Abstract
Purpose
The increasing use of amyloid PET in Alzheimer’s disease research and clinical trials has motivated efforts to standardize methodology. We compared retention of the 11C radiotracer Pittsburgh Compound B (PiB) and that of two 18F amyloid radiotracers (florbetapir and flutemetamol) using two study populations. We also examined the feasibility of converting between tracer-specific measures, using PiB as the common link between the two 18F tracers.
Methods
One group of 40 subjects underwent PiB and flutemetamol imaging sessions and a separate group of 32 subjects underwent PiB and florbetapir imaging sessions. We compared cortical and white matter retention for each 18F tracer relative to that of PiB, as well as retention in several reference regions and image analysis methods. Correlations between tracer pairs were used to convert tracer-specific threshold values for amyloid positivity between tracers.
Results
Cortical retention for each pair of tracers was strongly correlated regardless of reference region (PiB–flutemetamol, ρ = 0.84–0.99; PiB–florbetapir, ρ = 0.83–0.97) and analysis method (ρ = 0.90–0.99). Compared to PiB, flutemetamol had higher white matter retention, while florbetapir had lower cortical retention. Two previously established independent thresholds for amyloid positivity were highly consistent when values were converted between tracer pairs.
Conclusion
Despite differing white and grey matter retention characteristics, cortical retention for each 18F tracer was highly correlated with that of PiB, enabling conversion of thresholds across tracer measurement scales with a high level of internal consistency. Standardization of analysis methods and measurement scales may facilitate the comparison of amyloid PET data obtained using different tracers.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Amyloid PET radiotracers are increasingly being used in Alzheimer’s disease (AD) research and clinical trials of fibrillar amyloid-targeting treatments. The US Food and Drug Administration (FDA) has approved the 18F radiotracers flutemetamol (Vizamyl) and florbetapir (Amyvid) for clinical use, and approval by the European Medicines Agency (EMA) has been established for florbetapir and is under review for flutemetamol.
The first amyloid PET imaging agent, the research tracer 11C Pittsburgh Compound B (PiB), has frequently been used to aid in validation of the more recently developed 18F radiotracers [1–6]. Because each tracer has slightly different characteristics that influence quantification, the use of multiple tracers poses challenges for comparing results of studies using different tracers. This issue has also impeded the development of important measures such as thresholds for a significant amount of fibrillar brain amyloid deposition (amyloid positivity). For example, some reports suggest greater nonspecific white than grey matter retention of both 18F amyloid PET tracers [2, 3, 7] and of PiB [8, 9], a finding attributed to slower tracer clearance [10, 11]. This has led to uncertainty about white matter retention differences between tracers and their possible influence on cortical retention estimates [12]. Another potential source of variability is the use of different image processing pipelines that may involve heterogeneous spatial normalization techniques and/or strategies for defining reference regions and cortical regions of interest.
A worthwhile strategy for directly comparing two or more tracers is to examine data from a cohort scanned with both agents. Recent studies have used this strategy to examine relationships between PiB and flutemetamol [2] and PiB and florbetapir [6, 4]. Here, we linked separate datasets from these studies (PiB–flutemetamol, PiB–florbetapir) and used the tracer in common (PiB) as a means to perform comparisons among the three tracers.
To investigate the possibility that cortical retention and nonspecific white matter retention differ between tracers, we investigated cortical grey matter and subcortical white matter retention ratios for each 18F tracer relative to PiB. We also investigated the influence of different reference regions (brainstem/pons, cerebellar grey matter, whole cerebellum) on the cortical retention estimates for each pair of tracers. In addition, while our primary analysis used a common image processing pipeline, a secondary analysis investigated the influence of different image processing methods (calculating tracer retention in native space versus using a template-based transformation). Finally, we examined the feasibility of converting previously established tracer-specific thresholds for amyloid positivity to other tracers. The reliable conversion of thresholds from one tracer to another is an important step for establishing greater methodological uniformity across amyloid PET studies.
It is important to note that our two study populations were not matched for amyloid status or other important subject and image acquisition characteristics, so we did not aim to compare tracer measurements between these study populations directly. Rather, we sought to examine characteristics of each 18F tracer relative to those of PiB for each study population separately, as a step towards developing standardization approaches.
Materials and methods
Participants
Our study populations consisted of 40 participants (20 with mild cognitive impairment [MCI], 20 with AD) who underwent structural MRI and PET imaging sessions with PiB and flutemetamol within a period of less than 40 days at several centres in Europe as described previously [2]. The other sample consisted of 32 cognitively normal, MCI and AD participants from the Alzheimer’s Disease Neuroimaging Initiative (ADNI, see below) who underwent structural MRI and PET imaging sessions with PiB and florbetapir approximately 1.5 years apart as described previously [6]. All participants gave written informed consent that was approved by the Internal Review Board of each participating institution.
ADNI
The ADNI is a longitudinal observational biomarker and clinical study supported by the NIH, private pharmaceutical companies and nonprofit organizations. Approximately 50 medical centre and university sites across the US and Canada participate (http://adni.loni.usc.edu/about). The first ADNI protocol, ADNI1, enrolled approximately 800 participants (volunteers with normal cognition, MCI and AD). Subjects underwent multiple longitudinal biomarker and cognitive measurements at intervals of 6–12 month for 2–4 years. An add-on study enabled longitudinal PiB scanning in a subset of approximately 100 participants. Florbetapir scanning began with ADNI GO and has continued in ADNI 2, which is ongoing. While new subjects have been enrolled in ADNI GO/2, some ADNI1 subjects continued to participate. The ADNI participants included in the present study comprised a subset of 32 individuals who underwent at least one PiB scan and a subsequent florbetapir scan.
Tracer synthesis and PET imaging
Tracer synthesis and image acquisition details are described in detail elsewhere for the PiB–flutemetamol population [13, 11, 2] and the PiB–florbetapir population [6] (http://adni-info.org). Briefly, for the PiB–flutemetamol study, PiB images (six 5-min frames acquired from 40 to 70 min after injection) and flutemetamol images (six 5-min frames acquired from 85 to 115 min after injection) were realigned and smoothed to a common resolution of 6 mm FWHM to account for variations in data acquisition across scanners. For the ADNI PiB–florbetapir cohort, PiB and florbetapir images (four 5-min frames acquired from 50 to 70 min after injection) were realigned and smoothed to a common resolution of 8 mm FWHM.
Cortical retention ratio quantification
Subsequent image processing with FreeSurfer proceeded in parallel for our two datasets. Structural T1 images acquired concurrently with the PET images were used as a structural template to define cortical regions of interest and reference regions in native space for each subject using FreeSurfer (version 4.5.0; http://surfer.nmr.mgh.harvard.edu) as described previously [6, 14, 15].
PET scans were coregistered to the structural MR scans, which were segmented and parcellated into individual cortical regions with FreeSurfer. They were subsequently used to extract mean PiB, flutemetamol and florbetapir cortical retention ratios from grey matter within lateral and medial frontal, anterior and posterior cingulate, lateral parietal and lateral temporal cortices. Retention in four large regions of interest (frontal, cingulate, parietal and temporal cortices) was averaged to create a single cortical composite region. Finally, the mean retention in the brainstem, cerebellar grey matter and whole cerebellum (grey and white matter) reference regions was computed. Retention ratios were generated by dividing each of the five cortical regions of interest (frontal, cingulate, parietal, temporal and cortical composite) by each of the three reference regions (brainstem/pons, cerebellar grey matter, whole cerebellum), resulting in 15 retention ratios for each subject per tracer. We also examined retention in a FreeSurfer-defined subcortical white matter region relative to the cerebellar grey matter.
Comparison of image analysis methods
The FreeSurfer method analyses the data in native space. To investigate whether a standard space method that does not rely on structural images would generate similar results, we processed all scans with a fully automated PET-only adaptive template method that has been described recently [16]. Briefly, this method involves spatial normalization of PET images to an amyloid PET adaptive template in Montreal Neurological Institute (MNI) space that is generated using flutemetamol scans and can accommodate scans ranging from low to high tracer cortical retention. The adaptive template fitting method proceeds by iteratively optimizing the template appearance and spatial transformation to converge on an optimal set of parameters for a particular subject’s scan. Each pair of 18F and PiB images were coregistered, and the adaptive template method was used to spatially normalize the PIB scan to MNI template space. Normalization of the PiB scan (rather than 18F tracer scan) to the template made it possible to avoid bias since the template is based on flutemetamol images. The subject-specific transformation was then used to transform each coregistered 18F scan to MNI template space. Retention ratios were computed for a cortical composite and reference regions (pons, cerebellar grey matter and whole cerebellum) that are analogous to the native-space versions used with the FreeSurfer method, except that FreeSurfer generates a brainstem region including the pons and adjacent structures while the adaptive template method generates a smaller pons-only segment of the brainstem.
Statistical analyses
Wilcoxon signed-ranks tests were used to compare mean white matter retention for each pair of tracers (PiB and flutemetamol, PiB and florbetapir), and α = 0.05 was Bonferroni-corrected to adjust for the four comparisons per tracer pair, resulting in α = 0.012. Correlations between cortical retention for each pair of tracers and for image processing methods were assessed using Spearman’s rho (ρ) because the datasets were not normally distributed. Slope and intercept values from the linear regression equations representing correlations between pairs of tracers were used to convert threshold values between radiotracer units.
Results
Study populations
While we did not perform direct comparisons between our unmatched study populations because they were unmatched in several ways, demographic information is summarized for the two study populations in Table 1. The PiB–florbetapir population (the ADNI participants) were slightly older, had a higher proportion of men, and had a slightly higher education level compared with the PiB–flutemetamol subjects. The primary difference, however, between the two populations was that due to study design features, the pair of PET scans was concurrent for the PiB–flutemetamol population (<40 days between sessions) whereas the PiB–florbetapir scans were separated by an average of 1.5 ± 0.7 years. Because of this longer time interval, the diagnosis in 7 of 32 subjects (22 %) changed between their initial (PiB) scan and their subsequent (florbetapir) scan (see Table 1 footnote for details).
Subcortical white matter retention
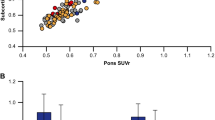
The pattern of retention in the subcortical white matter relative to the cerebellar grey matter differed for each 18F tracer relative to PiB (Table 2, Fig. 1). In the PiB–flutemetamol group (Fig. 1a), white matter retention was higher for flutemetamol (2.03 ± 0.26) than for PiB (1.81 ± 0.27, p < 0.001). White matter retention in the PiB–florbetapir group (Fig. 1b) did not meet our corrected statistical threshold (see Statistical analyses) but was marginally higher (p = 0.04) for PiB (1.87 ± 0.29) compared to florbetapir (1.77 ± 0.27).
Bar plots comparing the retention of each tracer in the subcortical white matter and the cortical composite region relative to the brainstem/pons, cerebellar grey matter and whole cerebellum for (a) the flutemetamol-PiB group and (b) the florbetapir-PiB group. Asterisks represent significant differences for each pair of regional associations
Cortical retention
In the PiB–flutemetamol group, cortical retention of PiB was higher than that of flutemetamol using brainstem and whole cerebellum normalization (p ≤ 0.001), but there was no difference in PiB and flutemetamol cortical retention with cerebellar grey matter normalization. In the PiB–florbetapir group, cortical retention was consistently higher for PiB than for florbetapir across all reference regions (p ≤ 0.01; Table 2, Fig. 1).
Composite cortical retention associations for the PiB–flutemetamol and PiB–florbetapir groups are shown for all three reference regions in Fig. 2. Spearman’s rho and slopes representing the linear regression for each tracer pair across cortical regions of interest (frontal, cingulate, parietal, temporal, and composite summary region) and with all three reference regions (brainstem, cerebellar grey matter, whole cerebellum) are also shown in the Supplementary table. The range of slopes for PiB–florbetapir associations was similar across all cortical regions of interest and reference regions (slope 0.59 – 0.66), indicating a narrower range of cortical retention values for florbetapir than for PiB. For PiB–flutemetamol correlations, on the other hand, slopes were closer to unity for the cerebellar grey matter-normalized (slope 0.79–0.83) and whole cerebellum-normalized values (slope 0.72 – 0.79), indicating similar cortical retention of PiB and flutemetamol when cerebellar reference regions were used.
Correlations between summary cortical composite retention ratios plotted for (a) subjects who received flutemetamol and PiB scans and (b) subjects who received florbetapir and PiB scans. Three reference regions (left brainstem/pons, centre cerebellar grey matter, right whole cerebellum) were used. Linear regression equations and Spearman’s rho are given for each correlation
Correlations between each pair of tracers were high (p < 0.001 in all cases) across all individual cortical regions examined, and no cortical region consistently showed the highest or lowest correlation. Correlations were highest with brainstem normalization (PiB–flutemetamol, ρ = 0.98 – 0.99 across cortical regions of interest; PiB–florbetapir, ρ = 0.95 – 0.97), compared to the correlations with cerebellar grey matter normalization (PiB–flutemetamol, ρ = 0.84 – 0.93; PiB–florbetapir, ρ = 0.83 – 0.86) and whole cerebellum normalization (PiB–flutemetamol, ρ = 0.88 – 0.95; PiB–florbetapir, ρ = 0.86 – 0.90). However, brainstem normalization also resulted in the lowest slopes overall (PiB–flutemetamol, slope 0.64 – 0.67; PiB–florbetapir, slope 0.61 – 0.66), indicating a reduced range of flutemetamol and florbetapir cortical retention relative to PiB.
Image processing methods
Comparing correlations between FreeSurfer and adaptive template processing indicated that cortical composite averages were highly correlated within each tracer. Correlations between composite values measured using the FreeSurfer (native space) processing method and the adaptive template method ranged from 0.90 to 0.99, and were highest for the brainstem-normalized data (Table 3). In addition, correlations between image processing methods were comparable across tracers (Fig. 3).
Example correlations between the two image processing methods (see complete list of correlations across tracers and reference regions in Table 3). Linear regression equations and values of Spearman’s rho are shown for the correlations between the FreeSurfer-based and adaptive template-based cortical composite values for (a) flutemetamol and (b) florbetapir, each with brainstem/pons and whole cerebellum normalization
Conversion of thresholds between radiotracers
Using the FreeSurfer-based datasets, we used the linear correlations between pairs of composite retention ratios to convert amyloid positivity threshold values established for one tracer to corresponding values for another, as we have shown previously for florbetapir and PiB [6]. This method enables direct comparison of independently derived thresholds, using PiB as the common tracer linking the 18F tracers. Specifically, we used composite summary measures from our PiB–flutemetamol and PiB–florbetapir datasets to calculate linear regression equations representing the correlation between pairs of tracers (analysed either with the same or with different reference regions). These regression equations were then used to convert threshold values from one tracer to another.
Using this strategy, we examined the feasibility of converting a previously validated PiB value of 1.47 (based on cerebellar grey matter normalization), which was originally derived from a receiver operating characteristic analysis of AD and cognitively normal ADNI participants [17]. This PiB threshold was transformed to a threshold for florbetapir (1.13) and for flutemetamol (1.21), both with whole cerebellum normalization (Fig. 4a). Next, we examined a previously validated florbetapir threshold, which was derived from the upper bound of the 95 % confidence interval above the mean uptake in a population of young, cognitively normal individuals [18] and further validated by histopathology data in a separate population [19]. This threshold was subsequently converted to a value compatible with the FreeSurfer processing stream [20], resulting in a value of 1.11 (based on whole cerebellum normalization). This was converted to a value of 1.27 for PiB (whole cerebellum normalization), which was in turn converted to a value of 1.20 for flutemetamol (whole cerebellum normalization) (Fig. 4b). Using this approach, we were able to derive comparative thresholds between florbetapir and flutemetamol of 1.11 and 1.20, respectively, despite the fact that these two tracers were not directly compared in the same subjects.
Two previously established thresholds for cortical amyloid positivity can be converted between radiotracers using PiB as the link between flutemetamol and florbetapir. Two examples are shown. a Using the linear regression equations, a PiB-based threshold of 1.47 (cerebellar grey matter normalization) was converted to 1.13 for florbetapir (whole cerebellum normalization) and to 1.21 for flutemetamol (whole cerebellum normalization). b A florbetapir-based threshold of 1.11 (whole cerebellum normalization) was converted to 1.27 for PiB (whole cerebellum normalization), and this new PIB threshold was in turn converted to flutemetamol units, resulting in a value of 1.20 (whole cerebellum normalization) that is almost identical to the flutemetamol threshold calculated from the first threshold (a flutemetamol threshold 1.21)
Discussion
We compared amyloid PET measurements in two different study populations, one that received both PiB and flutemetamol scans, and another that received PiB and florbetapir scans. To our knowledge, flutemetamol and florbetapir have not previously been compared to one another. Overall, PiB–florbetapir and PiB–flutemetamol correlations were high. Using a common image processing pipeline for both datasets, and using PiB as the link between the two datasets, we compared PiB–flutemetamol and PiB–florbetapir correlations and used them to examine the reproducibility of two independently-derived thresholds for amyloid positivity. We converted these two thresholds (1.47 for PiB and 1.11 for florbetapir) into flutemetamol thresholds using linear regression equations, which resulted in almost identical values of 1.21 and 1.20 (Fig. 4a right and b right). Similarly, the 1.47 PiB threshold was converted to a florbetapir threshold of 1.13 which is very similar to an autopsy-validated florbetapir threshold of 1.11. The fact that these thresholds were derived from independent populations, tracers and criteria, yet were internally consistent when converting between tracers, indicates strong reliability of the linear correlation between tracers despite the use of relatively small and unmatched populations (PiB–flutemetamol group, 40 subjects; PiB–florbetapir group, 32 subjects) to define these relationships.
Selection of cut-off values that are consistent across studies and tracers is increasingly important as amyloid PET imaging becomes more widely used. We see the current study as a proof of concept for the general approach to comparing amyloid PET tracers on the same scale (e.g. 0–100), and there are currently efforts underway to formalize this approach (with units called “centiloids”). Ideally, a tracer-specific threshold should be compared to a population of young normal control subjects. Since this is not possible for every study, the method proposed here is an alternative that enables conversion of thresholds between tracers and reference regions with reasonable consistency. Nonetheless, selection of a threshold should depend on the goals of the trial or study, since a more liberal or more conservative threshold will have different consequences depending on the clinical population, research question, or clinical trial it is applied to.
We also examined differences in cortical and white matter retention between the tracer pairs. Cortical retention was higher for PiB than for florbetapir regardless of the reference region; similarly, it was higher for PiB than for flutemetamol in two out of three reference regions. There was substantial white matter retention of all three tracers, but the pattern was different for each tracer relative to PiB. Specifically, white matter retention of flutemetamol was higher than that of PiB (Fig. 1, Table 2). On the other hand, white matter retention florbetapir and PIB was comparable (in fact, it was marginally higher for PiB than for florbetapir, although the difference did not meet our corrected statistical threshold).
White matter retention introduces a number of challenges in making accurate cortical measurements and comparing tracers. For example, the difference in white matter retention between tracers influences the measurement of cortical retention when using a reference region with white matter content. When normalizing cortical values by the brainstem (the reference region with the highest proportion of white matter) the higher white matter retention of flutemetamol compared to florbetapir reduces the cortical retention ratios more for flutemetamol, resulting in nearly identical slopes for PiB–flutemetamol and PiB–florbetapir. Another important concern is the influence of white matter retention on grey matter retention and vice versa due to partial volume effects. The high white matter retention we observed for PiB may have been due to the influence of higher PiB grey matter retention on neighbouring white matter voxels. This possibility is consistent with a recent report that high cortical flutemetamol retention results in higher estimated white matter retention for the same reason [12]. While the partial volume effect would be expected to influence the PiB–flutemetamol dataset as well, the fact that we did not observe this may be due to a disproportionate amount of nonspecific retention of flutemetamol. In other words, the effect of flutemetamol’s high white matter retention may have exceeded the partial volume effect of high cortical PiB on PiB white matter retention.
There was high concordance between cortical retention ratios generated using the FreeSurfer method, which relies on a coregistered structural MR image and quantifies tracer retention in the subject’s native space, and the adaptive template method, which relies on warping amyloid PET data to a template without the use of a structural image. There was strong agreement between these image processing methods across pairs of tracers and reference regions, suggesting that amyloid PET measurements are robust against image warping and different methods for defining cortical and reference regions. These findings are consistent with our previous findings comparing PiB and florbetapir using the FreeSurfer-based method and a different PET template approach [6]. However, the actual threshold level may vary depending on how target and reference regions are defined. The use of the entire brainstem (FreeSurfer) versus a pons-only region (adaptive template) did produce substantially different results since correlations between processing methods were highest for this reference region as a whole (Supplementary table).
The correlation and slope measurements derived from the PiB–flutemetamol and PiB–florbetapir associations provide complementary information. The correlation indicates how precisely the retention of one tracer in an individual can be estimated from another tracer’s retention. We found that correlations between each pair of radiotracers were highest with brainstem normalization and lower with cerebellar grey matter or whole cerebellum normalization (Fig. 2, Supplementary table). One possible explanation is that use of the brainstem reduces white matter retention variability between tracers and between individuals that is independent of the level of cortical retention (since brainstem white matter retention, unlike subcortical white matter retention, is not influenced by partial volume effects on cortical retention). The magnitude of the slope, on the other hand, indicates the degree of similarity between the range of cortical retention values for one tracer relative to another. Florbetapir cortical retention was lower than that of PiB across all reference regions (slope 0.61 to 0.65), whereas for PiB–flutemetamol, the slope varied: it was lowest for brainstem-normalized data (Fig. 1; 0.65) and higher for whole cerebellum-normalized and cerebellar grey matter-normalized data (0.72 and 0.81, respectively; Fig. 1). When normalizing cortical values by the brainstem (the reference region with the highest proportion of white matter) the higher white matter retention of flutemetamol compared to florbetapir reduces the cortical retention ratios more for flutemetamol, resulting in nearly identical slopes for PiB–flutemetamol and PiB–florbetapir.
This study had a number of limitations which were primarily related to the combination of study designs and populations. While we focused on variability between tracer pair correlations due to characteristics of the tracers themselves (e.g. differences in non-specific uptake), there are a number of possible sources of variability in tracer pair correlations such as differing proportions of subjects with high versus low amyloid in our two populations. Slightly lower correlations for the PiB–florbetapir group than for the PiB–flutemetamol group may also have been due to the longer time between scans. The longer time between the PiB and florbetapir scans also meant that in about one-fifth of subjects the diagnosis changed between scans. However, we have previously found that subjects whose diagnosis changes do not appear to show a greater discrepancy in cortical retention between PiB and florbetapir compared with those whose diagnosis remains stable [6]. The two populations also differed with respect to smoothing and the time window used to calculate tracer retention ratios. The PiB–florbetapir scans were smoothed to an 8 × 8 × 8 mm resolution while the PiB–flutemetamol scans were smoothed to a 6 × 6 × 6 mm resolution. Partial volume effects were therefore slightly greater in the PiB–florbetapir group. However, it is unclear whether this influenced the group differences observed since the group differences were probably due to a combination of partial volume effects and tracer characteristics. In the PiB–flutemetamol group, PiB images were acquired from 40 to 70 min after injection and flutemetamol images were acquired from 85 to 115 min, whereas in the PiB–florbetapir group, both PiB and florbetapir were acquired from 50 to 70 min. These differing time windows could have influenced the relative cortical and white matter effects observed if, for example, white to grey matter retention increases over time. Finally, test–retest error accounts for some of the between-tracer variability observed. In previous studies have test–retest errors in cortical retention associated with repeated amyloid PET scans in the same individuals have been found to be in the range of 1–9 % [18, 21, 2].
To summarize, we found that despite differing cortical and white matter retention characteristics, amyloid PET imaging tracers can be compared to one another using cohorts scanned with multiple tracers and processed using a common image analysis pipeline. Thresholds for categorizing subjects as amyloid-positive or amyloid-negative can be converted between tracers with a high level of consistency. The use of cohorts scanned with more than one tracer, including a common tracer such as PiB, enables flexible comparison of thresholds using linear correlations representing the relationships between cortical retention ratios. The availability of validated cross-tracer thresholds will be of benefit in the use of amyloid PET in clinical research, including trials of potential therapeutics.
References
Rowe CC, Ackerman U, Browne W, Mulligan R, Pike KL, O'Keefe G, et al. Imaging of amyloid beta in Alzheimer's disease with 18F-BAY94-9172, a novel PET tracer: proof of mechanism. Lancet Neurol. 2008;7(2):129–35. doi:10.1016/S1474-4422(08)70001-2.
Vandenberghe R, Van Laere K, Ivanoiu A, Salmon E, Bastin C, Triau E, et al. 18F-flutemetamol amyloid imaging in Alzheimer disease and mild cognitive impairment: a phase 2 trial. Ann Neurol. 2010;68(3):319–29. doi:10.1002/ana.22068.
Villemagne VL, Mulligan RS, Pejoska S, Ong K, Jones G, O'Keefe G, et al. Comparison of 11C-PiB and 18F-florbetaben for Abeta imaging in ageing and Alzheimer's disease. Eur J Nucl Med Mol Imaging. 2012;39(6):983–9. doi:10.1007/s00259-012-2088-x.
Wolk DA, Zhang Z, Boudhar S, Clark CM, Pontecorvo MJ, Arnold SE. Amyloid imaging in Alzheimer's disease: comparison of florbetapir and Pittsburgh compound-B positron emission tomography. J Neurol Neurosurg Psychiatry. 2012;83(9):923–6. doi:10.1136/jnnp-2012-302548.
Cselenyi Z, Jonhagen ME, Forsberg A, Halldin C, Julin P, Schou M, et al. Clinical validation of 18F-AZD4694, an amyloid-beta-specific PET radioligand. J Nucl Med. 2012;53(3):415–24. doi:10.2967/jnumed.111.094029.
Landau SM, Breault C, Joshi AD, Pontecorvo M, Mathis CA, Jagust WJ, et al. Amyloid-beta imaging with Pittsburgh compound B and florbetapir: comparing radiotracers and quantification methods. J Nucl Med. 2013;54(1):70–7. doi:10.2967/jnumed.112.109009.
Wong DF, Rosenberg PB, Zhou Y, Kumar A, Raymont V, Ravert HT, et al. In vivo imaging of amyloid deposition in Alzheimer disease using the radioligand 18F-AV-45 (florbetapir [corrected] F 18). J Nucl Med. 2010;51(6):913–20. doi:10.2967/jnumed.109.069088.
Forsberg A, Engler H, Almkvist O, Blomquist G, Hagman G, Wall A, et al. PET imaging of amyloid deposition in patients with mild cognitive impairment. Neurobiol Aging. 2008;29(10):1456–65. doi:10.1016/j.neurobiolaging.2007.03.029.
Rowe CC, Ng S, Ackermann U, Gong SJ, Pike K, Savage G, et al. Imaging beta-amyloid burden in aging and dementia. Neurology. 2007;68(20):1718–25. doi:10.1212/01.wnl.0000261919.22630.ea.
Fodero-Tavoletti MT, Rowe CC, McLean CA, Leone L, Li QX, Masters CL, et al. Characterization of PiB binding to white matter in Alzheimer disease and other dementias. J Nucl Med. 2009;50(2):198–204. doi:10.2967/jnumed.108.057984.
Nelissen N, Van Laere K, Thurfjell L, Owenius R, Vandenbulcke M, Koole M, et al. Phase 1 study of the Pittsburgh compound B derivative 18F-flutemetamol in healthy volunteers and patients with probable Alzheimer disease. J Nucl Med. 2009;50(8):1251–9. doi:10.2967/jnumed.109.063305.
Thomas BA, Erlandsson K, Modat M, Thurfjell L, Vandenberghe R, Ourselin S, et al. The importance of appropriate partial volume correction for PET quantification in Alzheimer's disease. Eur J Nucl Med Mol Imaging. 2011;38(6):1104–19. doi:10.1007/s00259-011-1745-9.
Klunk WE, Engler H, Nordberg A, Wang Y, Blomqvist G, Holt DP, et al. Imaging brain amyloid in Alzheimer's disease with Pittsburgh Compound-B. Ann Neurol. 2004;55(3):306–19. doi:10.1002/ana.20009.
Landau SM, Marks SM, Mormino EC, Rabinovici GD, Oh H, O'Neil JP, et al. Association of lifetime cognitive engagement and low beta-amyloid deposition. Arch Neurol. 2012;69(5):623–9. doi:10.1001/archneurol.2011.2748.
Mormino EC, Kluth JT, Madison CM, Rabinovici GD, Baker SL, Miller BL, et al. Episodic memory loss is related to hippocampal-mediated beta-amyloid deposition in elderly subjects. Brain. 2009;132(Pt 5):1310–23. doi:10.1093/brain/awn320.
Lundqvist R, Lilja J, Thomas BA, Lotjonen J, Villemagne VL, Rowe CC, et al. Implementation and validation of an adaptive template registration method for 18F-flutemetamol imaging data. J Nucl Med. 2013;54(8):1472–8. doi:10.2967/jnumed.112.115006.
Jagust WJ, Landau SM, Shaw LM, Trojanowski JQ, Koeppe RA, Reiman EM, et al. Relationships between biomarkers in aging and dementia. Neurology. 2009;73(15):1193–9. doi:10.1212/WNL.0b013e3181bc010c.
Joshi AD, Pontecorvo MJ, Clark CM, Carpenter AP, Jennings DL, Sadowsky CH, et al. Performance characteristics of amyloid PET with florbetapir F 18 in patients with alzheimer's disease and cognitively normal subjects. J Nucl Med. 2012;53(3):378–84. doi:10.2967/jnumed.111.090340.
Clark CM, Pontecorvo MJ, Beach TG, Bedell BJ, Coleman RE, Doraiswamy PM, et al. Cerebral PET with florbetapir compared with neuropathology at autopsy for detection of neuritic amyloid-beta plaques: a prospective cohort study. Lancet Neurol. 2012;11(8):669–78. doi:10.1016/S1474-4422(12)70142-4.
Landau SM, Mintun M, Joshi A, Koeppe RA, Petersen RC, Aisen P, et al. Amyloid deposition, hypometabolism, and longitudinal cognitive decline. Ann Neurol. 2012;72(4):578–86. doi:10.1002/ana.23650.
Tolboom N, Yaqub M, Boellaard R, Luurtsema G, Windhorst AD, Scheltens P, et al. Test-retest variability of quantitative [11C]PIB studies in Alzheimer's disease. Eur J Nucl Med Mol Imaging. 2009;36(10):1629–38. doi:10.1007/s00259-009-1129-6.
Acknowledgments
We thank Robert Koeppe, Abhinay Joshi, and Cindee Madison for helpful discussions and assistance with image analysis.
B.T. acknowledges the support of GlaxoSmithKline (GSK) and also that UCL/UCLH receives a portion of its research funding from the UK Department of Health Biomedical Research Centre’s funding scheme.
Data collection and sharing for this project was funded by the ADNI (National Institutes of Health grant U01 AG024904). ADNI is funded by the National Institute on Aging, and the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Abbott; Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Amorfix Life Sciences Ltd.; AstraZeneca; Bayer HealthCare; BioClinica, Inc.; Biogen Idec Inc.; Bristol-Myers Squibb Company; Eisai Inc.; Elan Pharmaceuticals Inc.; Eli Lilly and Company; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; GE Healthcare; Innogenetics, N.V.; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Medpace, Inc.; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Servier; Synarc Inc.; and Takeda Pharmaceutical Company. The Canadian Institutes of Health Research provides funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of California, Los Angeles. This research was also supported by NIH grants P30 AG010129 and K01 AG030514.
Conflicts of interest
S.M. Landau has previously consulted for Avid Radiopharmaceuticals, Inc., Janssen Alzheimer Immunotherapy, and Biogen Idec.
B.A. Thomas has performed research under a grant partially supported by GE Healthcare.
L. Thurfjell is an employee of GE Healthcare.
M. Schmidt is an employee of Janssen Research and Development.
R. Margolin is an employee of Janssen Alzheimer Immunotherapy R&D LLC.
M. Pontecorvo and M. Mintun are employees of Avid Radiopharmaceuticals, Inc.
W.J. Jagust collaborates with Avid Radiopharmaceuticals, Inc. through participation in the ADNI. He has previously consulted for GE Healthcare, Genentech, and Elan/Janssen Alzheimer Immunotherapy, and is currently a consultant to F. Hoffman-LaRoche and Synarc.
Author information
Authors and Affiliations
Consortia
Corresponding author
Additional information
Data used in the preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (http://adni.loni.usc.edu/). As such, the ADNI investigators contributed to the design and implementation of ADNI and/or provided data, but did not participate in the analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/about/governance
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary table
(DOCX 64 kb)
Rights and permissions
About this article
Cite this article
Landau, S.M., Thomas, B.A., Thurfjell, L. et al. Amyloid PET imaging in Alzheimer’s disease: a comparison of three radiotracers. Eur J Nucl Med Mol Imaging 41, 1398–1407 (2014). https://doi.org/10.1007/s00259-014-2753-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00259-014-2753-3