Abstract
The incidence of cancer is increasing worldwide; likewise, the emergence of antibiotic-resistant biofilm-forming pathogens has led to a tremendous increase in morbidity and mortality. This study aimed to evaluate the probiotic properties of bacteriocin-producing Enterococcus sp. with a focus on their anti-biofilm and anticancer activities. Three of 79 Enterococcus isolates (FM43, FM65, FM50) were identified as producers of broad-spectrum bioactive molecules and were molecularly characterized as Enterococcus faecium by 16S rRNA sequencing. Phenotypic and genotypic screening for potential virulence factors revealed no factors known to promote pathogenicity. Treatment with proteinase K resulted in diminished antimicrobial activity; PCR-based screening for bacteriocin genes suggested the presence of both entA and entB genes that encode enterocins A and B, respectively. Maximum antimicrobial activity was detected during the early stationary phase, while activity disappeared after 24 h in culture. Bacteriocins from these isolates were stable at high temperatures and over a wide range of pH. Interestingly, crude supernatants of Ent. faecium FM43 and Ent. faecium FM50 resulted in significant destruction (80% and 48%, respectively; P < 0.05) of Streptococcus mutans ATCC 25175–associated preformed biofilms. Moreover, in vitro cytotoxicity assays revealed that extracts from Ent. faecium isolates FM43, FM65, and FM50 inhibited Caco-2 cell proliferation by 76.9%, 70%, and 85.3%, respectively. Taken together, the multifunctional capabilities of the microbial-derived proteins identified in our study suggest potentially important roles as alternative treatments for biofilm-associated infections and cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Enterococcus is one of the main genera belonging to the lactic acid bacteria (LAB) group. Pathogenic and commensal strains of Enterococcus have recently become the subject of extensive research [1, 2]. This is largely due to the fact that, despite their pathogenic potential, commensal enterococci colonize the gastrointestinal tracts of humans and numerous animal species; these microorganisms are minimally virulent and have been used safely for decades as probiotics for both humans and animals [3]. Furthermore, enterococci have been shown to promote several desirable health benefits, including their role in reducing cholesterol levels [4] and strengthening the immune system [5].
Bacteriocins offer a promising alternative to traditional antibiotics and may be used as part of an overall strategy to combat the rise in antibiotic resistance [6]. The antimicrobial activity of bacteriocins with respect to both food-borne pathogens and bacteria that promote food spoilage has suggested that they may be applied to promote food preservation [7]. Bacteriocin production has been detected widely among the food-borne enterococci; notable among that are the strains of Ent. faecium and Ent. faecalis [8] that produce antimicrobial peptides known as enterocins. Many of these enterocins have been shown to have activity against Listeria monocytogenes, Staphylococcus aureus, Clostridium botulinum, and Vibrio cholerae [9]. Of note, Ent. faecium strains are among the most frequent producers of bacteriocins of the group known as fecal LAB microbiota [10]. However, the use of enterococci for clinical and industrial purposes must be carefully evaluated due to the emergence of virulence and resistance traits associated with these bacteria [11].
Dental caries are a commonplace and chronic complication that typically results from a homoeostatic imbalance between the host and the oral microbiome [12]. Alpha-hemolytic strains of Streptococcus mutans and Strep. sobrinus, which belong to the mutans group of streptococci [13], have been identified as major contributors to the development of dental caries; these bacteria are detected on the tooth surface in the form of a biofilm [14]. Bacterial biofilms are highly resistant to antibiotics and host defense mechanisms and as such pose a great challenge with respect to treatment and eradication of infection [15]. As such, attempts are underway to promote reduction or complete prevention of bacterial adherence and biofilm formation using a variety of biologically based intervention strategies, including the use of bacteriophages, phytochemicals, matrix-degrading enzymes, and probiotics [16,17,18]. Among these approaches, the use of bacteriocins for microbial biofilm control is a relatively new area of research [19]. Likewise, with the growing popularity of peptide-based therapeutics, several bacteriocins have been identified that are selectively toxic toward cancer cells [20]. Bacteriocin-mediated cytotoxicity and their capacity to target cancer cells may be attributed to structural features that include hydrophobicity, positive net charge, and size, as well as their capacity to oligomerize and to generate amphipathic structures, similar to features reported for antimicrobial peptides [21]. The eukaryotic cell membrane is a likely primary target; as such, the enhanced expression of negatively charged cell surface molecules on cancer cells provides greater vulnerability to the cytotoxic activity of these positively charged peptide mediators [22].
Given these perspectives, our aim in this study was to isolate bacteriocin-producing enterococci from various food and clinical samples in Egypt; this was followed by screening for virulence and antibiotic resistance traits so as to determine their safety for use in clinical applications. Toward this end, we examined the impact of bacteriocins from these strains with respect to their potential to prevent biofilm formation and to limit proliferation of cancer cells.
Materials and Methods
Indicator Strains and Culture Conditions
The indicator strains used in this study and associated culture conditions are presented in Table 1. All bacterial strains used in this study were stored in 40% (v/v) glycerol at − 80 °C.
Isolate Purification and Identification
Seventy-nine isolates were recovered from food and stool slurries. Enterococci were identified by colony morphology, Gram-stain, catalase test, and growth on bile esculin agar (Himedia, Mumbai, Maharashtra, India; Online Resource 1); the latter test is scored as positive when organisms can grow in the presence of 4% bile and are capable of hydrolyzing esculin to esculetin that reacts with Fe3+ to form a dark brown to black precipitate on the agar plate. All samples collected were inoculated into tubes containing 5 mL GM17 broth (Oxoid, Basingstoke, Hampshire, UK); these cultures were incubated aerobically at 37 °C for 24 h. A one loopful of culture was used to generate a streak on each bile esculin agar plate; these plates were then incubated aerobically at 37 °C for 24 h. To eliminate possible contamination with other aerobic microorganisms, isolated Enterococcus strains were streaked on De Man, Rogosa, and Sharpe (MRS) agar (Online Resource 1) and incubated under anaerobic conditions that facilitate growth of facultatively anaerobic Enterococcus species. All plates were incubated in anaerobic jars (Oxoid gas system) at 37 °C for 48 h.
Screening for Antimicrobial Activity
The spot-on-lawn assay as described by Dundaret et al. [23] was performed for preliminary screening for antimicrobial activity targeting Lactobacillus sakei LMG 2313. Briefly, Enterococcus colonies grown overnight on MRS agar (Lab M, Heywood, Lancashire, UK) were overlaid with 8 mL soft agar containing 80 μL of Lact. sakei LMG 2313 and incubated for 18 h at 30 °C. Antimicrobial activities of the isolates were detected as clear zones around the colonies.
The antimicrobial spectrum of candidate isolates was also evaluated using the agar well technique [24] targeting Strep. mutans ATCC 25175, Staph. aureus AO760 (Clinical sample), Micrococcus lysodeikticus ATCC 4698, Escherichia coli ATCC 5087, and Salmonella typhi ATCC 35664 (Table 1). Briefly, a fully isolated enterococcal colony was cultured in MRS broth at 30 °C for 18 h. The culture was then subjected to centrifugation at 6000×g for 15 min at 4 °C; the cell-free supernatant (CFS) was used to screen for antimicrobial activity. One hundred microliters of the CFS was placed in wells of ~ 10 mm in diameter created in agar plates that were pre-seeded with the indicator strains as above (1% v/v; 1.0 × 106 CFU/mL). After pre-diffusion at 4 °C for 30 min, the plates were then incubated at 37 °C for 24 h; the diameters of the inhibition zones around each well were then measured. Clear inhibition zones of 12 mm or more were recorded as positive antibacterial activity for each of the isolates tested.
Isolates with broad-spectrum activity against all of the aforementioned indicator strains were subjected to further evaluation with Strep. mutans ATCC 25175 as the indicator strain. The inhibitory activities, represented as arbitrary units (AU) per milliliter of supernatants from each of the isolates, were evaluated using the agar well diffusion method. Crude preparations of enterococcal-derived bacteriocins were subjected to serial twofold dilution, and the diameters of the zones of inhibition generated by each dilution were measured. The AU for each preparation was calculated as the reciprocal of the highest dilution resulting in a zone of inhibition of 12 mm or more [25].
Molecular Identification Using 16S rRNA Gene Sequencing
Genomic DNA was extracted from the microorganisms of interest using the GenElute DNA Bacterial Genomic DNA Kit (Sigma-Aldrich, St. Louis, MO, USA). Taxonomical classifications of these isolates were assessed based on the DNA sequences of the specific regions of the genes encoding 16S rRNA amplified using the universal primer pair 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′) (IDT, USA) [26]. The final volume of each reaction mixture was 25 μL and included 12.5 μL of Dream Taq Green PCR Master Mix (Thermo Scientific, Wilmington, DE, USA), 1 μL of each primer (10 μM), 2 μL of template genomic DNA (100 ng/μL), and 8.5 μL of nuclease-free water. Each amplification cycle included an initial denaturation step of 94 °C for 3 min, followed by denaturation at 94 °C for 30 s, and annealing at 50 °C for 30 s, and extension at 72 °C for 2 min. This cycle was repeated 35 times, followed by a final extension step at 72 °C for 5 min. Amplifications were performed using the Biometra TAdvanced thermal cycler (Biometra, Göttingen, Germany). After cycling, 10 μL of each PCR reaction was evaluated on a 1.5% (w/v) agarose gel that was stained with ethidium bromide (0.5 mg/mL), and visualized under a Gel Doc EZ Imager (Bio-Rad Laboratories, USA). Amplification products were sequenced by Macrogen® (Gasan-dong, Geumcheon-gu, Seoul, South Korea) using capillary electrophoresis. The sequences obtained were assembled using BioEdit software v.7.0.5.3. Each sequence was used to query the GenBank database to retrieve the most closely matching sequences, using the mega BLAST tool available online at the National Center for Biotechnology Information (NCBI at https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE_TYPE=BlastSearch). Multiple sequence alignments including those amplified here and those identified as closely related obtained from the NCBI database were carried out, and phylogenetic analysis was performed using MEGA6 software [27] using the neighbor-joining tree method [28]; evolutionary distances were calculated using the maximum composite likelihood method [29].
Pathogenicity and Antibiotic Susceptibility Testing
The hemolytic activities of the selected isolates were assessed by growing them on Columbia blood agar (Oxoid, Basingstoke, Hampshire, UK; Online Resource 1) containing 5% defibrinated blood. Gelatinase activity was evaluated as previously described by Harrigan et al. [39]. Detection of specific virulence factors, including the collagen adhesion protein, endocarditis antigen, cytolysin, aggregation substance, enterococcal surface protein, and hyaluronidase, was performed by PCR amplification of their respective genes with primers listed in Table 2 and using thermocycler settings previously described by Baker et al. [40]; Ent. faecalis ATCC 700802 was included as a positive control. Antimicrobial resistance patterns were determined using the disk diffusion method as described in the guidelines provided by the Clinical Laboratory Standard Institute (CLSI) [41].
Physicochemical Characterization of Bioactive Products
CFSs prepared as described above were subjected to physicochemical characterization, including testing with various degrading enzymes, including proteinase K and α-chymotrypsin, each at a final concentration of 1 mg/mL. CFSs with or without enzyme were incubated at 37 °C for 60 min. Subsequently, enzyme activity was terminated by heating at 100 °C for 5 min. The stability of antimicrobial activity identified in each CFS was also evaluated after incubation at elevated temperatures (60, 80, 100, and 121 °C) and at different pH ranging from 3 to 11 (adjusted with 1 N HCL or 5 N NaOH, respectively). Residual antimicrobial activity that remained after each of the aforementioned interventions was determined via comparisons of the diameters of their respective zones of inhibition.
Bacteriocin Production Kinetics
Selected Enterococcus isolates were used to inoculate 100 mL aliquots of MRS broth followed by incubation at 30 °C for 24 h; 5 μL was withdrawn at intervals, including 6, 8, 10, 12, 16, and 24 h. After centrifugation at 6000×g for 10 min, the pellets were discarded and the supernatants were used to evaluate antimicrobial activity by measuring the diameters of the zones of inhibition as previously described. Additionally, growth of each isolate was measured spectrophotometrically at OD = 620 nm at each 1-h interval for 24 h using a microtiter plate reader (Tecan Sunrise, Austria) [42].
Protein Precipitation by Ammonium Sulfate (Partial Purification)
About 1 mL of an overnight broth culture prepared for each of the selected isolates was inoculated into 100 mL sterile MRS broth and incubated for 12 h at 30 °C. Following incubation, entire cultures were subjected to centrifugation at 13,000×g for 15 min at 4 °C in a cooling centrifuge (Sigma 3-30K, Germany). The CFS was then mixed with ammonium sulfate to 75% saturation [43, 44] followed by shaking for 30 min at 5 °C. This step was followed by centrifugation at 10,000×g for 10 min at 4 °C to collect the precipitated proteins, which were dissolved in 2 mL of distilled water to obtain a 50× concentrated protein solution containing bacteriocins.
The Anti-biofilm Activities of the Produced Bacteriocins
The anti-biofilm activities were evaluated as previously described by Merritt et al. [45]. Briefly, wells of flat-bottomed 96-well microtiter plates were inoculated with Strep. mutans ATCC 25175 (1% v/v; 1.0 × 106 CFU/mL; 100 μL per well) in BHI media supplemented with 0.2% sucrose to enhance the biofilm formation; these plates were incubated for 18 h at 37 °C. The resulting biofilms were washed twice with phosphate-buffered saline (PBS) to remove any non-adherent cells. CFS (100 μL) from cultures of Ent. faecium FM43, Ent. faecium FM50, and Ent. faecium FM65 were added to each well, at total protein concentrations of 3.9 mg/mL, 5.9 mg/mL, and 3.4 mg/mL respectively that were equivalent to planktonically active bacteriocin units of 640 AU/mL, 2560 AU/mL, and 2560 AU/mL respectively. The plates were then incubated at 37 °C for 24 h. The samples were then sonicated for 30 s using a bath sonicator (Sonix TV ss-series ultrasonicator, Sonix IV Ultrasonic Cleaning Systems, North Charleston, SC, USA); the viable bacterial count (VC) was determined as previously described by Kadouri et al. [46]; bacterial isolates exposed to the same condition but without exposure to CFSs containing bacteriocins served as the positive control. Percentage inhibition of biofilm formation was calculated according to the following equation: \( \mathsf{Inhibition}\%=\mathsf{100}-\left(\mathsf{VC}\ \mathsf{sample}/\mathsf{VC}\ \mathsf{control}\right)\times \mathsf{100}.\mathsf{All}\ \mathsf{experiments}\ \mathsf{were}\ \mathsf{performed}\ \mathsf{in}\ \mathsf{triplicate}. \)
Scanning Electron Microscopy
Ten-microliter aliquots of overnight cultures of Strep. mutans ATCC 25175 matched to 0.5 McFarland standard (1.0 × 106 CFU/mL) were placed in the center of isopore polycarbonate sterile membrane filters (Sigma-Aldrich, St. Louis, MO, USA) and permitted to settle for 24 h. Each filter was then loaded onto a tryptone soya agar plate (TSA; Oxoid, UK) that has been covered with 100 μL of crude CFSs from each of the three Ent. faecium isolates; the plates were incubated at 37 °C for 24 h. After incubation, membrane filters were washed three times with PBS (pH 7.4), fixed with 2.5% (v/v) glutaraldehyde (Oxford, India) for 2 h at 4 °C, washed twice with PBS, and then dehydrated for 10 min with an ethanol gradient including 30%, 50%, 70%, 90%, and 100% (v/v) [47]. The preparations were then coated with gold and examined with a JSM-6510 electron microscope (JEOL, Akishima, Tokyo, Japan) at a voltage of 30 kV and at magnifications ranging from × 5000 to × 15,000. Samples of bacteriocin-treated cells were analyzed by SEM for any morphological changes using untreated cells as a reference.
PCR Screening of Bacteriocin-Related Structural Genes
Screening for genes encoding bacteriocins that are characteristic of Enterococcus sp. was performed by PCR using the primers listed in Table 2. PCR amplification of these genes was performed as previously described by Dundar et al. [23]. Briefly, each cycle included an initial denaturation step at 94 °C for 5 min, followed by 40 cycles that include denaturation at 94 °C for 30 s, annealing at 49–55 °C (based on primer pairs, see Table 2) for 60 s, and extension at 72 °C for 2 min and a final extension step at 72 °C for 10 min. The amplification products were evaluated by gel electrophoresis as previously described by Sambrook et al. [48] using 1.5% (w/v) agarose gel and 100-bp and 1-kb ladders (GeneRuler, Thermo Fisher Scientific, USA) to demarcate molecular size.
Assessment of Cytotoxicity on HepG2 and Caco-2 Cell Lines
Partially purified bacteriocin preparations were evaluated for their cytotoxic effects against cells of the HepG2 human liver carcinoma and the Caco-2 human colorectal adenocarcinoma cell lines; both were obtained from the VACSERA cell line bank (Cairo, Egypt). Cytotoxicity was measured using the MTT (3-[4,5-dimethylthiazole-2-yl]-2,5-diphenyltetrazolium bromide) assay as an essential test of ongoing cell viability. Cell lines were maintained in T-75 tissue culture flasks containing 20 mL of Roswell Park Memorial Institute (RPMI) 1640 Medium (Sigma-Aldrich, St. Louis, MO, USA) supplemented with a 1% antibiotic-antimycotic solution (10,000 units penicillin, 10 mg streptomycin, and 25 μg amphotericin B/mL) and 10% v/v fetal bovine serum. The medium was changed at 48-h intervals and cells were detached with a trypsin solution (0.25% in PBS). The cytotoxicity of the bacteriocins under investigation was evaluated as previously described [49]. Briefly, cells were seeded in a 96-well microtiter plate (100 μL/well) at a concentration of 4 × 104 cells/cm2; cells were then cultured overnight to facilitate their attachment to the plate. The cell monolayer was then exposed to crude bacteriocin preparations using a twofold serial dilution technique; protein concentrations of dilutions prepared in 1% Dulbecco’s modified Eagle’s medium (DMEM) ranged from 4.88 μg/mL to 10 mg/mL; cultures were incubated for 24 h at 5% CO2 and 37 °C. Each experiment was performed in triplicate; three wells containing the same volume of DMEM but without diluted bacteriocins were used as a positive control. Cultures were examined after incubation under a phase-contrast inverted microscope (LEICA DMI 3000 B); changes in cell morphology associated with the agents undergoing testing were recorded. The cells were then incubated in medium containing 0.5 mg/mL MTT in a CO2 incubator (Jouan, France) at 37 °C for 4 h. Following incubation, the medium was aspirated; formazan product was solubilized with dimethyl sulfoxide (DMSO) and absorbance was measured at 570 nm using a BioTek® Flx 800 microtiter plate reader (BioTek Instruments, Inc., Winooski, VT, USA). The relationship between cell survival and extract concentration was plotted in order to generate a survival curve for each tumor cell line together with the concentration of crude bacteriocin preparation that resulted in a 50% reduction of absorbance compared with the control value (IC50). Percentage of cytotoxicity and cell viability were calculated using the following equations [50]: %cytotoxicity = 1 − (mean absorbance of treated cells/mean absorbance of negative control) and %viability = 100 − % cytotoxicity.
Statistical Analysis
Data are presented as mean ± standard deviation of triplicate assays. These values were compared with controls using the non-parametric Mann-Whitney test (U test) in GraphPad Prism (V.7.00, San Diego, CA, USA). Data visualization was achieved using the R statistical platform (https://www.r-project.org) with ggplot2 package. Values of P ≤ 0.05 were considered to be statistically significant.
Results
Isolation of Enterococci and Screening for Antimicrobial Activity
All of the 79 isolates initially recovered appeared as shiny black colonies on bile esculin agar with the characteristic odor of Enterococcus sp.; the microorganisms were Gram-positive diplococci that were catalase-negative. Twenty-four isolates (~ 30% of those recovered) were identified as positive for antimicrobial activity against Lact. sakei LMG 2313 using the spot-on-lawn assay; secondary screening revealed further activity against at least two other indicator strains (Table 3). On the basis of these screening assays, three Enterococcus isolates, FM43, FM50, and FM65, were selected for further characterization and investigation for their biological activities.
Molecular Characterization of the Selected Isolates
The 16S rRNA gene sequences of each of the three selected isolates revealed high similarity with sequences of many strains of Ent. faecium that had been reported to the NCBI GenBank database. Furthermore, 16S rRNA gene sequences from Ent. faecium FM43 and Ent. faecium FM65 were identical to one another (100% similarity), while that of Ent. faecium FM50 was slightly divergent (97.36% similarity). The three sequences were aligned to those of 22 closely related species as determined from sequences deposited in the NCBI GenBank database; these are assembled for phylogenetic analysis as shown in Fig. 1. The partial 16S rRNA gene sequences of the Ent. faecium FM43, Ent. faecium FM65, and Ent. faecium FM50 strains were deposited in GenBank under the accession numbers MT012113, MT012114, and MT012115, respectively.
The phylogenetic tree of the 16S rRNA gene sequences of the Ent. faecium FM43, Ent. faecium FM65, and Ent. faecium FM50 isolates investigated in the current study (GenBank accession numbers MT012113 through MT012115). The phylogenetic tree was inferred using the neighbor-joining method [28]. The evolutionary distances were computed using the maximum composite likelihood method [29] and are in the units of the number of base substitutions per site. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches [51]. The analysis involved 25 nucleotide sequences. Codon positions included were 1st+2nd+3rd+Noncoding. All positions containing gaps or missing data were eliminated. Evolutionary analyses were conducted in MEGA6 [27]. The isolates investigated in the current study (Ent. faecium FM43, Ent. faecium FM65, and Ent. faecium FM50) are highlighted in gray
Pathogenicity and Antibiotic Sensitivity of the Enterococcus Isolates
Neither gelatinase activity nor β-hemolytic activity was detected in any of the three selected isolates. Similarly, PCR screening revealed none of the standard virulence-associated target genes. However, the susceptibility study revealed that all three of the selected Enterococcus isolates were sensitive to nearly all of the antibiotics tested (Table 4).
Physicochemical Characterization of the Antibacterial Compounds
Antibacterial activity detected in the CFSs of the selected isolates remained unaffected in response to α-chymotrypsin; by contrast, activity was lost completely in response to treatment with proteinase K. The observed antibacterial activity was also stable between pH 3 and pH 8; activity dropped slightly at pH 10 and disappeared completely at pH 11. Moreover, antibacterial activity of the CFSs was also maintained in response to temperatures up to 100 °C, although activity was lost after autoclaving at 121 °C for 15 min.
Kinetics of Growth and Antimicrobial Activity
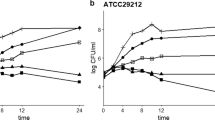
Antimicrobial activity and growth kinetics were monitored for 24 h. Antimicrobial activity was detected initially in cultures at the mid-logarithmic phase (6 h) and reached a maximum level of activity at early stationary phase (12 h); this was followed by a decline in antimicrobial activity which was not detected at 24 h (Fig. 2). Maximum activities at the 12-h time point included 640 AU/mL for Ent. faecium FM43 and 2560 AU/mL for both Ent. faecium FM50 and Ent. faecium FM65.
Comparison of bacteriocin production by selected isolates at different phases of growth curve. The growth curve analyses of Ent. faecium FM43 (a), Ent. faecium FM 65 (b), and Ent. faecium FM50 (c) indicated that the production of bacteriocin initiated at the mid-logarithmic phase (6 h) and reached the maximum level at the early stationary phase (12 h), followed by a decline in the antimicrobial activity, before it was lost at 24 h
Activity Against Bacterial Biofilms and SEM
Administration of total protein extracts that contain bacteriocins from Ent. faecium FM43 (3.9 mg/mL at 640 AU/mL) and Ent. faecium FM50 (5.9 mg/mL at 2560 AU/mL) resulted in significant reductions (80% and 48%, respectively) in Strep. mutans ATCC 25175–associated biofilm (P < 0.05; Fig. 3). By contrast, the total protein extract from Ent. faecium FM65 (3.4 mg/mL) had no statistically significant impact on the preformed biofilm. SEM images of the untreated control biofilm showed a nearly uniform thick layer of cells (Fig. 4a). By contrast, the treated biofilm was substantially less dense; individually, bacterial cells could be distinguished from one another (Fig. 4b).
Effect of the crude supernatants of Ent. faecium FM43, FM50, and FM65 on preformed biofilms of Strep. mutans ATCC 25175. The activity of crude supernatants of Ent. faecium FM43 (3.9 mg/mL), Ent. faecium FM50 (5.9 mg/mL), and Ent. faecium FM65 (3.4 mg/mL) on preformed biofilms of Strep. mutans ATCC 25175 was examined as detailed in the “Materials and Methods” section. Data, expressed as mean ± standard deviation of triplicate assays, are compared with the non-treated control using the non-parametric Mann-Whitney test (U test)
Scanning electron microscopy images of untreated control Strep. mutans ATCC 25175 (a) and after a 24-h treatment with the CFS fraction of the Ent. faecium FM43 strain (b). Magnification × 20,000. Note that while the untreated controls showed the growth of a uniformly thick biofilm model (a), the CFS-treated biofilm was much less dense and individually formed cells could be distinguished (b)
PCR Screening of Some Bacteriocin-Encoding Genes
PCR amplification of genes associated with bacteriocin biosynthesis suggested the presence of both entA and entB in the genomes of all three Ent. faecium isolates selected.
The Cytotoxic Effects of Bacteriocins on HepG2 and Caco-2 Cell Lines
Cytotoxicity resulting from the administration of partially purified bacteriocins to cultures of HepG2 and Caco-2 cancer cells was expressed as an IC50 value; this value represents the bacteriocin concentration required to reduce the viability of the target cells by 50%. Analysis of cytotoxicity revealed significant inhibition of Caco-2 cell proliferation, at approximately 77%, 70%, and 85% for 10 mg/mL proteins isolated from Ent. faecium FM43, Ent. faecium FM65, and Ent. faecium FM50, respectively (Fig. 5); the cytotoxicity observed was dose-dependent. The IC50 values for the total protein extracts containing bacteriocins produced by Ent. faecium FM43, Ent. faecium FM65, and Ent. faecium FM50 were calculated at 3.114, 4.232, and 1.875 mg/mL, respectively (Fig. 6). Interestingly, these protein extracts were not as effective at limiting proliferation of HepG2 cells.
The cytotoxic activity of crude bacteriocins from the selected isolates on Caco-2 and HepG2 cell lines. The cytotoxic effects of crude bacteriocins from Ent. faecium FM43, Ent. faecium FM65, and Ent. faecium FM50 were determined using the MTT assay in Caco-2 and HepG2 cell lines upon 24-h treatment with the crude bacteriocin at 10 mg/mL. Values represent the mean ± standard error (SE) of percent cell viability with respect to the untreated control. Data correspond to three independent experiments performed in triplicate
The antitumor survival curves of total protein extracts of the tested bacteriocins against HepG2 and Caco2 cell lines. The cytotoxic effects of bacteriocins obtained from a Ent. Faecium FM43, b Ent. Faecium FM50, and c Ent. Faecium FM65 on HepG2 (black filled circle) and Caco2 (black filled square) cell lines were determined using the MTT assay. The cells were initially maintained in a humidified incubator at 37 °C for 24 h, and the tested bacteriocins were added at different concentrations. The percentage of viable cells was calculated as described in the “Materials and Methods” section. d The IC50 values of the tested bacteriocins on Caco2 cancer cell line. The results are shown as means ± standard error (SE) of three independent experiments performed in triplicate
Discussion
Enterococci are natural inhabitants of the gastrointestinal tract. Although they are considered to be indicators of fecal pollution of water and foods, enterococci are often used as additives to starter cultures and are used in strategies for preservation of fermented foods, most notably cheese and fermented vegetable products [52].
A total of 79 isolates were recovered from different foods and from clinical samples; we identified the isolates as enterococci based on colony characteristics, microscopic examination, and the results of culture and testing on bile esculin agar. These are standard methods and are consistent with those used in several other studies that have also reported the isolation of Enterococcus sp. from various sources [53, 54]. Of note, Chuard and Reller [55] reported that the bile esculin test works well to facilitate sensitive (> 99%), economical, and rapid separation of enterococci and group D streptococci from non-group D viridans streptococci.
Three enterococcal isolates with broad-spectrum antimicrobial activity were identified for further assessment against Strep. mutans ATCC 25175, a pathogen that is well-known to promote dental caries [13]. In a previous study, Ent. faecium GM-1 had broad-spectrum activity against a variety of pathogenic indicator strains [56]. In the same context, another study has concluded that bacteriocins produced by the Ent. faecium strains exhibited a broader spectrum of activity compared with those from the Ent. faecalis [57].
Virulence factors, which are carried mainly by Ent. faecalis and to a lesser extent by Ent. faecium, have been a cause of substantial concern in studies featuring these species as probiotics [58]. Sensitivity against commonly used antibiotics and hemolytic activity have been considered factors reflecting the relative safety of bacterial strains selected as probiotics [59, 60]. Furthermore, gelatinase has the capacity to hydrolyze collagen, casein, fibrin, and other peptides; this enzyme may damage host tissue and lead to bacterial migration and spread, thereby increasing the virulence of positive strains of enterococci [2, 5]. Of note, none of the enterococcal strains selected for evaluation in the present study had gelatinase or β-hemolytic activities. In addition, all three isolates revealed a high antimicrobial susceptibility to typical antibiotics, including vancomycin, which has the major concern if they are essential for safety evaluation of enterococci as probiotics [25]. These results suggested that one or more of the three strains of Ent. faecium identified in this study might be developed and marketed for probiotic applications.
The bacteriocins produced by the selected isolates were stable over a broad range of pH and temperature; likewise, these antibacterial agents were confirmed as proteins by treatment with proteinase K. Taken together, these results suggest potential future use of these bacteriocins as starter/probiotic cultures for a variety of fermented food products [61]. Similar results were previously reported for bacteriocin EF478 [62] and bacteriocin KT11 [63].
Maximum antimicrobial activity for all three strains was observed at 12 h, during the early stationary phase of growth. This finding is consistent with those reported in a previous study in which a large number of characterized bacteriocins were synthesized during the logarithmic phase of bacterial growth [64]. Moreover, a previous study also revealed that the maximum bacteriocin activity detected in several Ent. faecium isolates was reached between 12 and 15 h [61]. In our study, the bioactivity was not detected after 24 h which might be attributed to bacteriocin aggregation, adsorption to the bacterial cell surface, or destruction secondary to release of proteolytic enzymes by the bacteriocin-producing strains [65, 66].
Bacterial biofilms are involved in 80% of human bacterial infections [67]; as such, we need to have some understanding of the role of antimicrobial peptides and their respective activity against preformed biofilms. Bacteriocins can be used either independently or in combination with existing antimicrobials to address problems associated with widespread resistance of biofilms to conventional antibiotics [68]. In the present study, two of the isolates (Ent. faecium FM43 and Ent. faecium FM50) promoted significant destruction of preformed biofilms generated by Strep. mutans ATCC 25175; these results suggested that one or both of these isolates might be developed for use in oral health maintenance, similar to what has been described for Ent. faecium WB2000 [69].
Enterocin A and enterocin B are widespread among enterococcal strains and play significant roles in controlling the growth of pathogens and other undesirable bacteria typically detected in fermented food products [70]. In the current study, entA and entB genes were identified in all selected isolates. These findings are consistent with results reported by Sonsa-Ard et al. [71], who reported that most Ent. faecium strains harbored at least one structural gene encoding enterocin; the two enterocin structural genes entA and entB are detected most frequently in these strains. Additionally, some studies have described multi-enterocin-producing strains [72, 73]. Interestingly, Ibarguren et al. [74] described a synergistic interaction between entB and entA in all strains of Ent. faecium C1, Mori1, M2d, and M1b.
Probiotics are health-promoting agents with various therapeutic uses; their potential use to combat cancer is currently the focus of significant attention and research [75]. However, to the best of our knowledge, there are only a few reports that have described the impact of bacteriocin on cancer or cancer cells. In this study, we identified dose-dependent bacteriocin-mediated cytotoxicity against cancer cells; these findings are in agreement with results presented in a previous study [76]. Specifically, we found that partially purified bacteriocin-containing preparations from all three of the selected Ent. faecium isolates were active against the cells of the Caco-2 cell line; unexpectedly, they were inactive in experiments targeting HepG2 cells. Similarly, another recent study described the cytotoxic activity of the Ent. faecalis bacteriocin Oe-342 in experiments targeting the Caco-2 cell line, although higher concentrations than those used in this study were required [77]. Another previous study focused on the cytotoxic effects associated with administration of total metabolites from Lact. plantarum that resulted in 33% inhibition of the Caco-2 cell proliferation using concentrations analogous to those used in this study [78]. Of note, no bacteriocins isolated from any strain of Ent. faecium have been reported to have cytotoxic activity against HepG2 cells [79].
Conclusions
In this study, we isolated three enterocin-producing strains of Ent. faecium and characterized their various bioactivities; the results of this study suggest that one or more of these bacteriocins may ultimately be useful as probiotics in the food industry. Interestingly, Enterococcus strains Ent. faecium M74 and Ent. faecium SF-68 have already been identified as safe and effective for use as food supplements in some probiotic preparations [80, 81]. Furthermore, the results of our study suggest that the broad-spectrum activity associated with each of these three selected isolates might relate to synergistic activities associated with both enterocin A and enterocin B; the actions of these enterocins may cover activity against the majority of Gram-positive and Gram-negative bacteria. Moreover, as this study focused on activity against Strep. mutans, the main cause for dental caries, activity of the bacteriocins associated with Ent. faecium FM43 and Ent. faecium FM50 against preformed biofilm generated by Strep. mutans ATCC 25175 may provide a valuable resource toward developing one or both of these strains for local application as a means to treat oral infections or as probiotics in oral pharmaceutical preparations. Finally, the cytotoxic effects promoted by partially purified bacteriocin preparations specifically on cells of the Caco-2 cell line suggest their potential utility for the treatment of colon cancer; bacteriocins may be introduced alone or in combination with existing anticancer drugs to enhance their efficacy and/or selectivity, while minimizing their associated adverse events. Also, the observed stability of different bacteriocins over a wide range of pH and temperature suggests that they may be introduced as preservatives in pharmaceutical formulations that undergo multiple, chemically harsh processing steps.
References
Leavis HL, Bonten MJ, Willems RJ (2006) Identification of high-risk enterococcal clonal complexes: global dispersion and antibiotic resistance. Curr Opin Microbiol 9:454–460. https://doi.org/10.1016/j.mib.2006.07.001
Franz CMAP, Huch M, Abriouel H, Holzapfel W, Gálvez A (2011) Enterococci as probiotics and their implications in food safety. Int J Food Microbiol 151:125–140. https://doi.org/10.1016/j.ijfoodmicro.2011.08.014
Arias CA, Murray BE (2012) The rise of the Enterococcus: beyond vancomycin resistance. Nat Rev Microbiol 10:266–278. https://doi.org/10.1038/nrmicro2761
Zhang F, Qiu L, Xu X, Liu Z, Zhan H, Tao X, Shah NP, Wei H (2017) Beneficial effects of probiotic cholesterol-lowering strain of Enterococcus faecium WEFA23 from infants on diet-induced metabolic syndrome in rats. J Dairy Sci 100:1618–1628. https://doi.org/10.3168/jds.2016-11870
Ben Braïek O, Smaoui S (2019) Enterococci: between emerging pathogens and potential probiotics. Biomed Res Int 2019:5938210–5938213. https://doi.org/10.1155/2019/5938210
Fahim HA, Khairalla AS, El-Gendy AO (2016) Nanotechnology: a valuable strategy to improve bacteriocin formulations. Front Microbiol 7:1385. https://doi.org/10.3389/fmicb.2016.01385
Pisoschi AM, Pop A, Georgescu C, Turcuş V, Olah NK, Mathe E (2018) An overview of natural antimicrobials role in food. Eur J Med Chem 143:922–935. https://doi.org/10.1016/j.ejmech.2017.11.095
Javed A, Masud T, Ul Ain Q et al (2011) Enterocins of Enterococcus faecium, emerging natural food preservatives. Ann Microbiol 61:699–708. https://doi.org/10.1007/s13213-011-0223-8
García de Fernando G (2011) Lactic acid bacteria: Enterococcus in milk and dairy products. Encycl Dairy Sci Second Ed 153–159. https://doi.org/10.1016/B978-0-12-374407-4.00528-8
Ness IF, Diep DB, Ike Y (2014) Enterococcal bacteriocins and antimicrobial proteins that contribute to niche control. In: Gilmore MS, Clewell DB, Ike Y, Shankar N (eds) Enterococci: from commensals to leading causes of drug resistant infection. Massachusetts Eye and Ear Infirmary, Boston, MA
Klein G (2003) Taxonomy, ecology and antibiotic resistance of enterococci from food and the gastro-intestinal tract. Int J Food Microbiol 88:123–131. https://doi.org/10.1016/S0168-1605(03)00175-2
Takahashi N, Nyvad B (2008) Caries ecology revisited: microbial dynamics and the caries process. Caries Res 42:409–418. https://doi.org/10.1159/000159604
Ben Taheur F, Kouidhi B, Fdhila K, Elabed H, Ben Slama R, Mahdouani K, Bakhrouf A, Chaieb K (2016) Anti-bacterial and anti-biofilm activity of probiotic bacteria against oral pathogens. Microb Pathog 97:213–220. https://doi.org/10.1016/j.micpath.2016.06.018
Abranches J, Miller JH, Martinez AR, Simpson-Haidaris PJ, Burne RA, Lemos JA (2011) The collagen-binding protein Cnm is required for Streptococcus mutans adherence to and intracellular invasion of human coronary artery endothelial cells. Infect Immun 79:2277–2284. https://doi.org/10.1128/IAI.00767-10
Høiby N, Ciofu O, Johansen HK, Song ZJ, Moser C, Jensen PØ, Molin S, Givskov M, Tolker-Nielsen T, Bjarnsholt T (2011) The clinical impact of bacterial biofilms. Int J Oral Sci 3:55–65. https://doi.org/10.4248/IJOS11026
Kaplan JB (2010) Biofilm dispersal: mechanisms, clinical implications, and potential therapeutic uses. J Dent Res 89:205–218. https://doi.org/10.1177/0022034509359403
Chhibber S, Nag D, Bansal S (2013) Inhibiting biofilm formation by Klebsiella pneumoniae B5055 using an iron antagonizing molecule and a bacteriophage. BMC Microbiol 13:174. https://doi.org/10.1186/1471-2180-13-174
Kumar A, Karig D, Acharya R, Neethirajan S, Mukherjee PP, Retterer S, Doktycz MJ (2013) Microscale confinement features can affect biofilm formation. Microfluid Nanofluidics 14:895–902. https://doi.org/10.1007/s10404-012-1120-6
Cotter PD, Ross RP, Hill C (2013) Bacteriocins-a viable alternative to antibiotics? Nat Rev Microbiol 11:95–105. https://doi.org/10.1038/nrmicro2937
Kaur S, Kaur S (2015) Bacteriocins as potential anticancer agents. Front Pharmacol 6:272. https://doi.org/10.3389/fphar.2015.00272
Gaspar D, Salomé Veiga A, Castanho MARB (2013) From antimicrobial to anticancer peptides. A review Front Microbiol 4:294. https://doi.org/10.3389/fmicb.2013.00294
Zhao H, Sood R, Jutila A, Bose S, Fimland G, Nissen-Meyer J, Kinnunen PKJ (2006) Interaction of the antimicrobial peptide pheromone Plantaricin A with model membranes: implications for a novel mechanism of action. Biochim Biophys Acta Biomembr 1758:1461–1474. https://doi.org/10.1016/j.bbamem.2006.03.037
Dundar H, Brede DA, La Rosa SL et al (2015) The fsr quorum-sensing system and cognate gelatinase orchestrate the expression and processing of proprotein EF_1097 into the mature antimicrobial peptide enterocin O16. J Bacteriol 197:2112–2121. https://doi.org/10.1128/JB.02513-14
Ahmad MS, El-Gendy AO, Ahmed RR et al (2017) Exploring the antimicrobial and antitumor potentials of Streptomyces sp. AGM12-1 isolated from Egyptian soil. Front Microbiol 8:438. https://doi.org/10.3389/fmicb.2017.00438
El-Ghaish S, El-Baz A, Hwanhlem N et al (2015) Bacteriocin production and safety evaluation of non-starter Enterococcus faecium IM1 and Enterococcus hirae IM1 strains isolated from homemade Egyptian dairy products. Eur Food Res Technol 240:1211–1223. https://doi.org/10.1007/s00217-015-2424-z
Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703. https://doi.org/10.1128/jb.173.2.697-703.1991
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. https://doi.org/10.1093/molbev/mst197
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. https://doi.org/10.1093/oxfordjournals.molbev.a040454
Tamura K, Nei M, Kumar S (2004) Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci U S A 101:11030–11035. https://doi.org/10.1073/pnas.0404206101
Vankerckhoven V, Van Autgaerden T, Vael C et al (2004) Development of a multiplex PCR for the detection of asaI, gelE, cylA, esp, and hyl genes in enterococci and survey for virulence determinants among European hospital isolates of Enterococcus faecium. J Clin Microbiol 42:4473–4479. https://doi.org/10.1128/JCM.42.10.4473-4479.2004
Martín-Platero AM, Valdivia E, Maqueda M, Martínez-Bueno M (2009) Characterization and safety evaluation of enterococci isolated from Spanish goats’ milk cheeses. Int J Food Microbiol 132:24–32. https://doi.org/10.1016/j.ijfoodmicro.2009.03.010
Tomita H, Fujimoto S, Tanimoto K, Ike Y (1997) Cloning and genetic and sequence analyses of the bacteriocin 21 determinant encoded on the Enterococcus faecalis pheromone-responsive conjugative plasmid pPD1. J Bacteriol 179:7843–7855. https://doi.org/10.1128/jb.179.24.7843-7855.1997
Aymerich T, Holo H, Håvarstein LS, Hugas M, Garriga M, Nes IF (1996) Biochemical and genetic characterization of enterocin A from Enterococcus faecium, a new antilisterial bacteriocin in the pediocin family of bacteriocins. Appl Environ Microbiol 62:1676–1682. https://doi.org/10.1128/aem.62.5.1676-1682.1996
Nilsen T, Nes IF, Holo H (2003) Enterolysin A, a cell wall-degrading bacteriocin from Enterococcus faecalis LMG 2333. Appl Environ Microbiol 69:2975–2984. https://doi.org/10.1128/AEM.69.5.2975-2984.2003
Gilmore MS, Segarra RA, Booth MC, Bogie CP, Hall LR, Clewell DB (1994) Genetic structure of the Enterococcus faecalis plasmid pAD1-encoded cytolytic toxin system and its relationship to lantibiotic determinants. J Bacteriol 176:7335–7344. https://doi.org/10.1128/jb.176.23.7335-7344.1994
Cintas LM, Casaus P, Holo H, Hernandez PE, Nes IF, Håvarstein LS (1998) Enterocins L50A and L50B, two novel bacteriocins from Enterococcus faecium L50, are related to staphylococcal hemolysins. J Bacteriol 180:1988–1994. https://doi.org/10.1128/jb.180.8.1988-1994.1998
Cintas LM, Casaus P, Håvarstein LS, Hernández PE, Nes IF (1997) Biochemical and genetic characterization of enterocin P, a novel sec- dependent bacteriocin from Enterococcus faecium P13 with a broad antimicrobial spectrum. Appl Environ Microbiol 63:4321–4330. https://doi.org/10.1128/aem.63.11.4321-4330.1997
Birri DJ, Brede DA, Tessema GT, Nes IF (2013) Bacteriocin production, antibiotic susceptibility and prevalence of haemolytic and gelatinase activity in faecal lactic acid bacteria isolated from healthy Ethiopian infants. Microb Ecol 65:504–516. https://doi.org/10.1007/s00248-012-0134-7
Harrigan WF, McCance ME (1966) Laboratory methods in microbiology. Academic Press, USA
Baker GC, Smith JJ, Cowan DA (2003) Review and re-analysis of domain-specific 16S primers. J Microbiol Methods 55:541–555. https://doi.org/10.1016/j.mimet.2003.08.009
Clsi (2013) Performance standards for antimicrobial susceptibility testing. Institute, Clinical and Laboratory Standards
Khodaei M, Sh SN (2018) Isolation and molecular identification of bacteriocin-producing enterococci with broad antibacterial activity from traditional dairy products in Kerman province of Iran. Korean J Food Sci Anim Resour 38:172–179. https://doi.org/10.5851/kosfa.2018.38.1.172
Wang Y, Qin Y, Xie Q, Zhang Y, Hu J, Li P (2018) Purification and characterization of plantaricin LPL-1, a novel class IIa bacteriocin produced by Lactobacillus plantarum LPL-1 isolated from fermented fish. Front Microbiol 9:2276. https://doi.org/10.3389/fmicb.2018.02276
Feliatra F, Muchlisin ZA, Teruna HY, et al (2018) Potential of bacteriocins produced by probiotic bacteria isolated from tiger shrimp and prawns as antibacterial to vibrio, pseudomonas, and aeromonas species on fish. F1000Research 7:. https://doi.org/10.12688/F1000RESEARCH.13958.1
Merritt JH, Kadouri DE, O’Toole GA (2005) Growing and analyzing static biofilms. Curr Protoc Microbiol https://doi.org/10.1002/9780471729259.mc01b01s00
Kadouri D, Venzon NC, O’Toole GA (2007) Vulnerability of pathogenic biofilms to Micavibrio aeruginosavorus. Appl Environ Microbiol 73:605–614. https://doi.org/10.1128/AEM.01893-06
Vahedi Shahandashti R, Kasra Kermanshahi R, Ghadam P (2016) The inhibitory effect of bacteriocin produced by Lactobacillus acidophilus ATCC 4356 and Lactobacillus plantarum ATCC 8014 on planktonic cells and biofilms of Serratia marcescens. Turkish J Med Sci 46:1188–1196. https://doi.org/10.3906/sag-1505-51
Sambrook J, Russel DW (2001) Molecular cloning: A laboratory manual. Cold spring harbor 3 2100
Skehan P, Storeng R, Scudiero D, Monks A, McMahon J, Vistica D, Warren JT, Bokesch H, Kenney S, Boyd MR (1990) New colorimetric cytotoxicity assay for anticancer-drug screening. J Natl Cancer Inst 82:1107–1112. https://doi.org/10.1093/jnci/82.13.1107
Valiyari S, Baradaran B, Delazar A, Pasdaran A, Zare F (2012) Dichloromethane and methanol extracts of Scrophularia oxysepala induces apoptosis in MCF-7 human breast cancer cells. Adv Pharm Bull 2:223–231. https://doi.org/10.5681/apb.2012.034
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution (N Y) 39:783–791. https://doi.org/10.2307/2408678
Ben Braïek O, Ghomrassi H, Cremonesi P, Morandi S, Fleury Y, le Chevalier P, Hani K, Bel Hadj O, Ghrairi T (2017) Isolation and characterisation of an enterocin P-producing Enterococcus lactis strain from a fresh shrimp (Penaeus vannamei). Antonie van Leeuwenhoek. Int J Gen Mol Microbiol 110:771–786. https://doi.org/10.1007/s10482-017-0847-1
Izquierdo E, Marchioni E, Aoude-Werner D et al (2009) Smearing of soft cheese with Enterococcus faecium WHE 81, a multi-bacteriocin producer, against Listeria monocytogenes. Food Microbiol 26:16–20. https://doi.org/10.1016/j.fm.2008.08.002
Khan H, Flint S, Yu PL (2010) Enterocins in food preservation. Int J Food Microbiol 141:1–10. https://doi.org/10.1016/j.ijfoodmicro.2010.03.005
Chuard C, Reller LB (1998) Bile-esculin test for presumptive identification of enterococci and streptococci: effects of bile concentration, inoculation technique, and incubation time. J Clin Microbiol 36:1135–1136. https://doi.org/10.1128/jcm.36.4.1135-1136.1998
Kang JH, Lee MS (2005) Characterization of a bacteriocin produced by Enterococcus faecium GM-1 isolated from an infant. J Appl Microbiol 98:1169–1176. https://doi.org/10.1111/j.1365-2672.2005.02556.x
Du Toit M, Franz CMAP, Dicks LMT, Holzapfel WH (2000) Preliminary characterization of bacteriocins produced by Enterococcus faecium and Enterococcus faecalis isolated from pig faeces. J Appl Microbiol 88:482–494. https://doi.org/10.1046/j.1365-2672.2000.00986.x
Morandi S, Silvetti T, Brasca M (2013) Biotechnological and safety characterization of Enterococcus lactis, a recently described species of dairy origin. Antonie van Leeuwenhoek, Int J Gen Mol Microbiol 103:239–249. https://doi.org/10.1007/s10482-012-9806-z
FAO/WHO (2002) Guidelines for evaluation of probiotic in food. Rep Jt FAO/WHO Work Gr Draft Guidel Eval Probiotic Food
Parte AC (2018) LPSN - list of prokaryotic names with standing in nomenclature (Bacterio.net), 20 years on. Int J Syst Evol Microbiol 68:1825–1829. https://doi.org/10.1099/ijsem.0.002786
Bagci U, Ozmen Togay S, Temiz A, Ay M (2019) Probiotic characteristics of bacteriocin-producing Enterococcus faecium strains isolated from human milk and colostrum. Folia Microbiol (Praha) 64:735–750. https://doi.org/10.1007/s12223-019-00687-2
Phumisantiphong U, Siripanichgon K, Reamtong O, Diraphat P (2017) A novel bacteriocin from Enterococcus faecalis 478 exhibits a potent activity against vancomycin-resistant enterococci. PLoS One 12:e0186415. https://doi.org/10.1371/journal.pone.0186415
Abanoz HS, Kunduhoglu B (2018) Antimicrobial activity of a bacteriocin produced by Enterococcus faecalis kt11 against some pathogens and antibiotic-resistant bacteria. Korean J Food Sci Anim Resour 38:1064–1079. https://doi.org/10.5851/kosfa.2018.e40
Lozo J, Vukasinovic M, Strahinic I, Topisirovic L (2004) Characterization and antimicrobial activity of bacteriocin 217 produced by natural isolate Lactobacillus paracasei subsp. paracasei BGBUK2-16. J Food Prot 67:2727–2734. https://doi.org/10.4315/0362-028X-67.12.2727
Taheri P, Samadi N, Reza Ehsani M et al (2012) An evaluation and partial characterization of a bacteriocin produced by Lactococcus lactis subsp lactis ST1 isolated from goat milk. Braz J Microbiol 43:1452–1462. https://doi.org/10.1590/S1517-83822012000400029
Zamfir M, Stefan IR, Stancu MM, Grosu-Tudor SS (2016) Production, mode of action and sequencing of the corresponding gene of a bacteriocin produced by Lactococcus lactis 19.3. Int J Food Sci Technol 51:2164–2170. https://doi.org/10.1111/ijfs.13196
Harro JM, Peters BM, O’May GA et al (2010) Vaccine development in Staphylococcus aureus: taking the biofilm phenotype into consideration. FEMS Immunol Med Microbiol 59:306–323. https://doi.org/10.1111/j.1574-695X.2010.00708.x
Mathur H, Field D, Rea MC, et al (2018) Fighting biofilms with lantibiotics and other groups of bacteriocins. npj Biofilms Microbiomes 4:1-13. https://doi.org/10.1038/s41522-018-0053-6
Suzuki N, Yoneda M, Hatano Y, Iwamoto T, Masuo Y, Hirofuji T (2011) Enterococcus faecium WB2000 inhibits biofilm formation by oral cariogenic streptococci. Int J Dent 2011:1–5. https://doi.org/10.1155/2011/834151
Ennahar S, Asou Y, Zendo T, Sonomoto K, Ishizaki A (2001) Biochemical and genetic evidence for production of enterocins A and B by Enterococcus faecium WHE 81. Int J Food Microbiol 70:291–301. https://doi.org/10.1016/S0168-1605(01)00565-7
Sonsa-Ard N, Rodtong S, Chikindas ML, Yongsawatdigul J (2015) Characterization of bacteriocin produced by Enterococcus faecium CN-25 isolated from traditionally Thai fermented fish roe. Food Control 54:308–316. https://doi.org/10.1016/j.foodcont.2015.02.010
Mojsova S, Krstevski K, Dzadzovski I et al (2015) Phenotypic and genotypic characteristics of enterocin producing enterococci against pathogenic bacteria. Maced Vet Rev 38:209–216. https://doi.org/10.14432/j.macvetrev.2015.08.052
Huang Y, Ye K, Yu K, Wang K, Zhou G (2016) The potential influence of two Enterococcus faecium on the growth of Listeria monocytogenes. Food Control 67:18–24. https://doi.org/10.1016/j.foodcont.2016.02.009
Ibarguren C, Raya RR, Apella MC, Audisio MC (2010) Enterococcus faecium isolated from honey synthesized bacteriocin-like substances active against different Listeria monocytogenes strains. J Microbiol 48:44–52. https://doi.org/10.1007/s12275-009-0177-8
Wang SM, Zhang LW, Fan RB, Han X, Yi HX, Zhang LL, Xue CH, Li HB, Zhang YH, Shigwedha N (2014) Induction of HT-29cells apoptosis by lactobacilli isolated from fermented products. Res Microbiol 165:202–214. https://doi.org/10.1016/j.resmic.2014.02.004
Ankaiah D, Esakkiraj P, Perumal V, Ayyanna R, Venkatesan A (2017) Probiotic characterization of Enterococcus faecium por1: cloning, over expression of enterocin-A and evaluation of antibacterial, anti-cancer properties. J Funct Foods 38:280–292. https://doi.org/10.1016/j.jff.2017.09.034
Al-Fakharany OM, Aziz AAA, El-Banna TE-S, Sonbol FI (2018) Immunomodulatory and anticancer activities of enterocin Oe-342 produced by Enterococcus feacalis isolated from stool. J Clin Cell Immunol 9:558. https://doi.org/10.4172/2155-9899.1000558
Er S, Koparal AT, Kivanç M (2015) Cytotoxic effects of various lactic acid bacteria on Caco-2 cells. Turkish J Biol 39:23–30. https://doi.org/10.3906/biy-1402-62
Paiva AD, Breukink E, Mantovani HC (2011) Role of lipid II and membrane thickness in the mechanism of action of the lantibiotic bovicin HC5. Antimicrob Agents Chemother 55:5284–5293. https://doi.org/10.1128/AAC.00638-11
Economou V, Sakkas H, Delis G, Gousia P (2017) Antibiotic resistance in Enterococcus spp. friend or foe? Foodborne Pathog Antibiot Resist 365–395. https://doi.org/10.1002/9781119139188.ch16
Olvera-García M, Sanchez-Flores A, Quirasco Baruch M (2018) Genomic and functional characterisation of two Enterococcus strains isolated from Cotija cheese and their potential role in ripening. Appl Microbiol Biotechnol 102:2251–2267. https://doi.org/10.1007/s00253-018-8765-3
Acknowledgments
We are so grateful to Dr. Tarek Dishisha and Dr. Mohamed Sebak for their kind help, suggestions, and recommendation during the current study.
Author information
Authors and Affiliations
Contributions
A.O.E. and A.S.K. designed all preliminary experiments. A.O.E. and A.F.A. revised the microbiology experiment design and organized and analyzed the data. F.M., A.O.E., and A.F.A. performed all the study experiments and interpreted the project results. F.M. wrote the initial draft of the manuscript. A.O.E., A.S.K., A.F.A., and E.A.G revised the manuscript, and S.M.A. did final revision of the manuscript prior to its submission.
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Ethical Approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 16 kb)
Rights and permissions
About this article
Cite this article
Molham, F., Khairalla, A.S., Azmy, A.F. et al. Anti-Proliferative and Anti-Biofilm Potentials of Bacteriocins Produced by Non-Pathogenic Enterococcus sp.. Probiotics & Antimicro. Prot. 13, 571–585 (2021). https://doi.org/10.1007/s12602-020-09711-1
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12602-020-09711-1