proceeded for normalizing the same voxel size (3 × 3 × 3 mm3) and resizing 80 × 80 × 60 matrix dimensions. The image resize was performed by the nearest-neighbor-like algorithm for scaling-down. These resliced native-space PET images were the input of the 3D CNN model with convolutional layers 32, filter size of 3 × 3 × 3, rectified linear unit (ReLU) activation layer, and a stride size of 2 × 2 × 2. After each convolutional layer, max-pooling layers were applied with a pool size of 2 × 2 × 2 and a stride size of 2 × 2 × 2. 3D convolutional layers and followed max-pooling layers are applied three times. Additionally, 3D convolutional layers were performed with 128 convolutional layers, a filter size of 5 × 5 × 5, and ReLU activation layers. Consequently, these multiple convolutional layers and max-pooling layers produced 128 feature vectors. The 128 features were related to four outputs. The four outputs corresponded to normalized SUVRs of four cortical subregions. The training/validation set for florbetapir PET was trained to minimize the loss function, mean absolute error (MAE) defined by \( \mathrm{L}\left({\mathrm{y}}_i,{\hat{y}}_i\right)=\frac{1}{N}\sum_{i=1}^N\left|{\mathrm{y}}_i-{\hat{y}}_i\right| \), where yi and \( {\hat{y}}_i \) are the regional SUVR, respectively. For the training of weights, Adam optimizer with learning rate = 0.0001 was used. The performance was independently tested by two test sets, florbetapir PET and florbetaben PET data. This training was conducted by Tensorflow (version 1.12) on Python (version 3.6).
Statistics
The assessment of the similarity between conventional MR-based SUVRs and deep learning-based predicted SUVRs were assessed by multiple metrics. Four subregional SUVRs were obtained by mean SUV of the whole cerebellum. The composite SUVRs were calculated by the mean value of normalized four subregional SUVRs. MAEs and intraclass correlation coefficients (ICCs) of the composite SUVR were estimated for each of the training/validation set and test sets. ADNI subjects can be categorized as amyloid positive or negative by the predefined cutoff value of 1.11 [20]. The cutoff of 1.11 applies to the composite SUVRs to obtain Cohen’s kappa, accuracy, sensitivity, and specificity for the training/validation set and test sets of florbetapir and florbetaben PET, respectively. The Bland-Altman plots show the means of the conventional MR-based SUVRs and deep learning-based predicted SUVRs plotted against their differences to assess agreement two methods for each test set.
Results
We developed the model to quantify the amyloid deposition using amyloid PET images. The SUVR could be calculated regardless of multicenter images and various radiotracers. The MAEs of the composite SUVR of training/validation and test sets for florbetapir PET were 0.04 and 0.06. The MAE of the composite SUVR of florbetaben PET as an independent test set was 0.05. The ICCs were 0.98, 0.96, and 0.97 for the training/validation set and test sets of florbetapir and florbetaben PET, respectively. Figure 3 presents the loss (MAE) according to the epochs. The MAEs and ICCs of quantitative SUVRs by conventional MR-based and deep learning-based methods for each subject of four cortical subregions are shown in Tables 1 and 2. Scatterplots show SUVRs calculated by the conventional MR-based method and deep learning-based predicted method for four cortical subregions (Fig. 4).
Scatterplots of SUVRs calculated by different quantification methods for four cortical subregions. SUVRs measured by the conventional MR-based method were compared with the deep learning-based predicted method. The conventional MR-based SUVR quantification method was highly correlated with the deep learning-based SUVR predicted quantification method. The training set of florbetapir (a) was drawn four cortical subregions for frontal, anterior/posterior cingulate, lateral parietal, and lateral regions, respectively. Histograms were incorporated into the scatter plot of the training dataset to visualize the distribution of SUVRs. The test sets of florbetapir (b) and florbetapet PET (c) were drawn four cortical subregions for frontal, anterior/posterior cingulate, lateral parietal, and lateral regions, respectively. (AD: Alzheimer’s disease, MCI: mild cognitive impairment, CN: cognitively normal
Samples were divided into amyloid-positive and amyloid-negative groups using a predefined cutoff value of 1.11. The agreement of amyloid positivity measured by Cohen’s kappa of two quantification methods, based on structural MR and deep learning, was 0.93, 0.87, and 0.89 for the training/validation set and test sets of florbetapir and florbetaben PET, respectively. When conventional MR-based SUVRs with the cutoff defined as an amyloid positivity, the accuracy, sensitivity, and specificity of deep learning-based SUVRs of a training/validation set and test sets for florbetapir and florbetaben PET are shown in Table 3. The Bland-Altman plots were also drawn to the agreement without bias for the quantification of four cortical subregions (Fig. 5).
Discussion
The quantification of amyloid deposition is challenging because multiple preprocessing steps and MRI scans are needed [5, 9, 21]. We developed the end-to-end deep learning model that could quantify a native-space amyloid PET [22]. The quality of our deep learning model was highly reliable and comparable with the conventional MR-based method. Our model was validated in the independent test sets for florbetaben PET as a different radiotracer as well as florbetapir PET.
The main advantage of our approach is that the model can provide rapid and accurate one-step quantification results of amyloid PET with native-space PET regardless of different machines, reconstruction algorithms, and types of F-18 radiotracers for amyloid PET. It could overcome the variability of the visual interpretation of amyloid PET in the clinical setting. Amyloid PET is clinically performed to estimate amyloid deposition in patients with cognitive impairment who need to differentiate AD from other reasons for cognitive disorders [5, 7]. In addition to amyloid positivity based on the visual interpretation, the quantification of amyloid deposition is important in predicting future cognitive decline in patients with mild cognitive impairment and early AD [5, 7]. Thus, the simple and rapid quantification method which could be acceptable in a busy clinical setting would be helpful for the interpretation of amyloid PET as well as evaluating the prognosis of patients with mild cognitive patients and early AD.
Due to the diversity of amyloid radiotracers, which cause variable quantitative measures of cortical radiotracer retention, Centiloid quantification was recently proposed for harmonizing multiple radiotracers of amyloid PET [12]. Even though amyloid PET quantification based on multiple tracers showed a good correlation, there is still a limitation to directly quantify the amyloid deposition using a simple linear equation [12]. As our approach could quantify the amyloid PET of multiple tracers, such scaling of multiple tracers could be achieved by a fine-tuning of the deep learning model. Furthermore, PET-only quantification methods are proposed such as variable atlas approaches [23] and MR-generating method [24] when MRI is not available. Even though these methods have shown promising results of high accuracy compared with MR-based methods, data preprocessing is still required. Thus, as a less complicated method of qualification, deriving SUVR directly from the deep learning model could serve as a breakthrough in managing the quantification of amyloid PET.
Our approach showed a good agreement with the MR-based quantification of SUVR. The MAE of the SUVRs of the one-step deep learning-based method was less than 0.06 in amyloid PET regardless of multicenter and various radiotracer images. Considering all agreement indices including ICCs, accuracy, accuracy, sensitivity, and specificity show very high scores that indicate high reliability of the model. Bland-Altman plots also showed that SUVR measured by the deep learning-based method was similar to SUVR of the conventional MR-based method. However, according to the Bland-Altman plots, SUVRs estimated by the deep learning-based methods are relatively underestimated in subjects who showed low SUVR. This pattern might be caused by the left-skewed distribution of amyloid SUVR. This underestimation may rarely affect the clinical implications of quantification considering a deep learning-based approach showed high ICC. Nevertheless, further modification methods in training including loss functions could solve this minor problem in the quantification as future work. The one-step deep learning method of amyloid quantification can be used for clinical trials without structural MRI scan and multiple preprocessing steps, and it can be used clinically as a biomarker for predicting the cognitive outcome using the model.
In spite of the high reliability of our model, there are some limitations. Although the model could calculate SUVRs of the one-step deep learning-based method, the model ignored patient’s characteristics such as age, baseline clinical diagnosis data, and cortical atrophy status [25,26,27]. We have tested ADNI data and with two radiotracers amyloid PET data. In the future, we should develop the modified deep learning model which considers other clinical information and should be validated in independent test sets including other radiotracers and another independent amyloid PET data [25, 28]. We will continue further study to validate the deep learning model using independent multicenter data obtained from routine clinical use.
Conclusion
We developed a deep learning model based on end-to-end training for the quantification of amyloid PET. We suggest a one-step quantification of amyloid PET without a structural MRI scan as well as multiple preprocessing steps. Our model was validated by florbetaben PET images as an independent dataset. Our approach directly provides amyloid PET quantification results from native-space PET images of florbetaben as well as florbetapir radiotracers. As an automated system, it could overcome problems in the quantification of amyloid PET such as the requirement of structural MRI and several preprocessing steps. Thus, we expect that our method may be easily applied to routine clinical practices. In the future, further modification and validation will be needed to be extended to multicenter data in the real clinical setting and various F-18-based radiotracers.
References
Bacskai BJ, Kajdasz ST, Christie RH, Carter C, Games D, Seubert P, et al. Imaging of amyloid-beta deposits in brains of living mice permits direct observation of clearance of plaques with immunotherapy. Nat Med. 2001;7:369–72.
Morris JC, Roe CM, Xiong C, Fagan AM, Goate AM, Holtzman DM, et al. APOE predicts amyloid-beta but not tau Alzheimer pathology in cognitively normal aging. Ann Neurol. 2010;67:122–31.
Jack CR Jr, Barrio JR, Kepe V. Cerebral amyloid PET imaging in Alzheimer’s disease. Acta Neuropathol. 2013;126:643–57.
Trojanowski JQ, Vandeerstichele H, Korecka M, Clark CM, Aisen PS, Petersen RC, et al. Update on the biomarker core of the Alzheimer’s disease neuroimaging initiative subjects. Alzheimers Dement. 2010;6:230–8.
Clark CM, Schneider JA, Bedell BJ, Beach TG, Bilker WB, Mintun MA, et al. Use of florbetapir-PET for imaging beta-amyloid pathology. JAMA. 2011;305:275–83.
Lopresti BJ, Klunk WE, Mathis CA, Hoge JA, Ziolko SK, Lu X, et al. Simplified quantification of Pittsburgh compound B amyloid imaging PET studies: a comparative analysis. J Nucl Med. 2005;46:1959–72.
Sperling RA, Aisen PS, Beckett LA, Bennett DA, Craft S, Fagan AM, et al. Toward defining the preclinical stages of Alzheimer's disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7:280–92.
Veitch DP, Weiner MW, Aisen PS, Beckett LA, Cairns NJ, Green RC, et al. Understanding disease progression and improving Alzheimer’s disease clinical trials: recent highlights from the Alzheimer’s disease neuroimaging initiative. Alzheimers Dement. 2019;15:106–52.
Landau SM, Fero A, Baker SL, Koeppe R, Mintun M, Chen K, et al. Measurement of longitudinal beta-amyloid change with 18F-florbetapir PET and standardized uptake value ratios. J Nucl Med. 2015;56:567–74.
Clark CM, Pontecorvo MJ, Beach TG, Bedell BJ, Coleman RE, Doraiswamy PM, et al. Cerebral PET with florbetapir compared with neuropathology at autopsy for detection of neuritic amyloid-beta plaques: a prospective cohort study. Lancet Neurol. 2012;11:669–78.
Jagust WJ, Landau SM, Koeppe RA, Reiman EM, Chen K, Mathis CA, et al. The Alzheimer’s disease neuroimaging initiative 2 PET Core: 2015. Alzheimers Dement. 2015;11:757–71.
Klunk WE, Koeppe RA, Price JC, Benzinger TL, Devous MD Sr, Jagust WJ, et al. The Centiloid project: standardizing quantitative amyloid plaque estimation by PET. Alzheimers Dement. 2015;11(1):1–15 e1-4.
Landau SM, Thomas BA, Thurfjell L, Schmidt M, Margolin R, Mintun M, et al. Amyloid PET imaging in Alzheimer’s disease: a comparison of three radiotracers. Eur J Nucl Med Mol Imaging. 2014;41:1398–407.
Choi H. Deep learning in nuclear medicine and molecular imaging: current perspectives and future directions. Nucl Med Mol Imaging. 2018;52:109–18.
Choi H, Jin KH. Alzheimer’s disease neuroimaging I. predicting cognitive decline with deep learning of brain metabolism and amyloid imaging. Behav Brain Res. 2018;344:103–9.
Spasov S, Passamonti L, Duggento A, Lio P, Toschi N. Alzheimer’s disease neuroimaging I. a parameter-efficient deep learning approach to predict conversion from mild cognitive impairment to Alzheimer’s disease. Neuroimage. 2019;189:276–87.
Choi H, Ha S, Kang H, Lee H, Lee DS. Alzheimer’s disease neuroimaging I. deep learning only by normal brain PET identify unheralded brain anomalies. EBioMedicine. 2019;43:447–53.
Weiner MW, Veitch DP, Aisen PS, Beckett LA, Cairns NJ, Cedarbaum J, et al. 2014 update of the Alzheimer’s disease neuroimaging initiative: a review of papers published since its inception. Alzheimers Dement. 2015;11:e1–120.
Weiner MW, Veitch DP, Aisen PS, Beckett LA, Cairns NJ, Green RC, et al. Recent publications from the Alzheimer’s disease neuroimaging initiative: reviewing progress toward improved AD clinical trials. Alzheimers Dement. 2017;13:e1–e85.
Joshi AD, Pontecorvo MJ, Clark CM, Carpenter AP, Jennings DL, Sadowsky CH, et al. Performance characteristics of amyloid PET with florbetapir F 18 in patients with Alzheimer’s disease and cognitively normal subjects. J Nucl Med. 2012;53:378–84.
Hurko O, Black SE, Doody R, Doraiswamy PM, Gamst A, Kaye J, et al. The ADNI publication policy: commensurate recognition of critical contributors who are not authors. Neuroimage. 2012;59:4196–200.
LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436–44.
Bourgeat P, Villemagne VL, Dore V, Brown B, Macaulay SL, Martins R, et al. Comparison of MR-less PiB SUVR quantification methods. Neurobiol Aging. 2015;36(Suppl 1):S159–66.
Choi H, Lee DS. Alzheimer’s disease neuroimaging I. generation of structural MR images from amyloid PET: application to MR-less quantification. J Nucl Med. 2018;59:1111–7.
Becker JA, Hedden T, Carmasin J, Maye J, Rentz DM, Putcha D, et al. Amyloid-beta associated cortical thinning in clinically normal elderly. Ann Neurol. 2011;69:1032–42.
Ripolles P, Marco-Pallares J, de Diego-Balaguer R, Miro J, Falip M, Juncadella M, et al. Analysis of automated methods for spatial normalization of lesioned brains. Neuroimage. 2012;60:1296–306.
Reig S, Penedo M, Gispert JD, Pascau J, Sanchez-Gonzalez J, Garcia-Barreno P, et al. Impact of ventricular enlargement on the measurement of metabolic activity in spatially normalized PET. Neuroimage. 2007;35:748–58.
Rowe CC, Ng S, Ackermann U, Gong SJ, Pike K, Savage G, et al. Imaging beta-amyloid burden in aging and dementia. Neurology. 2007;68:1718–25.
Funding
This work has supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (NRF-2019R1F1A1061412). Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012).
ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.;Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd. and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
Author information
Authors and Affiliations
Consortia
Corresponding author
Ethics declarations
Conflict of Interest
Ji-Young Kim, Hoon Young Suh, Hyun Gee Ryoo, Donkyu Oh, Hongyoon Choi, Jin Chul Paeng, Gi Jeong Cheon, Keon Wook Kang, Dong Soo Lee, and for the Alzheimer’s Disease Neuroimaging Initiative declare that they have no conflict of interest.
Ethical Approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed Consent
Informed consent to clinical testing and neuroimaging prior to participation of the ADNI cohort was obtained, approved by the institutional review boards (IRB) of all participating institutions.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Data used in preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in the analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf
Rights and permissions
About this article
Cite this article
Kim, JY., Suh, H.Y., Ryoo, H.G. et al. Amyloid PET Quantification Via End-to-End Training of a Deep Learning. Nucl Med Mol Imaging 53, 340–348 (2019). https://doi.org/10.1007/s13139-019-00610-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13139-019-00610-0