Abstract
Given that Staphylococcus aureus, an opportunistic pathogen, is one of the main etiological agents that causes various hospital and community infections associated with the production of virulence factors, emerging treatment strategies target to attenuate the activity of these factors can be promising to combat antibiotic-resistant strains. In this perspective, we investigated the antipathogenic potential against three S. aureus strains of chloroform extracts of cell-free culture supernatant from the probiotic bacteria Lactobacillus casei and Lactobacillus acidophilus, as well as its suitability as an alternative antimutagenic agent. Both extracts did not display antibacterial activity but significantly reduced the bacterial biofilm formation at different stages 3 h (up to 73%), 6 h (up to 45%), and 24 h (up to 46%). Moreover, the extracts decrease the virulence factors production, hemolysin (up to 67%), and coagulase (delayed coagulation), as well as the cell metabolism in the biofilm (up to 65%), disrupting a preformed biofilm (up to 46%), all devoid of affecting its growth suggesting that the inhibition could be mediated by Quorum sensing (QS). The extract’s effect on biofilm disruption and metabolic activity seems to be strain dependent. The 2,5-diketopiperazines present in the extracts showed the ability to bind to the QS regulatory proteins SarA and AgrA in molecular docking studies. In the mutagenicity assay, both probiotic bacteria were able to remove the mutagen, and this capacity increased with the bacteria concentration.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Staphylococcus aureus, an opportunistic pathogen, is one of the primary pathogens that cause various community and hospital-acquired infections associated with medical care [1]. This bacterial pathogen remains a leading cause of mild to severe and sometimes life-threatening infections. Its pathogenicity is associated with the secretion of an impressive collection of virulence factors such as toxins, enzymes, and biofilm regulated by bacterial Quorum sensing (QS) [1, 2]. Infectious capacity is dependent on the bacterial ability to form a biofilm, which is a significant source of hospital infections due to inadequate disinfection of medical devices [3]. Additionally, S. aureus is the bacteria mostly found in contaminant biofilms on surfaces, usually in contact with food, even on stainless steel [4].
Bacterial biofilms are complex communities of bacteria embedded in a self-produced matrix and attached to inert or living surfaces [5]. These communities of microorganisms are more resistant to the immune system and antibiotics than planktonic or free-floating bacteria. The emergence of S. aureus strains resistant to most antibiotics is becoming a serious public health threat, stimulating the development of novel antibacterial agents [6]. A new approach is needed for antibiotic therapy. Several studies proposed an indirect attack on bacteria as an antimicrobial strategy by interfering with their communication, known as QS [7]. Microbial communication coordinates many of the virulence and pathogenesis pathways through these systems. Therefore, QS inhibitors could be the key to controlling the pathogen [8] and improving therapeutic results when used together with existing antibiotics lines. In addition to that, considerable attention is currently addressed to probiotics, prebiotics, or their combined use as symbiotics, naturally to improve human health. FAO/WHO defines probiotics as “live microorganisms that, when administered in adequate amounts, confer a health benefit on the host” [9]. Human probiotics should include human origin bacteria with several desirable properties and present no health risks [10].
Probiotics are considered an ideal option as new antivirulence agents. Unlike conventional antibiotics, they do not possess the ability to induce an intense selective pressure on resistant bacteria, and they are also less cytotoxic than the QS suppressing agents used [11]. Probiotics or their derivatives have been reported to prevent QS, biofilm formation, and survival of biofilm pathogens, interfere with biofilm integrity/quality, and ultimately lead to biofilm eradication. Some of these mechanisms include the secretion of antagonistic substances such as bacteriocins, exopolysaccharides, surfactants, organic acids, lactic acid, fatty acids, enzymes (amylase, lipase), and hydrogen peroxide [11]. Recently, we reported that metabolites of L. casei and L. acidophilus (diketopiperazines) could combat the pathogenicity of P. aeruginosa by inhibiting the biofilm biomass and biofilm metabolic activity and also the production of the virulence factors (elastase and pyocyanin) as well as by interfering with the QS and disrupting a preformed biofilm [12]. However, the effect of these metabolites on the virulence of S. aureus, a Gram-positive bacterium, has not been studied.
Another thing, the human diet may contain a wide range of natural mutagenic compounds due to contamination of the raw material or the formation of toxic metabolites produced by pathogenic bacteria (such as chloramines) [13] or during the processing, cooking, and storage of food [14]. The accumulation of these mutagens and pro-mutagens in the body can produce alterations or damage to the DNA, leading to a cancer mutation and initiation [15]. Lactic acid bacteria and bifidobacteria exhibit antimutagenic activities against heterocyclic-amines, N-nitroso compounds, sodium azide, benzo[a]pyrene, and aflatoxin B [16,17,18]. One possible mechanism for the antimutagenic properties of lactic acid bacteria involves a physical binding of the bacterial cell wall’s mutagenic compounds. Also, some of their products of fermentation are antimutagenic [19].
This work investigates whether supernatant extracts of lactic acid bacteria of human intestinal microbiota containing diketopiperazines as principal components could interfere with growth, biofilm formation and disruption, metabolic activity, and virulence factor production of S. aureus strains. Docking tests were also performed with the SarA and AgrA proteins (regulators of this bacterium’s QS). On the other hand, we also investigate the mutagen-binding ability of human probiotics against sodium azide. Two strains isolated from infant human feces, Lactobacillus casei CRL 431 and Lactobacillus acidophilus CRL 730, have been selected to study because of their widely documented probiotic properties [20].
2 Materials and Methods
2.1 Strains and Bacterial Growth Conditions
The bacterial strains S. aureus ATCC 6538, S. aureus LVP 90 (methicillin-resistant clinical isolate), and S. aureus LVP 95 were grown in Müller–Hinton (MH) broth at 37 °C under aerobic conditions at 150 rpm in a mechanical shaker. The probiotic strains L. casei CRL 431 and L. acidophilus CRL 730 (CRL: Culture Collection of Centro de Referencia para Lactobacilos, CERELA-CONICET, Tucumán, Argentina) isolated from feces of healthy children [20] were cultured at 37 °C in LAPTg broth (peptone, 15 g/L; tryptone, 10 g/L; yeast extract, 10 g/L; glucose, 10 g/L; Tween 80, 0.1%, v/v) under microaerophilic conditions.
Salmonella typhimurium TA 100 [21] was grown in Nutrient Broth II (Oxoid Australia, West Heidelberg, Australia) in the presence of 25 µg/mL of ampicillin. Tests of histidine requirement, rfa mutation, uvrB mutation, and R-factor were carried out to confirm the genotypes of S. typhimurium TA 100. Before each mutagenicity test, S. typhimurium cells were grown at 37 °C for 16 h until reaching 1.2 × 109 UFC/mL.
2.2 Preparation of Lactic Acid Bacteria Culture Supernatant Extracts
A previously described protocol was employed to prepare the chloroform extract from L. casei CRL 431 and L. acidophilus CRL 730 culture [12]. The obtained extracts (CELc and CELa) were kept at 4 °C in a caramel-colored flask until further experimental use.
Each extract was dissolved with dimethylsulphoxide (DMSO, Sigma-Aldrich) to obtain stock solutions (4 mg dry weight/mL) for the biological assays.
2.3 Bioassay of Antibacterial Activity
Bacterial growth inhibition was determined after 3, 6, and 24 h of incubation at 37 °C with and without the extracts, as stated previously by Díaz et al. [12]. Overnight cultures of the three S. aureus strains were diluted 1/100 v/v in Müller–Hinton (MH) fresh medium (OD560 nm: 0.08 = 1.2 × 106 CFU/mL). 195 µL of each culture was inoculated into the wells of a polystyrene microplate. Then, 5 µL of each extract solution (CELc and CELa) was added to the wells to reach final concentrations of 10 and 100 µg/mL. As growth control, a bacterial culture added with 5 µL of DMSO (2.5% final concentration in the well) instead of the extract was employed. The positive control was ciprofloxacin (5 µg/mL). Bacterial growth was monitored at OD 560 nm using a microplate spectrophotometric reader (Multiskan Go, Thermo).
2.4 Antibiofilm Activity
2.4.1 Biofilm Formation Assay
For biofilm quantification, a micromethod based on a protocol previously reported was employed [22]. Biofilms formed after 3-, 6-, and 24-h incubation of bacterial cultures in the presence of 10 and 100 µg/mL of CELc and CELa were determined as stated previously [12]. A cell suspension with DMSO at 2.5% was used as a negative control. Ciprofloxacin was used in the same bioassay as a positive control at 5 µg/mL.
2.4.2 Biofilm Disruption Assay
The wells were incubated with 200 µL of the three S. aureus strains (OD560 nm 0.08 ± 0.02) for 24 h. The biofilm formed after that time was washed twice and air-dried. Then, 5 µL of extracts (CELc or CELa) (10 and 100 µg/mL final concentrations in the wells, 2.5% DMSO) and 195 µL of phosphate-buffered saline (PBS) were added and incubated by 24 h at 37 °C. In the control wells, 5 µl DMSO (2.5% final concentration) or ciprofloxacin (5 µg/mL) was added instead of the extract. As the final step, the microplate was washed twice, and the total biofilm biomass was stained with crystal violet, as mentioned previously (see 2.4.1.).
2.4.3 Biofilm Metabolic Activity Assay
The metabolic activity of the bacteria in the biofilm was evaluated using the reduction assay of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) with some modifications [23]. The biofilm was developed on the polystyrene microplate as previously described (see 2.4.1.) and then was washed twice with distilled water to remove the planktonic cells. Then 5 µL of CELc and CELa (final concentrations in the well 10 and 100 μg/mL, 2.5% DMSO) was added into each well containing 195 µL of PBS (pH 6.5) and incubated for 24 h at 37 °C. As a final step, the microplate was rewashed, and 100 µL of MTT solution (500 μg/mL) was added to each well and incubated again for 6 h at 37 °C. The insoluble purple formazan dissolved in DMSO was measured at 570 nm using a microplate reader (Multiskan Go, Thermo).
2.5 Virulence Factors Inhibition
2.5.1 Hemolysin Assay
Hemolysis was quantified by red cell lysis, as described previously with some modifications [24]. All S. aureus strains were grown in MH broth for 24 h at 37 °C in the presence of different concentrations of the extracts (10, 100, and 500 µg/mL). A culture control with DMSO (1%) was also carried out (null hemolysis). Afterward, 200 µL of the cultures mixed with 25 µL of red blood cells (obtained from centrifugation of whole blood at 4 °C for 10 min at 3000 rpm and washed twice with PBS buffer) and 775 µL of PBS buffer (pH = 7.4) were incubated at 37 °C with shaking (150 rpm) for 1 h. Then the supernatants obtained by centrifugation at 3000 rpm for 10 min were measured spectrophotometrically at 450 nm. The positive control for hemolysis was 1% Triton X-100 (100% hemolysis).
2.5.2 Coagulase Assay
For coagulase, the method of Bae et al. [25] with slight modifications was used. Plasma coagulation capacity by the S. aureus strains was evaluated by first incubating the culture in MH broth for 24 h at 37 °C in the presence of the extracts (10, 100, and 500 µg/mL). A control culture was carried out with 1% DMSO. After 24 h of incubation, 500 µL of each culture was taken and added to tubes containing 500 µL of plasma (previously obtained by centrifugation whole blood at 3000 rpm for 10 min at 4 °C) that were incubated at 37 °C. Every half an hour, it was checked whether the clot formation occurred.
2.6 Molecular Docking Calculations
Autodock 4.1 [26] was used to perform the molecular docking studies of the 2,5 diketopiperazines with two S. aureus proteins, SarA and ArgA, which structures were downloaded from the PDB Database with identification codes 2FNP [27] and 3BS1 [28], respectively. Missing heavy atoms and hydrogens atoms were added, and titratable amino acid states were adjusted at pH 7.4 with PDB2PQR Server [29, 30]. AutodockTools was used to prepare proteins and compounds for docking.
Two types of docking calculations were performed: blind and regular docking. The blind docking involves constructing a large grid around the whole protein to explore the whole surface for putative binding sites. A grid spacing of 0.5 Å, and a medium number of energetic evaluations (2.5 million) was used. For the regular docking, a grid was constructed around a specific region with a spacing of 0.375 Å and a large number of energetic evaluations (25 million). A Lamarckian genetic algorithm (GA) was used to generate 200 docking poses for each compound. Conformations were clusterized according to a root-mean-square deviation value of 2 Å. The best-docked conformation was selected based on binding energy and cluster population.
PyMol (PyMOL) was used to visualize, analyze, and prepare figures of the docked structures.
2.7 Antimutagenic Activity
2.7.1 Bacterial Concentration Used for the Assay
The probiotic cultured solutions were centrifuged at 3500 rpm at 4 °C for 15 min. The cells were washed twice with sterile phosphate buffer (pH 7, 100 mM, 0.85% NaCl) and resuspended in the same buffer. The cell suspensions were adjusted to 0.1, 0.4, and 0.9 at 560 nm, which correspond to 1.2 × 106, 1.2 × 108, and 1.2 × 1011 CFU/mL, respectively.
2.7.2 Dose-Response Curves for Mutagen
Dilutions of sodium azide ranging from 0.25 to 5 µg/mL were tested to prepare the curves used to determine the appropriate concentration of mutagen for the Salmonella mutagenicity assay [21]. Based on dose–response curves, a concentration of 0.5 µg/plate of sodium azide can be used.
2.7.3 Assessing the Ability of Probiotics to Bind Mutagens
The antimutagenic activity of L. casei CRL 431 and L. acidophilus CRL 730 against sodium azide was determined as described previously [21], measuring the inhibition of S. typhimurium TA 100 mutation. One hundred µL of the probiotic bacterial suspensions (1 × 106, 1 × 108, and 1 × 1011 CFU/mL) was mixed with 100 µL of the mutagen solution (10 µg/mL final concentration). A positive control (100% revertants) was prepared with mutagen and without probiotic bacteria. Each suspension was incubated at 37 °C for 2 h and then was centrifuged at 5000 rpm at 4 °C. Then the supernatants containing the mutagen not bind to lactic acid bacteria (residual mutagen) were separated.
2.7.4 Salmonella Mutagenicity Assay
Aliquots of 100 µL from an overnight culture of S. typhimurium TA 100 (approximate 2 × 108 CFU/mL) were incubated with 100 µL of the supernatants obtained above at 150 rpm for 30 min at 37 °C in a shaker and, afterward, were mixed with 2 mL top agar (0.6%, w/v) supplemented with 0.5% (w/v) NaCl, 0.5 mM L-histidine (Sigma-Aldrich) and 0.5 mM D-biotin (Merck, Germany). The mixture was then gently mixed and finally poured onto a plate containing minimum glucose agar (glucose 2% w/v plus agar 1.5% w/v). The plates were incubated at 37 °C for 48 h, and HIS+ revertant colonies were counted.
Antimutagenic activity of each probiotic bacteria was determined as the reduction in the number of colonies on the Petri plates (plates prepared with mutagen solution treated with probiotic bacteria) compared to the control (plates prepared with mutagen and without probiotic bacteria). The antimutagenic activity was expressed as a percentage of inhibition of S. typhimurium TA 100 mutation [31]
A = Number of His+ revertants induced by the mutagen (positive control), B = number of His+ revertants with bacteria and mutagen, and C = number of spontaneous His+ revertants (negative control) without bacteria and mutagen.
2.8 Statistical Analysis
Results are means of at least three independent experiments. The data obtained were subjected to statistical treatment with the analytical software INFOSTAT for Windows program. The differences in mean absorbance values were evaluated by analysis of variance (ANOVA). The Tukey test was used to compare between pairs. The level of significance was p ≤ 0.05.
3 Results
3.1 Effect of Extracts on Bacterial Growth and Biofilm Formation
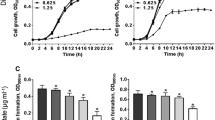
At both concentrations assayed, the bacterial culture supernatant extracts (CELc and CELa) did not modify significantly (p ˃ 0.05) the growth of any S. aureus strain at different incubation times (Fig. 1 Supplementary material).
Biofilm formation by Staphylococcus aureus ATCC 6538 (a), LVP 90 (b), and LVP 95 (c), at 3, 6, and 24 h of incubation in presence and absence of 10 and 100 µg/mL of chloroformic extracts: CELc (filled square) and CELa (square), and ciprofloxacin (5 µg/mL). Values represent the means ± SD of at least three independent experiments. *values are significantly different at p ≤ 0.05, compared to the respective DMSO control
Figure 1 shows the effect of 10 and 100 µg/mL of CELc and CELa on the biofilm formation of S. aureus ATCC 6538 (Fig. 1a), LVP 90 (Fig. 1b), and LVP 95 (Fig. 1c) at different incubation times. CELc and CELa had a significant potential to inhibit biofilm formation of all S. aureus strains at different contact times. In S. aureus ATCC 6538 and LVP 95, the biofilm inhibition was significant (p ≤ 0.05) for both extracts at 100 µg/mL in the three biofilm formation phases. In the adhesion stage (3 h), the reduction was similar to the antibiotic (between 61 and 73%) for both extracts. In the biofilm development stage (6 h), the inhibition values were between 31 and 45% for both extracts. Finally, in the third stage (24 h), the mature biofilms formed were 19 and 40% lesser than control (DMSO) for S. aureus ATCC 6538 and 31 and 33% lesser for S. aureus LVP 95 (Fig. 1a, c). CELc was the most active extract to inhibit the biofilm formation of S. aureus ATCC 6538 in a dose-dependent manner at different incubation times (Fig. 1a). However, S. aureus LVP90 is sensitive to both Lactobacillus extracts since the lowest concentration significantly inhibited (p ≤ 0.05) the three phases of biofilm formation (3, 6, and 24 h) (Fig. 1b).
3.2 Biofilm Disruption and Metabolic Activity of Cells in the Biofilm
The disruption of performed biofilm and the cell metabolic activity into biofilm in the presence and absence of the chloroformic extracts are shown in Fig. 2a, b. At 10 and 100 µg/mL, both extracts were able to decrease the established 24-h-old biofilms of S. aureus ATCC 6538 between 25 and 43%, of S. aureus LVP 90 between 33 and 53%, and S. aureus LVP 95 between 26 and 33% (Fig. 2a).
Biofilm disruption (square) and biofilm metabolic activity (filled square) of Staphylococcus aureus ATCC 6538, LVP 90, and LVP 95, at 24 h of incubation in the absence and presence of 10 and 100 µg/mL of chloroformic extracts (CELc and CELa), and ciprofloxacin (5 µg/mL). Values represent the means ± SD of at least three independent experiments. According to the Tukey test, different letters mean significant differences than the respective control (DMSO) (p ≤ 0.05)
Likewise, the extracts decreased the biofilm metabolic activity of the LVP 90 and LVP 95 strains. Values ranged from 25 to 40% for 10 µg/mL and between 48 and 58% for 100 µg/mL. However, the extracts were little active in the biofilm metabolic activity of the ATCC 6538 strain (Fig. 2b). The extract’s effect on biofilm disruption and metabolic activity seems to be strain dependent.
3.3 Effects of Supernatant Extracts Against Hemolysin Activity of S. Aureus Strains
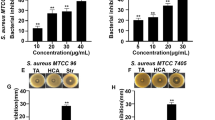
It was observed that CELc and CELa decreased the hemolysin activity of the three S. aureus strains in a dose-dependent manner (Fig. 3). S. aureus ATCC 6538 was the most sensible strain. At 100 μg/mL, both extracts blocked 50% or more of its production. Besides, the diminution of the hemolysin activity of all S. aureus strains tested against the probiotic culture extracts was between 42 to 67% at 500 µg/mL. It is important to highlight that bacterial growth was not affected in any condition.
Effect of lactic acid bacteria culture supernatant extracts on hemolysin activity by Staphylococcus aureus ATCC 6538 (square), LVP 90 (black square), and LVP 95 (grey square). Values (mean ± SD, n = 3) followed by a different letter are significantly different (ANOVA corrected by Tukey test, p ≤ 0.05). DMSO samples have been used as a control
3.4 Effects of Supernatant Extracts Against Coagulase Activity of S. aureus Strains
The inhibition of the coagulase activity of S. aureus strains is represented in Table 1. The ability to retard plasma clot formation was observed by CELc and CELa extracts compared to the control without extract. The time required for human plasma coagulation was higher in S. aureus ATCC 6538, observing a concentration-dependent effect but not extract dependent. The capacity of 100 µg/mL of the CELa and CELc extract to avoid complete plasma coagulation in all the strains studied is notable. In neither case, the bacterial growth was affected.
3.5 Molecular Docking of 2,5 Diketopiperazines to SarA and AgrA
Blind docking of the four 2,5 diketopiperazines into SarA revealed two major binding sites: the DNA-binding motifs (that includes the helix-turn-helix region and the beta-hairpin or winged region) and the divalent cation binding pocket (Fig. 4, Table 2).
The subsequent refinement of docking simulations centered on each major site showed that two binding subregions within the SarA DNA-binding region could be identified. DNA subregion 1, located between the helix-turn-helix of chain A and chain B’s interface (concave side), is more populated and presents docking poses with more negative binding energy. It is interesting to note that these compounds bind this subregion from the concave side, and there is an equivalent binding pocket that can be reached from the convex side of the dimer. On the other hand, DNA subregion 2, less represented, is located between the winged and helix-turn-helix motifs of chain A. The lower number of docking poses found for the latter region could be due to the reduced space limiting the interactions capable of stabilizing any bound ligand.
The docked molecule cyclo-Pro-Gly located within subregion 1 of the SarA DNA-binding region is mainly stabilized by three hydrogen bonds formed by side-chain amido moiety GLN-66A, the backbone carbonyl oxygen of GLN-61A, and the side chain of THR-17B. Within DNA subregion 2, only two hydrogen bonds are established with backbone NH of PHE-53A and backbone carbonyl of VAL-92A (Fig. 5).
Cyclo-Leu-Pro extends its leucine side chain toward hydrophobic residues (such as PHE-34A, LEU-13B, and LEU-60A) that help hold the monomers together as a dimer in subregion 1. Besides, three hydrogen bonds strengthen the interaction: side-chain amido moiety of GLN-66A, backbone carbonyl oxygen of LEU-60A, and the side chain of THR-17B. The docking cluster in subregion 2 is far less populated, and the molecule acquires an orientation with lower binding energy. It fits into a pocket with hydrophobic interactions from VAL-68A and VAL-92A reinforced by one hydrogen bond with backbone NH of VAL-92A (Fig. 5).
With regard to cyclo-Leu-Leu, the conformation and orientation adopted by this compound in subregion 1 are almost identical to cyclo-Leu-Pro diketopiperazine. In fact, the molecular superposition is large, and it presents the same hydrophobic and hydrogen bond interactions. The slight energy difference is due to a higher torsional free energy in cyclo-Leu-Leu. On the other hand, the binding complex in subregion 2 results more stable as one of the Leu side chains gets closer to the hydrophobic protein side, chains, and at least three hydrogen bonds are established (Fig. 5).
Cyclo-Phe-Pro presents the Phe side chain located within the same pocked as Leu side chain from the latter two compounds when bound in subregion 1. Amido group of GLN-66A and carboxylate group of ASP-20B interact with this diketopiperazine through hydrogen bonds. In addition, Pi-Pi interactions with PHE-34A and TYR-62A enhance the binding (Fig. 5).
The second major binding site is the SarA divalent cation binding pocket located at the convex side of the dimer, by the N terminus of chain B and C terminus of chain A where all bounded compounds belong to the most populated docking cluster. Cyclo-Pro-Gly and cyclo-Phe-Pro present a similar orientation deeper in the pocket towards TYR-18A, whereas the other two 2,5 diketopiperazines molecules are slightly displaced facing GLU-11A. Hydrogen bonds from the side chain of THR-4A and THR-4B and backbone atoms of the surrounding amino acids ILE-3A, THR-4A, and LYS-5A stabilize the docking complexes (Fig. 6).
On the other hand, docking of cyclic dipeptides into AgrA (PDB ID:3BS1) revealed a common binding site of all studied compounds located by a β hairpin motif that protrudes into the major groove of the double-stranded DNA (Fig. 7). All compounds present the same overall orientation that maximizes the interactions towards the protein and the nucleic acid. In addition to the hydrogen bond interactions collected in Table 3, the hydrophobic side chain of compounds (Leu and Phe) faces towards the base of the protein pocket, where nonpolar side chains of LEU-171 and LEU-186 provide a suitable environment to reinforce binding (Fig. 7).
3.6 Antimutagenic Activity of Probiotic Bacteria
The antimutagenic activity of L. casei CRL 431 and L. acidophilus CRL 730 against the mutagen sodium azide is shown in Table 4. As can be seen, the inhibition in the reversion of S. typhimurium by probiotics was strain and cell concentration dependent. The percentages of antimutagenicity at different cell concentrations were 89, 97, and 100% for L. casei and 31, 61, and 99% for L. acidophilus at 106, 108, and 1011 CFU/mL, respectively. It is essential to highlight that at a cell concentration in the order of 106, the L. casei showed a remarkable antimutagenic potential.
4 Discussion
Due to the increase in antibiotic resistance of human pathogens, the search for alternative strategies to combat them has begun. Among these alternatives, the attenuation/interference in the expression of virulence factors regulated by QS is an interesting new approach for developing a new generation of antipathogenic agents [23]. Unlike antibiotics, these agents target bacteria’s virulence factor rather than its planktonic growth [24]. S. aureus often exhibits antibiotic resistance and is responsible for numerous outbreaks of nosocomial infections. Besides antibiotics resistance, it produces an arsenal of virulence factors regulated by QS like hemolysins, staphyloxanthin, lipase, protease, leukocidin, coagulase, and biofilm, among others [25].
This work evaluated the antipathogenic potential of compounds present in chloroformic extracts from the supernatants of two lactic acid bacteria with known probiotic activity. The extracts CELc and CELa did not display antibacterial activity; however, they had antipathogenic capacity. The chloroformic extracts affected virulence factors (coagulase, hemolysin, and biofilm) without modifying planktonic bacterial growth. Furthermore, when the biofilm had already formed, the extracts were more effective in disrupting it and/or inhibiting sessile cell metabolism than the antibiotic used. All of these results suggested a QS inhibition mechanism.
Coagulase, also known as agglutination factor, which is part of what is known as microbial surface component recognition adhesive matrix molecules (MSCRAMM), is necessary for S. aureus to infect host tissue in the initial adhesion and subsequently in biofilm formation [7]. In the present study, both extracts could delay plasma clot formation compared to the control group. We also appreciate that the highest biofilm inhibition in our assays occurred in the initial adhesion stage (3 h) for all the strains tested, which could be correlated with the decrease observed in coagulase activity.
Hemolysin, another virulence factor, has strong pathogenicity; for example, S. aureus alpha-hemolysin can mediate the death of several different types of cells, including erythrocytes, endothelial cells, and an array of immunological cells such as T cells, B cells, and monocytes [32]. Accordingly, the development of drugs targeting hemolysin is a new way to treat S. aureus infection. This work demonstrated that CELc and CELa could inhibit hemolysin activity by S. aureus strains.
Previous studies have shown that Lactobacillus has antipathogenic potential against S. aureus and P. aeruginosa [33]; however, these properties were attributed to polar metabolites present in the cell-free supernatant like organic acids suggesting that the inhibition could be pH dependent. On the contrary, our work focuses on studying intermedium polarity metabolites (extracted with chloroform) secreted by probiotic Lactobacillus to recognize other metabolites responsible for the antipathogenic activity.
The gas chromatography-mass spectrometry (GC–MS) of these extracts showed that they are rich in four 2,5 diketopiperazines: cyclo-Pro-Gly, cyclo-Leu-Pro, cyclo-Leu-Leu, and cyclo-Phe-Pro [12]. Diketopiperazines are the smallest cyclic peptides known, commonly biosynthesized from amino acids by many organisms, including bacteria, fungi, and mammals [34]. Previous studies identified these compounds in lactic acid bacteria. They found that Lactobacillus reuteri RC-14 produces two diketopiperazines, cyclo-Phe-Pro (also present in our extract) and cyclo-Tyr-Pro, both capable of interfering with the staphylococcal agr QS system, a key regulator of virulence genes in S. aureus [35]. Also, cyclo-Leu-Pro (also present in our extract) was found to be the chief compound in the chloroform extract of the Bacillus amyloliquefaciens cell-free culture supernatant responsible for the significant reduction in biofilm formation and virulence genes expression by the cariogenic pathogen, Streptococcus mutans, without affecting its viability [36]. Thus, the cis-cycle dipeptide Leu-Tyr can also inhibit the biofilm formation in Staphylococcus epidermidis [37]. Gowrishankar et al. [7] found that the cyclic dipeptide-cyclo (L-leucyl- L-prolyl) secreted by B. amyloliquefaciens could inhibit biofilm production and virulence factors of methicillin-resistant Staphylococcus aureus and disrupt a mature biofilm.
The QS autoinducers in S. aureus are peptides, a type of macrocyclic thiolactone whose amino acid sequence is highly variable. The macrocycle structure is critical to autoinducer peptide (AIP) function since linear peptides do not activate QS [38, 39]. Several QS inhibitors have been described, with activity against S. aureus, with a cyclic structure or conjugated aromatic rings [39]. Given the cyclical structure of the diketopiperazines produced by the lactic acid bacteria under study and the inhibition of biofilm and virulence factors by the extracts containing them, our results agree with previous studies indicating that competitive inhibition of AIP constitutes a therapeutic approach that allows the attenuation of bacterial virulence. In line with this subject, Piewngam et al. [40] observed that the probiotic bacterium B. subtilis isolated from human feces could interfere with the colonization of S. aureus in the intestine. B. subtilis produces fengycin, a cyclic lipopeptide able to interfere with the QS of S. aureus, competing with the natural autoinducer for AgrC binding sites [33].
SarA, a 14.7-kDa protein (124 residues), is a global regulator that controls in a cell density-dependent manner that expresses several virulence factors and biofilm formation process and plays an essential role in antibiotic resistance of various strains of S. aureus and acts as a transcriptional activator of the genes encoding, among others, for fibronectin-binding proteins (fnbA and fnbB), hemolysins (hla, hld, hlgB, and hlgC), serine proteases (splA, splB, splD, and splF), and of the bap gene, which is essential for biofilm development in some strains [41, 42]. The docking studies carried out on the SarA protein revealed that the four 2,5-diketopiperazines interact with SarA protein in regions that should disturb its normal function either by preventing the DNA binding or by disrupting the association of multiples homodimers required for activation [27], which is observed in the reduction of biofilm production and other virulence factors such as hemolysin. This result falls in line with a recent study’s finding, indicating the significant down-regulation of the virulence gene sarA upon exposure to cyclo (L-leucyl- L-prolyl) secreted by mangrove rhizosphere Bacterium-B. amyloliquefaciens [7].
AgrA constitutes a two-component signaling system that responds to the autoinductive peptide presence that can positively regulate the expression of many virulence factors genes, such as hla [43]. Virulence and biofilm are differently regulated by agr system in S. aureus, and it is known that mutations in the agr quorum-sensing system are known to improve biofilm development [43]. Docking results showed that the four diketopiperazines could bind to both DNA and protein AgrA sites.
To our knowledge, this study is the first report of the interaction of diketopiperazines isolated from human probiotic bacteria with the SarA and AgrA receptors of Staphylococcus aureus through docking studies. The 2,5 diketopiperazines demonstrate interesting interactions with the studied targets, consistent with the observed antivirulence and antibiofilm activities.
Mutagens removal is an additional property that improves the food safety of the probiotic. Both probiotic strains assayed in this work were able to remove the mutagen. The percentage of antimutagenicity was rose significantly due to the increase in the number of L. casei or L. acidophilus cells. The correlation between lactic acid bacteria concentration and the mutagen removal ability is in concordance with previous reports. A mixture of goat probiotics formed by L. reuteri DDL 19, Lactobacillus alimentarius DDL 48, Enterococcus faecium DDE 39, and Bifidobacterium bifidum DDBA presented antimutagenic capacity against sodium azide and benzo[a]pyrene, and this detoxifying capacity increased when the concentration of probiotic bacteria was higher [14, 15]. Ambalam et al. [44] postulated that the antimutagenic capacity is due to the interaction between the mutagen and the low molecular weight glycopeptides of the lactic acid bacteria wall. The retention power could be specific for each mutagen and also strain dependent [31]. Besides, the dipeptide combination cyclo-Leu–Pro plus cyclo-Phe–Pro extracted from Streptomyces strains exerted significant antimutagenic activity in S. typhimurium TA98 and TA100, where a low concentration of the dipeptides (1 µg/plate) inhibited mutant colony formation by 42 and 51%, respectively, upon exposure to the mutagen 2-(2-furyl)-3-(5-nitro-2-furyl) acrylamide [45]. Besides, the dipeptides did not have mutagenic activity when used alone, even at high concentrations (around 10 mg/plate) [45].
This research determined that both probiotic strains studied have promising potential to remove the mutagens incorporated in the feed. Furthermore, it suggests the potential of the diketopiperazines secreted by Lactobacillus strains to combat S. aureus infections by inhibiting the formation of biofilm and virulence factors. These results prompt future preclinical and clinical investigations to fully demonstrate proof of principle.
References
Oliveira, D.; Borges, A.; Simões, M.: Staphylococcus aureus toxins and their molecular activity in infectious diseases. Toxins. 10, 252 (2018)
Sully, E.K.; Malachowa, N.; Elmore, B.O.; Alexander, S.M.; Femling, J.K.; Gray, B.M.; DeLeo, F.R.; Otto, M.; Cheung, A.L.; Edwards, B.S.; Sklar, L.A.; Horswill, A.R.; Hall, P.R.; Gresham, H.D.: Selective chemical inhibition of agr quorum sensing in Staphylococcus aureus promotes host defense with minimal impact on resistance. PLoS Pathog. 10, e1004174 (2014)
Doulgeraki, A.I.; Di Ciccio, P.; Ianieri, A.; Nychas, G.J.E.: Methicillin-resistant food-related Staphylococcus aureus: a review of current knowledge and biofilm formation for future studies and applications. Res. Microbiol. 168, 1–15 (2017)
Rodrigues, J.B.; Targino de Souza, N.; Scarano, J.O.A.; Targino de Sousa, J.M.; Lira, M.C.; Queiroz de Figuereido, R.C.B.; Leito de Souza, E.; Magnini, M.: Efficacy of using oregano essential oil and carvacrol to remove young and mature Staphylococcus aureus biofilms on food-contact surfaces of stainless steel. LWT Food Sci. Technol. 93, 293–299 (2018)
Tilahun, A.; Haddis, S.; Teshale, A.; Hadush, T.: Review on biofilm and microbial adhesion. Int. J. Microbiol. Res. 7, 63–73 (2016)
Okesola, A.O.: Community-acquired methicillin-resistant Staphylococcus aureus-a review of literature. Afr. J. Med. Med. Sci. 40, 97–107 (2011)
Gowrishankar, S.; Kamaladevi, A.; Ayyanar, S.; Balamurugan, K.; Pandian, S.K.: Bacillus amyloliquefaciens-secreted cyclic dipeptide- cyclo (L-Leucyl- L-Prolyl) inhibits biofilm and virulence in methicillin-resistant Staphylococcus aureus. RSC Adv. 116, 95788–95804 (2015)
Murray, E.J.; Crowley, R.C.; Truman, A.; Clarke, S.R.; Cottam, J.A.; Jadhav, G.P.; Steele, V.R.; O´Shea, P.; Lindholm, C.; Cockayne, A.; Chhabra, S.R.; Chan, W.C.; Williams, P.: Targeting Staphylococcus aureus quorum sensing with nonpeptidic small molecule inhibitors. J. Med. Chem. 57, 2813–2819 (2014)
Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Berni Canani, R.; Flint, H.J.; Salminen, S.; Calder, P.C.; Sanders, M.E.: Expert consensus document: the international scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 11, 506–514 (2014)
Linares, D.M.; Gómez, C.; Renes, E.; Fresno, J.M.; Tornadijo, M.E.; Ross, R.P.; Stanton, C.: Lactic acid bacteria and bifidobacteria with potential to design natural biofunctional health-promoting dairy foods. Front. Microbiol. 8, 846 (2017)
Barzegari, A.; Kheyrolahzadeh, K.; Khatibi, S.M.H.; Sharifi, S.; Memar, M.Y.; Vahed, S.Z.: The battle of probiotics and their derivatives against biofilms. Infect. Drug. Resist. 13, 659–672 (2020)
Díaz, M.A.; González, S.N.; Alberto, M.R.; Arena, M.E.: Human probiotic bacteria attenuate Pseudomonas aeruginosa biofilm and virulence by quorum-sensing inhibition. Biofouling 36, 1–13 (2020)
Osowski, A.; Pietrzak, M.; Wieczorek, Z.; Wieczorek, J.: Natural compounds in the human diet and their ability to bind mutagens prevents DNA–mutagen intercalation. Curr. Issues Intest. Microbiol. 7, 73–89 (2006)
Ling, Y.; Wang, H.; Yong, W.; Zhang, F.; Sun, L.; Yang, M.-L.; Yong-Ning, W.; Chu, X.-G.: The effects of washing and cooking on chlorpyrifos and its toxic metabolites in vegetables. Food Control 22, 54–58 (2011)
Shuker, D.E.: The enemy at the gates? DNA adducts as biomarkers of exposure to exogenous and endogenous genotoxic agents. Toxicol. Lett. 134, 51–56 (2002)
Apás, A.L.; González, S.N.; Arena, M.E.: Potential of goat probiotic to bind mutagens. Anaerobe 28, 8–12 (2014)
Utz, E.M.; Apás, A.L.; Díaz, M.A.; González, S.N.; Arena, M.E.: Goat milk mutagenesis is influenced by probiotic administration. Small Rumin. Res. 161, 24–27 (2018)
Shoukat, S.: Potential anti-carcinogenic effect of probiotic and lactic acid bacteria in detoxification of benzo[a]pyrene: a review. Trends. Food. Sci. Technol. 99, 450–459 (2020)
Mosallaie, F.; Jooyandeh, H.; Hojjati, M.; Fazlara, A.: Biological reduction of aflatoxin B1 in yogurt by probiotic strains of Lactobacillus acidophilus and Lactobacillus rhamnosus. Food. Sci. Biotechnol. 29, 793–803 (2019)
González, S.N.; Cardozo, R.; Apella, M.C.; Oliver, G.: Biotherapeutic role of fermented milk. Biotherapy 8, 129–134 (1995)
Maron, D.M.; Ames, B.N.: Revised methods for the Salmonella mutagenicity test. Mutat. Res. 113, 173–215 (1983)
O´Toole, G.A.; Kolter, R.: Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signaling pathways: a genetic analysis. Mol. Microbiol. 28, 449–461 (1998)
Jadhav, S.; Shah, R.; Bhave, M.; Palombo, E.A.: Inhibitory activity of yarrow essential oil on Listeria planktonic cells and biofilms. Food Control 29, 125–130 (2013)
Tang, F.; Li, L.; Meng, X.M.; Lia, B.; Wang, C.G.; Wang, S.Q.; Wang, T.-L.; Tian, Y.-M.: Inhibition of alpha-hemolysin expression by resveratrol attenuates Staphylococcus aureus virulence. Microb. Pathog. 127, 85–90 (2019)
Bae, W.Y.; Kim, H.Y.; Kim, K.T.; Paik, H.D.: Inhibitory effects of Inula britannica extract fermented by Lactobacillus plantarum KCCM 11613P on coagulase activity and growth of Staphylococcus aureus including methicillin-resistant strains. J. Food. Biochem. 43, e12785 (2019)
Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J.: AutoDock4 and AutoDocktools4: automated docking with selective receptor flexibility. J. Comput. Chem. 30, 2785–2791 (2009)
Liu, Y.; Manna, A.C.; Pan, C.H.; Kriksunov, I.A.; Thiel, D.J.; Cheung, A.L.; Zhang, G.: Structural and function analyses of the global regulatory protein SarA from Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 103, 2392–2397 (2006)
Sidote, D.J.; Barbieri, C.M.; Wu, T.; Stock, A.M.: Structure of the Staphylococcus aureus AgrA LytTR domain bound to DNA reveals a beta fold with an unusual mode of binding. Structure 16, 727–735 (2008)
Dolinsky, T.J.; Nielsen, J.E.; McCammon, J.A.; Baker, N.A.: PDB2PQR: an automated pipeline for the setup, execution, and analysis of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 32, W665–W667 (2004)
Dolinsky, T.J.; Czodrowski, P.; Hui, L.; Nielsen, J.E.; Jensen, J.H.; Klebe, G.; Baker, N.A.: PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 35, W522–W525 (2007)
Apás, A.L.; Ross, R.; González, S.N.; Arena, M.E.: Probiotic administration effect on fecal mutagenicity and microflora in the goat’s gut. J. Biosci. Bioeng. 110, 537–540 (2010)
Nygaard, T.K.; Pallister, K.B.; DuMont, A.L.; DeWald, M.; Watkins, R.L.; Pallister, E.Q.; Malone, C.; Griffith, S.; Horswill, A.L.; Torres, V.J.; Voyich, J.M.: Alpha-toxin induces programmed cell death of human T cells, B cells, and monocytes during USA300 infection. PLoS ONE 7, e36532 (2012)
Onbas, T.; Osmanagaoglu, O.; Kiran, F.: Potential properties of Lactobacillus plantarum F-10 as a bio-control strategy for wound infections. Probiotics Antimicrob. Proteins 11(4), 1110–1123 (2019)
Peres de Caravalho, M.P.; Wolf-Rainer, A.: Antimicrobial and biofilm inhibiting diketopiperazines. Curr. Med. Chem. 19, 3564–3577 (2012)
Li, J.; Wang, W.; Xu, S.X.; Magarvey, N.A.; McCormick, J.K.: Lactobacillus reuteri-produced cyclic dipeptides quench agr-mediated expression of toxic shock syndrome toxin-1 in staphylococci. Proc. Natl. Acad. Sci. USA 108, 3360–3365 (2011)
Gowrishankar, S.; Poornima, B.; Pandian, S.K.: Inhibitory efficacy of cyclo (l-leucyl-l-prolyl) from mangrove rhizosphere bacterium–Bacillus amyloliquefaciens (MMS-50) toward cariogenic properties of Streptococcus mutans. Res. Microbiol. 165, 278–289 (2014)
Scopel, M.; Abraham, W.R.; Henriques, A.T.; Macedo, A.J.: Dipeptide cis- cyclo(Leucyl-Tyrosyl) produced by sponge associated Penicillium sp. F37 inhibits biofilm formation of the pathogenic Staphylococcus epidermidis. Bioorg. Med. Chem. Lett. 23, 624–626 (2013)
Tan, L.; Li, S.R.; Jiang, B.; Hu, X.M.; Li, S.: Therapeutic targeting of the Staphylococcus aureus accessory gene regulator (agr) system. Front. Microbiol. 9, 55 (2018)
Ibrahim, Y.M.; Abouwarda, A.M.; Nasr, T.; Omar, F.A.; Bondock, S.: Antibacterial and anti-quorum sensing activities of a substituted thiazole derivative against methicillin-resistant Staphylococcus aureus and other multidrug-resistant bacteria. Microb. Pathog. 149, 104500 (2020)
Piewngam, P.; Zheng, Y.; Nguyen, T.H.; Dickey, S.W.; Soo Joo, H.; Villaruz, A.E.; Glose, K.A.; Fisher, E.M.; Hunt, R.L.; Li, B.; Chiou, J.; Pharkjaksu, S.; Khongthong, S.; Cheung, G.Y.C.; Kiratisin, P.; Otto, M.: Pathogen elimination by probiotic Bacillus via signaling interference. Nature 562, 532–537 (2018)
Chen, Q.; Xie, S.; Lou, X.; Cheng, S.; Liu, X.; Zheng, W.; Zheng, Z.; Wang, H.: Biofilm formation and prevalence of adhesion genes among Staphylococcus aureus isolates from different food sources. MicrobiologyOpen 9(1), e00946 (2020)
Balamurugan, P.; Krishna, V.P.; Bharath, D.; Vairaprakash, R.L.P.; Princy, S.A.: Staphylococcus aureus Quorum regulator SarA targeted compound, 2-[(Methylamino)methyl]phenol inhibits biofilm and down-regulates virulence genes. Front. Microbiol. 8, 1290 (2017)
Cheung, G.Y.C.; Wang, R.; Khan, B.A.; Sturdevant, D.E.; Otto, M.: Role of the accessory gene regulator agr in community-associated methicillin-resistant Staphylococcus aureus pathogenesis. Infect. Immun. 79, 1927–1935 (2011)
Ambalam, P.; Dave, J.M.; Nair, B.M.; Vyas, B.R.: In vitro mutagen binding and antimutagenic activity of human Lactobacillus rhamnosus. 231. Anaerobe 17, 217–222 (2011)
Rhee, K.H.: Cyclic dipeptides exhibit synergistic, broad spectrum antimicrobial effects and have antimutagenic properties. Int. J. Antimicrob. Agents. 24, 423–427 (2004)
Acknowledgements
The authors acknowledge the financial support from the SCAIT-UNT (PIUNT D26/638-1), the Agencia Nacional de Promoción Científica y Técnica ANPCyT (PICT 02071 and 02514), and the Consejo Nacional de Investigaciones Científicas y Técnicas, CONICET (PIP 662, PUE 0021). The authors also acknowledge the authorization granted to use the probiotic strains CRL 431 and CRL 730 of the Culture Collection of CERELA-CONICET, Tucumán, Argentina.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Díaz, M.A., Alberto, M.R., Vega-Hissi, E.G. et al. Interference in Staphylococcus Aureus Biofilm and Virulence Factors Production by Human Probiotic Bacteria with Antimutagenic Activity. Arab J Sci Eng 47, 241–253 (2022). https://doi.org/10.1007/s13369-021-05934-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13369-021-05934-8