Abstract
Microorganisms synthesize several compounds with antimicrobial activity in order to compete or defend themselves against others and ensure their survival. In this line, the cell wall is a major protective barrier whose integrity is essential for many vital bacterial processes. Probably for this reason, it represents a ‘hot spot’ as a target for many antibiotics and antimicrobial peptides such as bacteriocins. Bacteriocins have largely been recognized by their pore-forming ability that collapses the selective permeability of the cytoplasmic membrane. However, in the last few years, many bacteriocins have been shown to inhibit cell wall biosyntheis alone, or in a concerted action with pore formation like nisin. Examples of cell wall-active bacteriocins are found in both Gram-negative and Gram-positive bacteria and include a wide diversity of structures such as nisin-like and mersacidin-like lipid II-binding bacteriocins, two-peptide lantibiotics, and non-modified bacteriocins. In this review, we summarize the current knowledge on these antimicrobial peptides as well as the role, composition, and biosynthesis of the bacterial cell wall as their target. Moreover, even though bacteriocins have been a matter of interest as natural food antimicrobials, we propose them as suitable tools to provide new means to improve biotechnologically relevant microorganisms.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
All living organisms produce some kind of inhibiting substances as part of their defense or immune system in order to thrive in a competing niche. In particular, microbial antagonism is well documented, and the production of antimicrobials includes antibiotics, lytic enzymes, and low-molecular-weight metabolites such as organic acids, toxins, and bacteriocins.
Bacteriocins are ribosomally synthesized antimicrobial peptides or proteins produced by bacteria with inhibitory (bactericidal or bacteriostatic) activity against species closely related to the producer (narrow spectrum), or active beyond the genus boundary (broad spectrum). Although bacteriocin production is not an essential trait in bacteria, it is evolutionary maintained and widely distributed, even though it entails biological costs. Indeed, it is estimated that around 99 % of bacterial species produce bacteriocins, and even within a species, different bacteriocins can be synthesized [72, 113]. Some bacteriocins have been shown to be involved in competition among bacterial strains, niche colonization, and in quorum sensing and communication [32, 34, 51, 118].
Bacteriocins comprise a very heterogenous group regarding their primary structure and physicochemical properties. Likewise, there is a plethora of modes of action targeting DNA replication, transcription, translation, enzymatic reactions, cell wall biosynthesis, or the cytoplasmic membrane. Disruption of the selective permeability of the membrane by pore formation is a common inhibitory mechanism for cationic antimicrobial peptides, which are virtually produced by all forms of life [146], including many bacteriocins synthesized by Gram-positive bacteria (i.e. pore-forming bacteriocins). However, a striking feature of pore-forming bacteriocins, when compared to their eukaryotic counterparts, is their high specific activity at the nanomolar range and their relatively limited spectrum of activity. This is explained now by the increasing evidence that many of the pore-forming bacteriocins make target-mediated pores rather than being membrane disruptors. This holds true for several non-modified bacteriocins that use the membrane-associated component of the mannose-phosphotransferase system as a specific receptor [40, 72]. Another example is the use of the cell wall precursor lipid II as a docking molecule for pore formation, combining the inhibition of cell wall biosynthesis with the formation of pores [19, 23]. Moreover, several non-pore-forming bacteriocins targeting the cell wall have been described able to either hinder cell wall biosynthesis or hydrolyze pre-existing peptidoglycan.
Based on its essential nature, the biosynthetic pathway of the cell wall has been, and still is, a validated target in antibiotic development [25, 129]. In view of the need of novel antimicrobials, cell wall-active bacteriocins are foreseen as a source of novel structures and activities which may represent new leads for antibiotic development. This review will focus on cell wall-active bacteriocins synthesized by both Gram-positive and Gram-negative bacteria. Moreover, examples are given on how these bacteriocins may be used as tools to improve biotechnological processes by enhancing the performance of the microorganisms involved.
Bacteriocin Diversity and Classification: A Constant Debate
Bacteriocin production is a widespread trait among bacteria, and some species of Archaea also produce their own bacteriocin-like compounds called archaeocins [100, 113]. These are reported to be very resistant to extreme conditions with very promising applications. However, little is known about their mode of action and they will not be covered in this review.
Among bacteriocins synthesized by Gram-negative bacteria, colicins and microcins, produced by Escherichia coli and mostly active against Gram-negative microorganisms, have been extensively studied. Colicins are large proteins (25–80 kDa), usually plasmid-borne that encodes a toxin gene, an immunity gene, and, in some cases, a stress-induced lysis gene. The latter encodes a protein that lyses the producer cell to release the colicin into the environment. When this gene is absent, the bacteriocin is actively transported across the membrane. Expression of colicin genes is regulated by the DNA-damage SOS response, but it can also be influenced by other global regulatory networks [28]. Colicin production leads to the death of the producing cell and they are lethal for close-related strains which are recognized by the bacteriocin, being the producer strain immune to them. All colicins share as to their mode of action common steps that involve the recognition of their cognate receptors in the outer membrane of target cells, translocation across the outer membrane, and activation of their toxic effect. A great variety of killing mechanisms have been described such as membrane pore formation (e.g. colicin A), DNA nuclease activity (colicin E2), and the inhibition of the biosynthesis of proteins (colicins E3, E5, D), peptidoglycan, and the lipopolysaccharide O-antigen (colicin M) [5, 24, 28, 54, 113].
By contrast, microcins are smaller (<10 kDa), they are not lethal against the producing cell and their regulation is SOS-independent. Gene clusters are located either in plasmids or in the chromosome and consist of structural and immunity genes and those coding for transport or modification enzymes [95]. Two main sub-classes, I and II, have been established depending on their post-translational modifications, gene cluster organization, and the sequence of the leader peptide, showing great structural heterogeneity [24, 42, 54]. Their synthesis is activated under specific stress conditions, and, once synthesized, they are actively transported outside the cell either by microcin-specific or by general transporters. Some microcins recognize specific receptors in the target cell, usually involved in uptaking essential nutrients. Others are synthesized as harmless peptides which are further processed and activated within the target cell [43]. Their killing mechanism is not fully understood, but, in some cases, disruption of the cell membrane polarity [79], transcription inhibition by binding to RNA polymerases [39], or translation inhibition [93] have been documented.
On the other hand, bacteriocins produced by Gram-positive bacteria are usually small, heat-stable cationic peptides with high isoelectric points. They are active preferably against a wide panel of Gram-positive microorganisms, including food-borne and spoilage bacteria. This feature has been the main driving force of bacteriocin research, particularly of those produced by the food-related lactic acid bacteria as for their use as natural preservatives in food [33, 46]. Bacteriocin gene clusters can be plasmid or chromosomally encoded, and some are localized in transposons, as described for nisin [65]. The gene clusters basically consist of the structural gene and those involved in immunity. Genes specifying modification enzymes are also present in some cases. Moreover, Gram-positive microorganisms have evolved specific regulation mechanisms related to bacteriocin synthesis and transport [61, 76, 113].
Owing to the great variety of chemical and structural properties of bacteriocins, several classification schemes have been proposed based on different criteria such as molecular weight, producing microorganism, structure, or mode of action, although it still remains controversial and a universal classification is an issue for on-going discussions [36, 73, 97, 110]. The classification proposed by Heng and Tagg [62] integrates bacteriocins produced by both Gram-positive and Gram-negative bacteria and establishes four main classes.
Class I or lantibiotics encompasses small (<15 kDa), heat-stable, post-translational-modified peptides containing amino acids such as lanthionine, ß-methyl-lanthionine, and dehydrated amino acids, which form distinctive intramolecular structural rings. Class I is sub-classified in 3 sub-classes: the linear I-a, represented by cationic and pore-forming peptides; the globular I-b, non-cationic peptides that inhibit enzymatic reactions; and the multi-component I-c, composed by two peptides, both needed to be fully active. Class II bacteriocins are defined as small (<15 kDa), heat-stable, and non-post-translational-modified, usually amphiphilic and/or hydrophobic. They are sub-classified into three sub-classes: Pediocin-like II-a, characterized by a conserved N-terminal sequence YGNGV/L and a stabilizing disulfide bond, highly active against Listeria and other genera such as Enterococcus, Lactobacillus, Pediococcus or Clostridium, among others; sub-class II-b represents a miscellaneous group of linear non-pediocin-like peptides; and sub-class II-c comprises multi-component bacteriocins. Another group, class III, is composed by large (>30 kDa) heat-labile proteins, which are sub-divided into sub-class III-a or bacteriolytic proteins, and sub-class III-b or non-lytic, which are generally active on cytosolic targets. Finally, class IV includes circular proteins characterized by a head-to-tail covalent bond. Examples of cell wall-active bacteriocins are shown in Table 1.
The Bacterial Cell Wall
The cell wall represents the first barrier between the bacterial cells and their environment. It preserves the integrity of the cell by maintaining a defined and stable cell shape; it is needed to counteract the inner osmotic pressure and represents an assembly scaffold for other surface macrostructures such as polysaccharides, S-layers, flagella, and secretion systems [64, 124]. Moreover, the cell wall is a dynamic structure that participates also in cell growth and cell division [108, 142]. This structure is, therefore, crucial for survival, and as such, it is targeted by several antibiotics, bacteriophages, and bacteriocins.
The bacterial cell wall is composed basically by peptidoglycan (PG), anionic polymers, proteins, and polysaccharides. The PG structure is formed by a network of linear glycan strands made up of alternating N-acetylglucosamine (NAG) and N-acetylmuramic acid (NAM) covalently linked by β1-4 glycosidic bonds. Linked to NAM, there is a pentapeptide chain L-Ala–D-Glu–X–D-Ala–D-Ala, where X represents a diamino acid, usually a meso-diaminopimelic acid (DAP) in most Gram-negative or L-Lys in Gram-positive bacteria. The glycan strands are further cross-linked via the pentapeptide either directly or through a short peptide bridge, depending on the species. Since the peptides are localized helically along the sugar strand, cross-links are made in all directions, forming a multilayered three-dimensional network [116].
PG is present in all bacteria, except in Mycoplasmas, Planctomyces, and the genera Rickettsia and Chlamidiae [140]. A scheme of the CW structure of Gram-positive and Gram-negative bacteria is shown in Fig. 1. In Gram-positive bacteria, the PG represents the 90 % of the total cell wall. Anionic polymers such as teichoic acids (or lipoteichoic acids when anchored to the cytoplasmic membrane) are also major constituents. Proteins can be either covalently linked to PG or associated through cell wall-binding domains. In Gram-negative microorganisms, the PG is a thin layer that accounts only for the 10 % of the CW, being the external membrane the outermost structure. The lipopolysaccharide (LPS) is the major constituent that contributes to the structural integrity of the cell and acts as a protective barrier.
The biosynthesis of PG is a multistage process beginning in the cytoplasm where the UDP-NAG and UDP-NAM-pentapeptide units are synthesized. Still in the cytoplasm but on the membrane side, lipid I is synthesized by the addition of the UDP-NAM-pentapeptide unit to the lipid carrier undecaprenyl phosphate. Subsequently, UDP-NAG is transferred to lipid I, yielding lipid II [137]. The PG monomer is translocated through the membrane, and, once in the periplasm, it is polymerized into the growing PG chain by the penicillin-binding proteins (PBPs), with glycosyltransferase and/or transpeptidase activities [122]. The lipid carrier is recycled to be used in a new cycle of PG synthesis.
Despite the fact that the basic chemical architecture of the PG is similar in bacteria, no species keeps its PG in an unmodified state and may vary even within the same species depending on growth conditions [139]. Modification of the PG may occur along synthesis and maturation (e.g. degree of cross-linking) but also by the activity of particular enzymes, very often involved in resistance to antimicrobials. This is the case of PG O-acetylation accounting for resistance to lysozyme [8, 31, 50] or the presence of D-lactate, D-Ser or Gly instead of D-Ala in the last position of the pentapeptide in some vancomycin-resistant bacteria [112].
Lipid II-binding Bacteriocins
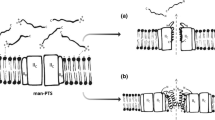
Several antibiotics are known to target the different steps of cell wall biosynthesis. Particularly, the cell wall precursor lipid II is very often targeted, and different recognition domains for several antibiotics have been described [18, 129]. Moreover, it has been recently described that some defensins, antimicrobial peptides produced by animals, plants, and fungi, also target lipid II to exert their antibacterial activity [126, 128]. In the case of bacteriocins, many have been described to bind to lipid II, including pore- and non-pore-forming lantibiotics, non-modified bacteriocins, two-peptide bacteriocins, and some Gram-negative colicins (Fig. 2). In particular examples, potent inhibitory activities comparable to leading antibiotics such as vancomycin have been described [69].
Mode of action of some cell wall-active bacteriocins. Square N-acetylglucosamine. Triangle N-acetylmuramic acid. CM, cytoplasmic membrane; PG, peptidoglycan. Within the PG, β1-4 glycosidic bonds (horizontal lines) and cross-bridges (vertical lines). Boxes detailed muropeptide composition of streptococcal (left) and staphylococcal (right) peptidoglycan
Nisin and Nisin-like Bacteriocins
The lantibiotic nisin A is the most studied Gram-positive bacteriocin. It is a 3.3 kDa, elongated, amphiphilic, and positively charged peptide with a wide spectrum of activity, inhibiting several Gram-positive and also some Gram-negative bacteria, provided that the external membrane is disrupted [133]. Several natural nisin variants, produced by Lactococcus lactis (nisin A, Z, F, and Q) and by Streptococcus uberis (nisin U), have been isolated. Early work on the mode of action of nisin already suggested that cell wall biosynthesis was inhibited [111], and, later on, it was experimentally confirmed that nisin makes use of lipid II as a docking molecule for pore formation [19, 23]. Lipid II is an integral part of the pore which is made up of 8 molecules of nisin and 4 of lipid II [59]. The combination of both inhibition of cell wall biosynthesis and pore formation is the basis of the potent activity of nisin. Binding to lipid II also implies that the cell wall precursor is delocalized from the sites where is needed for cell wall biosynthesis, interfering with cell growth and cell division [60, 68]. Moreover, binding to lipid II is also needed for effective membrane disruption in germinated spores to inhibit spore outgrowth from several pathogenic Gram-positive bacteria [56, 90]. Nisin has also been reported to activate cell wall hydrolytic enzymes, particularly in staphylococci at the septum level, by displacing them from the teichoic and lipoteichoic acids [12, 13].
The N-terminus of nisin, specifically rings A and B, forms an essential binding cage, which interacts with the pyrophosphate moiety of lipid II via hydrogen bonds, [67] and the C-terminus is essential for pore formation [145]. Their contribution to bacterial killing has been assessed by using mutated nisin versions [145]. Interestingly, specific mutations on the hinge region resulted in nisin variants with enhanced activity [45].
The nisin-like lipid II-binding structural motif is present in many pore-forming lantibiotics such as gallidermin and epidermin [15], mutacin 1140 [132], subtilin [105], and bovicin HC5 [104], whose interaction with lipid II has been experimentally demonstrated. Of note, the pore-forming ability of these nisin-like lipid II-binding peptides seems to be strongly dependent on membrane thickness and composition and determines the inhibitory spectra of these lantibiotics. Thereby, in order to evaluate the contribution of pore formation to the mode of action of these and other bacteriocins, special care must be taken when choosing a particular susceptible strain [138].
Mersacidin and Mersacidin-like Lantibiotics
Mersacidin is a small 1.8-kDa globular uncharged lantibiotic produced by Bacillus HIL Y-84,54728 [30]. It is active against a variety of Gram-positive bacteria and, remarkably, against methicillin-resistant Staphylococcus aureus (MRSA) with comparatively higher activity than vancomycin in animal models [30, 74, 98]. Mersacidin is not a pore-forming bacteriocin. Instead, it strongly inhibits the transglycosylation step during cell wall biosynthesis by binding to lipid II without affecting the biosynthesis of DNA, RNA, or proteins [21, 22]. The structure of the complex mersacidin–lipid II has not been solved. However, NMR analyses revealed that mersacidin possesses a dynamic conformation able to suffer structural changes depending on the environmental conditions, thus modulating the distribution of the charged residues, which affects the effective interaction with lipid II [66]. Moreover, mersacidin activity is enhanced by calcium ions in vivo, promoting its interaction with the negatively charged surface of the cytoplasmic membrane [16].
Mersacidin and related lantibiotics are characterized by the presence of an interwined tioether bridge within the conserved TxS/TxE/DC motif, being the glutamate residue essential for the inhibitory activity, as shown by site-directed mutagenesis [134]. This pattern is conserved in other lantibiotics, indicating a putative inhibitory activity of these bacteriocins based on lipid II-binding [14].
Among mersacidin-like lantibiotics, inhibition of cell wall biosynthesis has been demonstrated for plantaricin C and nukacin ISK-1 [70, 143]. Moreover, the lantibiotic planosporicin was discovered during a screening for inhibitors of peptidoglycan inhibitors [29]. There are several lantibiotics whose structure resembles that of plantaricin C and nukacin ISK-1 (i.e. the lacticin 481 group). They all have their particular inhibitory spectra, and their activity could rely also on lipid II-binding and pore formation [7, 14, 41].
Plantaricin C is a 3.5-kDa bacteriocin, synthesized by Lactobacillus plantarum LL441, which displays inhibitory activity against a wide range of Gram-positive bacteria, including food-borne pathogens, such as Staphylococcus, Streptococcus, Clostridium, or Bacillus [52]. It shares structural features with nisin and mersacidin; it has a highly positively charged N-terminus, which may facilitate the interaction with the negatively charged cytoplasmic membrane, and a ring and compact conformation in the C-terminus, resembling the mersacidin lipid II-binding motif [136]. Accordingly, plantaricin C is able to form pores in model membranes and intact cells of particular bacterial species but it is also a potent inhibitor of cell wall biosynthesis forming a tighter complex with lipid II when compared to mersacidin [53, 143].
Nukacin ISK-1 is a 2.9-kDa lantibiotic produced by Staphylococcus warneri ISK-1 with a N-terminal linear domain and a globular C-terminus, essential for the antimicrobial activity, which holds the mersacidin-like lipid II-binding motif TxS/TxD/EC [71, 121]. Nukacin ISK-1 is bacteriostatic and unable to form pores. Instead, it binds to lipid II leading to the accumulation of cell wall precursors inside the cell. Treated cells show a reduced thickness of the cell wall and incomplete septa [2]. Immunity has been associated with the cooperative role of the ABC transporter NukFEG and the lantibiotic-binding immunity protein NukH, which may block the bacteriocin before reaching its target [101].
Two-peptide Lantibiotics
A growing class of lipid II-binding molecules is constituted by two-peptide lantibiotics or class Ic according to the Heng and Tagg’s classification [62]. In these systems, two pre-peptides are ribosomally synthesized as inactive forms (LanA1 and LanA2), which are later enzymatically modified into their mature forms, the α-peptide carrying a mersacidin-like lipid II domain and an elongate positively charged ß-peptide involved in pore formation. Both peptides are encoded by their corresponding structural genes, act synergistically usually in a 1:1 ratio, and are required for full activity. This distribution of specialized killing mechanisms in two peptides has been described for lacticin 3147 [144] and haloduracin [102] and may hold true for other closely related two-peptide lantibiotics [99].
Lacticin 3147 is synthesized by L. lactis ssp. lactis DPC3147 and it is highly active against Gram-positive bacteria, including food-borne pathogens such as Listeria monocytogenes and Bacillus cereus or relevant pathogens such as MRSA [91, 119, 120]. Lacticin 3147 structure was resolved by Martin et al. [83] and it is composed by LtnA1 (3.3 kDa), the globular-type lantibiotic with a mersacidin-like lipid II-binding motif, and LtnA2 (2.8 kDa), a positively charged elongated-type lantibiotic. Both peptides are needed for high inhibitory activity at nanomolar concentration [96, 144]. LtnA1 has been shown to be a strong inhibitor of cell wall biosynthesis in in vitro assays. The interaction with lipid II appears to stabilize LtnA1 promoting its interaction with LtnA2, capable of pore formation [91, 144]. Potassium release assays showed that LtnA1 should be present before the final action of LtnA2, demonstrating that LtnA1 and LtnA2 act in a sequential manner in a 1:1 ratio, an event which has been frequently found for other two-peptide bacteriocins ([96] and references therein). Mutagenesis assays showed that the rings of LtnA1 involved in lipid II binding, the glutamate residue in this region, as well as the rings in LtnA2 involved in the interaction of the two peptides, are essential for activity [35]. In addition to this, the presence of calcium ions, or in general a positively charged environment, was proved to enhance the antimicrobial activity of lacticin 3147 as LtnA1 binds to whole cells only when it is surrounded by positive charges, either calcium or the positively charged partner LtnA2 [16]. It is worth-noting that the inhibitory activity is also strain-dependent [96].
Haloduracin is produced by the alkaliphilic Bacillus halodurans C-125, has inhibitory activity against several Gram-positive bacteria, and also inhibits spore outgrowth of Bacillus anthracis [80], being its effectiveness strain-dependent [103]. It is composed of two lantibiotic-type peptides, Halα (2.8 kDa) and Halβ (2.3 kDa) [92], which act synergistically at a nanomolar concentration. Halα contains several overlapping rings and the lipid II-binding motif present in mersacidin, actagardine, and LtnA of lacticin 3147, while Halβ shows a more elongated structure. Halα binds to lipid II and inhibits transglycosylation by PBP1 during PG biosynthesis. This complex promotes binding of Halβ that forms pores in the membrane [102]. This sequential mechanism of action, suggested to occur in a lipid II/Halα/Halβ (1:2:2) ratio, is similar to that of lacticin 3147 despite having significant structural differences [102, 103].
Lactococcin 972
Lactococcin 972 (Lcn972) is a 7.5 kDa, cationic and highly active bacteriocin synthesized by L. lactis IPLA972. So far, it is the first non-lantibiotic that does not target the cytoplasmic membrane, that is, it does not make pores [86]. Rather, it inhibits PG biosynthesis by binding specifically to lipid II. Lcn972 activity was antagonized in vivo by lipid II, and not by other cell wall precursors. Lcn972 also co-precipitated with micelles containing lipid II and interfered in vitro with the enzymatic reactions of PBP2 (polymerization of PG) and FemX, both enzymes that use lipid II as a substrate [84]. In contrast to other lipid II-binding bacteriocins, Lcn972 shows two particular features: first, it seems to block the incorporation of cell wall precursors at the septum; and second, it possesses a narrow activity spectrum, being active exclusively against lactococci under active division [85]. The existence of a putative co-target has been proposed to explain this targeted mode of action.
Another striking feature is that Lcn972 is easily inactivated by heat, anticipating a complex structure–function relationship. Lcn972 was claimed to be a homodimer based on the results of in-gel bioassays in which the inhibitory activity appeared to be linked to a 15-kDa band, instead of the expected 7.5 kDa [86]. However, recent experimental data revealed that this aberrant migration occurred only in the presence of glycerol that stabilizes the folded form of Lcn972 and preliminary NMR data further discard the presence of such a homodimer (Turner D., personal communication).
The lack of homology between Lcn972 and other lipid II-binding peptides suggests that Lcn972 carries a novel lipid II-binding motif. Interestingly, there are more than 100 hits in the public databases that are related to Lcn972, building the protein family Pfam09683.
Colicin M
Within Gram-negative bacteriocins, colicin M, a 29.5-kDa protein synthesized by E. coli, is a unique colicin that interferes with the biosynthesis of PG and the O-antigen of the LPS in susceptible E. coli strains [57, 58, 123]. Colicin M is imported in susceptible cells by recognizing the ferrichrome receptor FhuA and actively translocated into the periplasm to exert its antimicrobial activity [17, 130]. Its narrow activity spectrum is due to the species-specific condition of the receptors and the translocation mechanisms involved. It has a phosphoesterase activity which degrades lipid II, specifically cleaving the phosphodiester bond between the lipid moiety and the pyrophosphoryl group [44, 57], in a magnesium-dependent manner [4]. Consequently, the lipid carrier C-55 cannot be recycled in new PG synthesis rounds nor in the synthesis of the O-antigen. Interestingly, Patin et al. [106] have demonstrated that colicin M can hydrolyze in vitro and in vivo any lipid II molecule regardless the composition of the peptide side chain, thus representing a good candidate to study cell wall degradation in bacteria.
The structure of colicin M revealed a complex folded conformation challenging the identification of the typical colicin domains [147]. However, the high identity observed in the C-terminus of colicin M and other orthologs in several species of Burkholderia, Pectobacterium, and virulent Pseudomonas supports the notion that the catalytic domain resides in this region, while the N-terminus and the central region are likely involved in target receptor recognition and translocation, respectively [5, 109, 147].
Cell Wall-degrading Bacteriocins
Several large and heat-sensitive proteins synthesized by bacteria that hydrolyze the cell wall have been described. They are comprised into class IIIa (so-called bacteriolysins), although their classification as bacteriocins is controversial [36, 62]. Bacteriolysins are usually organized in modules bearing catalytic activities and target recognition domains, resembling those present in bacteriophage endolysins [63] and streptococcal fratricins [9]. Immunity proteins are encoded in the vicinity of the structural genes and they often encode FemABX-like proteins, peptidyl transferases, that catalyze the incorporation of amino acid(s) into the interchain peptide bridge of the PG [11, 48]. Examples of class IIIa bacteriolysins are listed in Table 1 and some examples are given below.
Lysostaphin
Lysostaphin may be the most studied bacteriolytic bacteriocin with regard to clinical applications. It is a 27-kDa metallo-enzyme firstly identified in Staphylococcus simulans [125] and highly specific against S. aureus [3, 55]. Its mature form displays an N-terminal domain responsible for the catalytic activity, and a C-terminal involved in binding to the target PG [3]. Lysostaphin is a glycylglycine endopeptidase targeting the pentaglycine cross-bridge of the PG in many staphylococci [55], cleaving specifically between the third and the fourth Gly residues [127]. Producer strains do not have specific immunity genes; instead, they are resistant due to the plasmid-encoded gene lif, involved in the addition of serine residues to the pentaglycine bridge that prevents hydrolysis by lysostaphin [38, 135]. Several authors have proposed lysostaphin as an effective therapeutic agent against S. aureus or as a tool to detect this pathogen in food matrices ([77] and references therein).
Zoocin A
Zoozin A is a 30-kDa endopeptidase synthesized by Streptococcus equi subsp. zooepidemicus 4881 that hydrolyzes the bond between the terminal d-alanine of the peptide side chain and the l-alanine of the cross-bridge in the PG of sensitive streptococci [49]. It carries two functional domains: the catalytic N-terminus, showing high similarity to other PG endopeptidases such as lysostaphin, and the C-terminal domain involved in PG recognition and binding [1]. Immunity relies on zif, involved in the addition of alanine residues to the cross-bridge, making it longer and more resistant to hydrolysis. Zif shows high similarity with FemABX-like proteins [48, 117].
Millericin B
Millericin B is a 28-kDa PG hydrolase synthesized by Streptococcus milleri NMSCC 061 with inhibitory activity against a wide range of Gram-positive bacteria. Its endopeptidase activity cleaves at both the peptide side chain and the cross-bridge [10]. Beukes and Hastings [11] have identified three genes putatively involved in immunity and export. milF and tRNAleu incorporate leucine residues into the cross-bridge that hinder hydrolysis. milT is homologous to ABC transporters and is likely involved in millericin export.
Pesticin
Within Gram-negatives, PG hydrolytic enzymes are also found. For example, pesticin is a 39.9-kDa bacteriocin synthesized by Yersinia pestis, which kills other Yersinia species and some E. coli strains [28]. The genetic organization and its three-domain structural architecture resemble that of colicins featuring an N-terminal translocation domain, a central receptor binding domain, and the C-terminal activity domain. The activity domain has a similar folding to lysozyme-related proteins such as the archetypal phage T4 lysin [107]. Although they differ in sequence, both T4 lysozyme and pesticin share the same enzymatic activity, cleaving the β1-4 glycosidic bond between NAM and NAG in the PG chain [141]. Like colicin M, pesticin’s receptor is the outer membrane iron-siderophore transporter FyuA which also plays a role as virulent factor of many pathogens. Immunity to pesticin relies on Pim, a protein which specifically recognizes a sequence located in the catalytic domain.
The modular organization of pesticin allowed designing a hybrid bacteriocin composed by the FyuA receptor domain and a foreign phage-related muramidase domain, not recognized by the immunity protein Pim. This hybrid bacteriocin is able to penetrate across the outer membrane and effectively kill pesticin producers and several Gram-negative bacteria [81].
Cell Wall-active Bacteriocins as Biotechnological Tools
Bacteriocins have mainly been considered as food biopreservatives, particularly those produced by lactic acid bacteria (LAB). In fact, more than 700 patents related to LAB bacteriocins have been registered and more than 400 are linked to the improvement of food quality, probiotics for animal feed, and mastitis treatment [46, 94]. The potential of LAB bacteriocins in food biopreservation relies basically on the traditional role of these bacteria as starters in food fermentations and their GRAS (Generally Regarded As Safe) status. Moreover, the bacteriocins they produce do not have a toxic effect on eukaryotic cells [46, 82] and they have a wider spectrum of activity compared to those produced by Gram-negative bacteria, impairing the development of food-borne pathogens. Accordingly, several LAB bacteriocins are commercialized in several countries as food biopreservatives. Their application as biopreservatives in food has been extensively reviewed elsewhere [46, 47, 94, 97].
From a clinical point of view, the development of pathogens such as vancomycin-resistant enterococci (VRE) and MRSA is of particular concern to animal and public health agencies worldwide. In this line, bacteriocins such as nisin, lacticin 3147, mersacidin, or lysostaphin have also been considered as an alternative to traditional antibiotics [6, 37, 74, 75, 131].
Besides the applications of bacteriocins based on their antibiotic activities, cell wall-active bacteriocins may be also very useful to study cell wall biology and, particularly, the response to cell envelope stress in relevant microorganisms. As outlined below, this approach has been exemplified by the role of the cell wall-active bacteriocin Lcn972 in understanding the molecular mechanisms that govern the response of the industrially relevant L. lactis to cell wall damaging. The generated knowledge has provided a basis for improving L. lactis performance in food fermentations (Fig. 3).
Lactococcus lactis is the main component of the mesophilic starter cultures used in cheese manufacturing, and robust strains are continuously demanded to improve the yield of industrial fermentations and guarantee the optimal characteristics of the final fermented product [20]. In the dairy environment, L. lactis must tolerate diverse stress conditions either during starter manufacture or along the fermentation process. Thus, the knowledge on how L. lactis monitors cell wall integrity and develops appropriate responses is, without a doubt, relevant to optimize its performance and robustness. In this context, the transcriptional response of L. lactis to Lcn972 helped to identify CesSR as the main two-component system (TCS) that orchestrates the primary line of defense to cell wall damage in this microorganism [87]. Several genes in L. lactis were identified as members of the CesR regulon, and among them, llmg0169 and the operon llmg2164-2163, encoding a putative membrane protein and a Psp-like protein, respectively, were highly induced. These genes could be further correlated to a higher survival of L. lactis under technological stress such as low pH, heat, or NaCl, among others, and have been proposed as biomarkers for strain robustness [114] (Fig. 3).
The characterization of resistant mutants to cell wall-active bacteriocins of industrially relevant microorganisms may also help to select more robust strains and reveal interesting mechanisms by which they become better adapted to their industrial use. For instance, a recent work has shown that L. lactis is able to shorten the peptide chain of its muropeptides to counteract the activity of Lcn972. Remarkably, these mutants were also insensitive to other dairy preservatives such as lysozyme and nisin, and to some bacteriophages which are a major threat in dairy fermentations [115].
There are as well examples of biotechnological applications derived from the current knowledge on cell wall-active bacteriocins. CesSR and its orthologues in other Gram-positives such as LiaSR in B. subtilis and VraSR in S. aureus were shown to be specifically induced by lipid II binders such as nisin, bacitracin, and vancomycin [78, 87, 88]. This feature has been further exploited to develop HTS (High-throughput screening) reporter systems to identify cell wall-active compounds and monitor the stress response they trigger. These methods were designed with the aim to be easy handling, quick and applicable to high amount of samples. A reporter B. subtilis strain was created by fusing the promoter of the TCS LiaSR to the lacZ gene, thus expressing beta-galactosidase as response to antibiotics interfering with the lipid II cycle [89]. Qualitative assays were performed with agar diffusion tests, and quantification was possible in liquid cultures in microtiter plates, thus enabling to screen and identify lipid II-interfering compounds [26]. Similarly, the promoter of llmg0169, the most up-regulated gene after CesSR activation in L. lactis, was fused to the gene encoding the green fluorescence protein (GFP), and a microtiter fluorescence-based assay was developed to monitor cell envelope response in L. lactis under specific conditions [27].
In view of these highlighted reports, cell wall-active bacteriocins are envisaged, not only as effective antimicrobials, but also as tools to get a deeper knowledge on the genetic and physiological consequences of cell wall damage in bacteria. Understanding bacterial regulatory mechanisms involved in stress responses may ultimately lead to a rational selection of industrially relevant microorganisms for specific applications.
References
Akesson M, Dufour M, Sloan GL, Simmonds RS (2007) Targeting of streptococci by zoocin A. FEMS Microbiol Lett 270(1):155–161
Asaduzzaman SM, Nagao J, Iida H, Zendo T, Nakayama J, Sonomoto K (2009) Nukacin ISK-1, a bacteriostatic lantibiotic. Antimicrob Agents Chemother 53(8):3595–3598
Baba T, Schneewind O (1996) Target cell specificity of a bacteriocin molecule: a C-terminal signal directs lysostaphin to the cell wall of Staphylococcus aureus. EMBO J 15(18):4789–4797
Barreteau H, Bouhss A, Fourgeaud M, Mainardi JL, Touzé T, Gérard F, Blanot D, Arthur M, Mengin-Lecreulx D (2009) Human- and plant-pathogenic Pseudomonas species produce bacteriocins exhibiting colicin M-like hydrolase activity towards peptidoglycan precursors. J Bacteriol 191(11):3657–3664
Barreteau H, El Ghachi M, Barneoud-Arnoulet A, Sacco E, Touzé T, Duché D, Gérard F, Brooks M, Patin D, Bouhss A, Blanot D, van Tilbeurgh H, Arthur M, Lloubès R, Mengin-Lecreulx D (2012) Characterization of Colicin M and its Orthologs Targeting Bacterial Cell Wall Peptidoglycan Biosynthesis. Microb Drug Resist. doi:10.1089/mdr.2011.0230
Bastos MCF, Coutinho BG, Coelho MLV (2010) Lysostaphin: a Staphylococcal bacteriolysin with potential clinical applications. Pharmaceuticals 3(4):1139–1161
Bauer R, Dicks LM (2005) Mode of action of lipid II-targeting lantibiotics. Int J Food Microbiol 101(2):201–216
Bera A, Herbert S, Jakob A, Vollmer W, Götz F (2005) Why are pathogenic staphylococci so lysozyme resistant? The peptidoglycan O-acetyltransferase OatA is the major determinant for lysozyme resistance of Staphylococcus aureus. Mol Microbiol 55(3):778–787
Berg KH, Biornstad TJ, Johnsborg O, Håvarstein LS (2012) Properties and biological role of streptococcal fratricins. Appl Environ Microbiol 78(10):3515–3522
Beukes M, Bierbaum G, Sahl HG, Hastings JW (2000) Purification and partial characterization of a murein hydrolase, millericin B, produced by Streptococcus milleri NMSCC 061. Appl Environ Microbiol 66(1):23–28
Beukes M, Hastings JW (2001) Self-protection against cell wall hydrolysis in Streptococcus milleri NMSCC 061 and analysis of the millericin B operon. Appl Environ Microbiol 67(9):3888–3896
Bierbaum G, Sahl HG (1985) Induction of autolysis of staphylococci by the basic peptide antibiotics Pep 5 and nisin and their influence on the activity of autolytic enzymes. Arch Microbiol 141(3):249–254
Bierbaum G, Sahl HG (1987) Autolytic system of Staphylococcus simulans 22: influence of cationic peptides on activity of N-acetylmuramoyl-l-alanine amidase. J Bacteriol 169(12):5452–5458
Bierbaum G, Sahl HG (2009) Lantibiotics: mode of action, biosynthesis and bioengineering. Curr Pharm Biotechnol 10(1):2–18
Bonelli RR, Schneider T, Sahl HG, Wiedemann I (2006) Insights into in vivo activities of lantibiotics from gallidermin and epidermin mode-of-action studies. Antimicrob Agents Chemother 50(4):1449–1457
Böttiger T, Schneider T, Martínez B, Sahl HG, Wiedemann I (2009) Influence of Ca(2+) ions on the activity of lantibiotics containing a mersacidin-like lipid II binding motif. Appl Environ Microbiol 75(13):4427–4434
Braun V, Patzer SI, Hantke K (2002) Ton-dependent colicins and microcins: modular design and evolution. Biochimie 84(5–6):365–380
Breukink E, de Kruijff B (2006) Lipid II as a target for antibiotics. Nat Rev Drug Discov 5(4):321–332
Breukink E, Wiedemann I, van Kraaij C, Kuipers OP, Sahl H, de Kruijff B (1999) Use of the cell wall precursor lipid II by a pore-forming peptide antibiotic. Science 286(5448):2361–2364
Bron PA, Kleerebezem M (2011) Engineering lactic acid bacteria for increased industrial functionality. Bioeng Bugs 2(2):80–87
Brötz H, Bierbaum G, Markus A, Molitor E, Sahl HG (1995) Mode of action of the lantibiotic mersacidin: inhibition of peptidoglycan biosynthesis via a novel mechanism? Antimicrob Agents Chemother 39(3):714–719
Brötz H, Bierbaum G, Reynolds PE, Sahl HG (1997) The lantibiotic mersacidin inhibits peptidoglycan biosynthesis at the level of transglycosylation. Eur J Biochem 246(1):193–199
Brötz H, Josten M, Wiedemann I, Schneider U, Götz F, Bierbaum G, Sahl HG (1998) Role of lipid-bound peptidoglycan precursors in the formation of pores by nisin, epidermin and other lantibiotics. Mol Microbiol 30(2):317–327
Budic M, Rijavec M, Petkovsek Z, Zgur-Bertok D (2011) Escherichia coli bacteriocins: antimicrobial efficacy and prevalence among isolates from patients with bacteraemia. PLoS ONE 6(12):e28769. doi:10.1371/journal.pone.0028769
Bugg TD, Braddick D, Dowson CG, Roper DI (2011) Bacterial cell wall assembly: still an attractive antibacterial target. Trends Biotechnol 29(4):167–173
Burkard M, Stein T (2008) Microtiter plate bioassay to monitor the interference of antibiotics with the lipid II cycle essential for peptidoglycan biosynthesis. J Microbiol Methods 75(1):70–74
Campelo AB, Rodríguez A, Martínez B (2010) Use of green fluorescent protein to monitor cell envelope stress in Lactococcus lactis. Appl Environ Microbiol 76(3):978–981
Cascales E, Buchanan SK, Duché D, Kleanthous C, Lloubès R, Postle K, Riley M, Slatin S, Cavard D (2007) Colicin biology. Microbiol Mol Biol Rev 71(1):158–229
Castiglione F, Cavaletti L, Losi D, Lazzarini A, Carrano L, Feroggio M, Ciciliato I, Corti E, Candiani G, Marinelli F, Selva E (2007) A novel lantibiotic acting on bacterial cell wall synthesis produced by the uncommon actinomycete Planomonospora sp. Biochemistry 46(20):5884–5895
Chatterjee S, Chatterjee DK, Jani RH, Blumbach J, Ganguli BN, Klesel N, Limbert M, Seibert G (1992) Mersacidin, a new antibiotic from Bacillus. In vitro and in vivo antibacterial activity. J Antibiot (Tokyo) 45(6):839–845
Clarke AJ, Dupont C (1992) O-acetylated peptidoglycan: its occurrence, pathobiological significance, and biosynthesis. Can J Microbiol 38(2):85–91
Claverys JP, Martin B, Håvarstein LS (2007) Competence-induced fratricide in streptococci. Mol Microbiol 64(6):1423–1433
Cleveland J, Montville TJ, Nes IF, Chikindas ML (2001) Bacteriocins: safe, natural antimicrobials for food preservation. Int J Food Microbiol 71(1):1–20
Corr SC, Li Y, Riedel CU, O’Toole PW, Hill C, Gahan CG (2007) Bacteriocin production as a mechanism for the antiinfective activity of Lactobacillus salivarius UCC118. Proc Natl Acad Sci USA 104(18):7617–7621
Cotter PD, Deegan LH, Lawton EM, Draper LA, O’Connor PM, Hill C, Ross RP (2006) Complete alanine scanning of the two-component lantibiotic lacticin 3147: generating a blueprint for rational drug design. Mol Microbiol 62(3):735–747
Cotter PD, Hill C, Ross RP (2005) Bacteriocins: developing innate immunity for food. Nat Rev Microbiol 3(10):777–788
Crispie F, Twomey D, Flynn J, Hill C, Ross P, Meaney W (2005) The lantibiotic lacticin 3147 produced in a milk-based medium improves the efficacy of a bismuth-based teat seal in cattle deliberately infected with Staphylococcus aureus. J Dairy Res 72(2):159–167
DeHart HP, Heath HE, Heath LS, LeBlanc PA, Sloan GL (1995) The lysostaphin endopeptidase resistance gene (epr) specifies modification of peptidoglycan cross bridges in Staphylococcus simulans and Staphylococcus aureus. Appl Environ Microbiol 61(4):1475–1479
Delgado MA, Rintoul MR, Farías RN, Salomón RA (2001) Escherichia coli RNA polymerase is the target of the cyclopeptide antibiotic microcin J25. J Bacteriol 183(15):4543–4550
Diep DB, Skaugen M, Salehian Z, Holo H, Nes IF (2007) Common mechanisms of target cell recognition and immunity for class II bacteriocins. Proc Natl Acad Sci USA 104(7):2384–2389
Dufour A, Hindré T, Haras D, Le Pennec JP (2007) The biology of lantibiotics from the lacticin 481 group is coming of age. FEMS Microbiol Rev 31(2):134–167
Duquesne S, Destoumieux-Garzón D, Peduzzi J, Rebuffat S (2007) Microcins, gene-encoded antibacterial peptides from enterobacteria. Nat Prod Rep 24(4):708–734
Duquesne S, Petit V, Peduzzi J, Rebuffat S (2007) Structural and functional diversity of microcins, gene-encoded antibacterial peptides from enterobacteria. J Mol Microbiol Biotechnol 13(4):200–209
El Ghachi M, Bouhss A, Barreteau H, Touzé T, Auger G, Blanot D, Mengin-Lecreulx D (2006) Colicin M exerts its bacteriolytic effect via enzymatic degradation of undecaprenyl phosphate-linked peptidoglycan precursors. J Biol Chem 281(32):22761–22772
Field D, Connor PM, Cotter PD, Hill C, Ross RP (2008) The generation of nisin variants with enhanced activity against specific gram-positive pathogens. Mol Microbiol 69(1):218–230
Gálvez A, Abriouel H, López RL, Ben Omar N (2007) Bacteriocin-based strategies for food biopreservation. Int J Food Microbiol 120(1–2):51–70
García P, Martínez B, Rodríguez A, Rodríguez L (2010) Food biopreservation: promising strategies using bacteriocins, bacteriophages and endolysins. Trends Food Sci Technol 21:373–382
Gargis SR, Gargis AS, Heath HE, Heath LS, LeBlanc PA, Senn MM, Berger-Bächi B, Simmonds RS, Sloan GL (2009) Zif, the zoocin A immunity factor, is a FemABX-like immunity protein with a novel mode of action. Appl Environ Microbiol 75(19):6205–6210
Gargis SR, Heath HE, Heath LS, Leblanc PA, Simmonds RS, Abbott BD, Timkovich R, Sloan GL (2009) Use of 4-sulfophenyl isothiocyanate labeling and mass spectrometry to determine the site of action of the streptococcolytic peptidoglycan hydrolase zoocin A. Appl Environ Microbiol 75(1):72–77
Giaouris E, Briandet R, Meyrand M, Courtin P, Chapot-Chartier MP (2008) Variations in the degree of D-Alanylation of teichoic acids in Lactococcus lactis alter resistance to cationic antimicrobials but have no effect on bacterial surface hydrophobicity and charge. Appl Environ Microbiol 74(15):4764–4767
Gillor O, Riley MA, Chavan MA (2007) Bacteriocins’ role in bacterial communication bacteriocins. Springer, Berlin, pp 135–145
González B, Arca P, Mayo B, Suárez JE (1994) Detection, purification, and partial characterization of plantaricin C, a bacteriocin produced by a Lactobacillus plantarum strain of dairy origin. Appl Environ Microbiol 60(6):2158–2163
Gonzalez B, Glaasker E, Kunji E, Driessen A, Suarez JE, Konings WN (1996) Bactericidal mode of action of plantaricin C. Appl Environ Microbiol 62(8):2701–2709
Gordon DM, O’Brien CL (2006) Bacteriocin diversity and the frequency of multiple bacteriocin production in Escherichia coli. Microbiology 152(Pt 11):3239–3244
Gründling A, Schneewind O (2006) Cross-linked peptidoglycan mediates lysostaphin binding to the cell wall envelope of Staphylococcus aureus. J Bacteriol 188(7):2463–2472
Gut IM, Blanke SR, van der Donk WA (2011) Mechanism of inhibition of Bacillus anthracis spore outgrowth by the lantibiotic nisin. ACS Chem Biol 6(7):744–752
Harkness RE, Braun V (1989) Colicin M inhibits peptidoglycan biosynthesis by interfering with lipid carrier recycling. J Biol Chem 264(11):6177–6182
Harkness RE, Braun V (1989) Inhibition of lipopolysaccharide O-antigen synthesis by colicin M. J Biol Chem 264(25):14716–14722
Hasper HE, de Kruijff B, Breukink E (2004) Assembly and stability of nisin-lipid II pores. Biochemistry 43(36):11567–11575
Hasper HE, Kramer NE, Smith JL, Hillman JD, Zachariah C, Kuipers OP, de Kruijff B, Breukink E (2006) An alternative bactericidal mechanism of action for lantibiotic peptides that target lipid II. Science 313(5793):1636–1637
Håvarstein LS, Diep DB, Nes IF (1995) A family of bacteriocin ABC transporters carry out proteolytic processing of their substrates concomitant with export. Mol Microbiol 16(2):229–240
Heng NCK, Tagg JR (2006) What’s in a name? Class distinction for bacteriocins. Nat Rev Microbiol 4(2). doi:10.1038/nrmicro1273-c1
Hermoso JA, García JL, García P (2007) Taking aim on bacterial pathogens: from phage therapy to enzybiotics. Curr Opin Microbiol 10(5):461–472
Höltje JV (1998) Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol Mol Biol Rev 62(1):181–203
Horn N, Swindell S, Dodd H, Gasson M (1991) Nisin biosynthesis genes are encoded by a novel conjugative transposon. Mol Gen Genet 228(1–2):129–135
Hsu ST, Breukink E, Bierbaum G, Sahl HG, de Kruijff B, Kaptein R, van Nuland NA, Bonvin AM (2003) NMR study of mersacidin and lipid II interaction in dodecylphosphocholine micelles. Conformational changes are a key to antimicrobial activity. J Biol Chem 278(15):13110–13117
Hsu ST, Breukink E, Tischenko E, Lutters MA, de Kruijff B, Kaptein R, Bonvin AM, van Nuland NA (2004) The nisin-lipid II complex reveals a pyrophosphate cage that provides a blueprint for novel antibiotics. Nat Struct Mol Biol 11(10):963–967
Hyde AJ, Parisot J, McNichol A, Bonev BB (2006) Nisin-induced changes in Bacillus morphology suggest a paradigm of antibiotic action. Proc Natl Acad Sci USA 103(52):19896–19901
Iancu C, Grainger A, Field D, Cotter PD, Hill C, Ross RP (2012) Comparison of the Potency of the Lipid II Targeting Antimicrobials Nisin, Lacticin 3147 and Vancomycin Against Gram-Positive Bacteria. Probiotics Antimicrob Proteins 4(2):116–121
Islam MR, Nishie M, Nagao J, Zendo T, Keller S, Nakayama J, Kohda D, Sahl HG, Sonomoto K (2012) Ring A of nukacin ISK-1: a lipid II-binding motif for type-A(II) lantibiotic. J Am Chem Soc 134(8):3687–3690
Islam MR, Shioya K, Nagao J, Nishie M, Jikuya H, Zendo T, Nakayama J, Sonomoto K (2009) Evaluation of essential and variable residues of nukacin ISK-1 by NNK scanning. Mol Microbiol 72(6):1438–1447
Kjos M, Borrero J, Opsata M, Birri DJ, Holo H, Cintas LM, Snipen L, Hernández PE, Nes IF, Diep DB (2011) Target recognition, resistance, immunity and genome mining of class II bacteriocins from Gram-positive bacteria. Microbiology 157(Pt 12):3256–3267
Klaenhammer TR (1988) Bacteriocins of lactic acid bacteria. Biochimie 70(3):337–349
Kruszewska D, Sahl HG, Bierbaum G, Pag U, Hynes SO, Ljungh A (2004) Mersacidin eradicates methicillin-resistant Staphylococcus aureus (MRSA) in a mouse rhinitis model. J Antimicrob Chemother 54(3):648–653
Kruszewska H, Zareba T, Tyski S (2004) Examination of antimicrobial activity of selected non-antibiotic drugs. Acta Pol Pharm 61(Suppl):18–21
Kuipers OP, Beerthuyzen MM, de Ruyter PG, Luesink EJ, de Vos WM (1995) Autoregulation of nisin biosynthesis in Lactococcus lactis by signal transduction. J Biol Chem 270(45):27299–27304
Kumar JK (2008) Lysostaphin: an antistaphylococcal agent. Appl Microbiol Biotechnol 80(4):555–561
Kuroda M, Kuroda H, Oshima T, Takeuchi F, Mori H, Hiramatsu K (2003) Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol Microbiol 49(3):807–821
Lagos R, Wilkens M, Vergara C, Cecchi X, Monasterio O (1993) Microcin E492 forms ion channels in phospholipid bilayer membrane. FEBS Lett 321(2–3):145–148
Lawton EM, Cotter PD, Hill C, Ross RP (2007) Identification of a novel two-peptide lantibiotic, haloduracin, produced by the alkaliphile Bacillus halodurans C-125. FEMS Microbiol Lett 267(1):64–71
Lukacik P, Barnard TJ, Keller PW, Chaturvedi KS, Seddiki N, Fairman JW, Noinaj N, Kirby TL, Henderson JP, Steven AC, Hinnebusch BJ, Buchanan SK (2012) Structural engineering of a phage lysin that targets Gram-negative pathogens. Proc Natl Acad Sci USA 109(25):9857–9862
Maher S, McClean S (2006) Investigation of the cytotoxicity of eukaryotic and prokaryotic antimicrobial peptides in intestinal epithelial cells in vitro. Biochem Pharmacol 71(9):1289–1298
Martin NI, Sprules T, Carpenter MR, Cotter PD, Hill C, Ross RP, Vederas JC (2004) Structural characterization of lacticin 3147, a two-peptide lantibiotic with synergistic activity. Biochemistry 43(11):3049–3056
Martínez B, Böttiger T, Schneider T, Rodríguez A, Sahl HG, Wiedemann I (2008) Specific interaction of the unmodified bacteriocin lactococcin 972 with the cell wall precursor lipid II. Appl Environ Microbiol 74(15):4666–4670
Martínez B, Rodríguez A, Suárez JE (2000) Lactococcin 972, a bacteriocin that inhibits septum formation in lactococci. Microbiology 146(Pt 4):949–955
Martínez B, Suárez JE, Rodríguez A (1996) Lactococcin 972: a homodimeric lactococcal bacteriocin whose primary target is not the plasma membrane. Microbiology 142(Pt 9):2393–2398
Martínez B, Zomer AL, Rodríguez A, Kok J, Kuipers OP (2007) Cell envelope stress induced by the bacteriocin Lcn972 is sensed by the Lactococcal two-component system CesSR. Mol Microbiol 64(2):473–486
Mascher T, Margulis NG, Wang T, Ye RW, Helmann JD (2003) Cell wall stress responses in Bacillus subtilis: the regulatory network of the bacitracin stimulon. Mol Microbiol 50(5):1591–1604
Mascher T, Zimmer SL, Smith TA, Helmann JD (2004) Antibiotic-inducible promoter regulated by the cell envelope stress-sensing two-component system LiaRS of Bacillus subtilis. Antimicrob Agents Chemother 48(8):2888–2896
Mazzotta AS, Crandall AD, Montville TJ (1997) Nisin resistance in Clostridium botulinum spores and vegetative cells. Appl Environ Microbiol 63(7):2654–2659
McAuliffe O, Ryan MP, Ross RP, Hill C, Breeuwer P, Abee T (1998) Lacticin 3147, a broad-spectrum bacteriocin which selectively dissipates the membrane potential. Appl Environ Microbiol 64(2):439–445
McClerren AL, Cooper LE, Quan C, Thomas PM, Kelleher NL, van der Donk WA (2006) Discovery and in vitro biosynthesis of haloduracin, a two-component lantibiotic. Proc Natl Acad Sci USA 103(46):17243–17248
Metlitskaya A, Kazakov T, Kommer A, Pavlova O, Praetorius-Ibba M, Ibba M, Krasheninnikov I, Kolb V, Khmel I, Severinov K (2006) Aspartyl-tRNA synthetase is the target of peptide nucleotide antibiotic Microcin C. J Biol Chem 281(26):18033–18042
Montalbán-López M, Sánchez-Hidalgo M, Valdivia E, Martínez-Bueno M, Maqueda M (2011) Are bacteriocins underexploited? Novel applications for old antimicrobials. Curr Pharm Biotechnol 12(8):1205–1220
Moreno F, González-Pastor JE, Baquero MR, Bravo D (2002) The regulation of microcin B, C and J operons. Biochimie 84(5–6):521–529
Morgan SM, O’Connor PM, Cotter PD, Ross RP, Hill C (2005) Sequential actions of the two component peptides of the lantibiotic lacticin 3147 explain its antimicrobial activity at nanomolar concentrations. Antimicrob Agents Chemother 49(7):2606–2611
Nes IF, Diep DB, Holo H (2007) Bacteriocin diversity in Streptococcus and Enterococcus. J Bacteriol 189(4):1189–1198
Niu WW, Neu HC (1991) Activity of mersacidin, a novel peptide, compared with that of vancomycin, teicoplanin, and daptomycin. Antimicrob Agents Chemother 35(5):998–1000
O’Connor EB, Cotter PD, O’Connor P, O’Sullivan O, Tagg JR, Ross RP, Hill C (2007) Relatedness between the two-component lantibiotics lacticin 3147 and staphylococcin C55 based on structure, genetics and biological activity. BMC Microbiol 7:24
O’Connor EM, Shand RF (2002) Halocins and sulfolobicins: the emerging story of archaeal protein and peptide antibiotics. J Ind Microbiol Biotechnol 28(1):23–31
Okuda K, Aso Y, Nakayama J, Sonomoto K (2008) Cooperative transport between NukFEG and NukH in immunity against the lantibiotic nukacin ISK-1 produced by Staphylococcus warneri ISK-1. J Bacteriol 190(1):356–362
Oman TJ, Lupoli TJ, Wang TS, Kahne D, Walker S, van der Donk WA (2011) Haloduracin alpha binds the peptidoglycan precursor lipid II with 2:1 stoichiometry. J Am Chem Soc 133(44):17544–17547
Oman TJ, van der Donk WA (2009) Insights into the mode of action of the two-peptide lantibiotic haloduracin. ACS Chem Biol 4(10):865–874
Paiva AD, Breukink E, Mantovani HC (2011) Role of lipid II and membrane thickness in the mechanism of action of the lantibiotic bovicin HC5. Antimicrob Agents Chemother 55(11):5284–5293
Parisot J, Carey S, Breukink E, Chan WC, Narbad A, Bonev B (2008) Molecular mechanism of target recognition by subtilin, a class I lanthionine antibiotic. Antimicrob Agents Chemother 52(2):612–618
Patin D, Barreteau H, Auger G, Magnet S, Crouvoisier M, Bouhss A, Touzé T, Arthur M, Mengin-Lecreulx D, Blanot D (2012) Colicin M hydrolyses branched lipids II from Gram-positive bacteria. Biochimie 94(4):985–990
Patzer SI, Albrecht R, Braun V, Zeth K (2012) Structural and mechanistic studies of pesticin, a bacterial homolog of phage lysozymes. J Biol Chem 287(28):23381–23396
Pérez-Núñez D, Briandet R, David B, Gautier C, Renault P, Hallet B, Hols P, Carballido-López R, Guédon E (2011) A new morphogenesis pathway in bacteria: unbalanced activity of cell wall synthesis machineries leads to coccus-to-rod transition and filamentation in ovococci. Mol Microbiol 79(3):759–771
Pilsl H, Glaser C, Gross P, Killmann H, Olschläger T, Braun V (1993) Domains of colicin M involved in uptake and activity. Mol Gen Genet 240(1):103–112
Rea MC, Ross RP, Cotter PD, Hill C, Drider D, Rebuffat S (2011) Classification of bacteriocins from Gram-positive bacteria. In: Drider D, Rebuffat S (eds) Prokaryotic antimicrobial peptides. Springer, New York, pp 29–53
Reisinger P, Seidel H, Tschesche H, Hammes WP (1980) The effect of nisin on murein synthesis. Arch Microbiol 127(3):187–193
Reynolds PE, Courvalin P (2005) Vancomycin resistance in enterococci due to synthesis of precursors terminating in D-alanyl-d-serine. Antimicrob Agents Chemother 49(1):21–25
Riley MA, Wertz JE (2002) Bacteriocins: evolution, ecology, and application. Annu Rev Microbiol 56:117–137
Roces C, Campelo AB, Veiga P, Pinto JP, Rodríguez A, Martínez B (2009) Contribution of the CesR-regulated genes llmg0169 and llmg2164-2163 to Lactococcus lactis fitness. Int J Food Microbiol 133(3):279–285
Roces C, Courtin P, Kulakauskas S, Rodríguez A, Chapot-Chartier MP, Martínez B (2012) Isolation of Lactococcus lactis Mutants Simultaneously Resistant to the Cell Wall-Active Bacteriocin Lcn972, Lysozyme, Nisin, and Bacteriophage c2. Appl Environ Microbiol 78(12):4157–4163
Rogers HJ, Perkins HR, Ward JB (1980) Microbial cell walls and membranes. Chapman and Hall, London
Rohrer S, Berger-Bächi B (2003) FemABX peptidyl transferases: a link between branched-chain cell wall peptide formation and beta-lactam resistance in gram-positive cocci. Antimicrob Agents Chemother 47(3):837–846
Ruiz-Barba JL, Cathcart DP, Warner PJ, Jiménez-Díaz R (1994) Use of Lactobacillus plantarum LPCO10, a bacteriocin producer, as a starter culture in Spanish-style green olive fermentations. Appl Environ Microbiol 60(6):2059–2064
Ryan MP, Meaney WJ, Ross RP, Hill C (1998) Evaluation of lacticin 3147 and a teat seal containing this bacteriocin for inhibition of mastitis pathogens. Appl Environ Microbiol 64(6):2287–2290
Ryan MP, Rea MC, Hill C, Ross RP (1996) An application in cheddar cheese manufacture for a strain of Lactococcus lactis producing a novel broad-spectrum bacteriocin, lacticin 3147. Appl Environ Microbiol 62(2):612–619
Sashihara T, Kimura H, Higuchi T, Adachi A, Matsusaki H, Sonomoto K, Ishizaki A (2000) A novel lantibiotic, nukacin ISK-1, of Staphylococcus warneri ISK-1: cloning of the structural gene and identification of the structure. Biosci Biotechnol Biochem 64(11):2420–2428
Sauvage E, Kerff F, Terrak M, Ayala JA, Charlier P (2008) The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol Rev 32(2):234–258
Schaller K, Höltje JV, Braun V (1982) Colicin M is an inhibitor of murein biosynthesis. J Bacteriol 152(3):994–1000
Scheurwater EM, Burrows LL (2011) Maintaining network security: how macromolecular structures cross the peptidoglycan layer. FEMS Microbiol Lett 318(1):1–9
Schindler CA, Schuhardt VT (1964) Lysostaphin: a new bacteriolytic agent for the Staphylococcus. Proc Natl Acad Sci USA 51:414–421
Schmitt P, Wilmes M, Pugnière M, Aumelas A, Bachère E, Sahl HG, Schneider T, Destoumieux-Garzón D (2010) Insight into invertebrate defensin mechanism of action: oyster defensins inhibit peptidoglycan biosynthesis by binding to lipid II. J Biol Chem 285(38):29208–29216
Schneewind O, Fowler A, Faull KF (1995) Structure of the cell wall anchor of surface proteins in Staphylococcus aureus. Science (New York, NY) 268(5207):103–106
Schneider T, Kruse T, Wimmer R, Wiedemann I, Sass V, Pag U, Jansen A, Nielsen AK, Mygind PH, Raventós DS, Neve S, Ravn B, Bonvin AM, De Maria L, Andersen AS, Gammelgaard LK, Sahl HG, Kristensen HH (2010) Plectasin, a fungal defensin, targets the bacterial cell wall precursor Lipid II. Science 328(5982):1168–1172
Schneider T, Sahl HG (2010) An oldie but a goodie–cell wall biosynthesis as antibiotic target pathway. Int J Med Microbiol 300(2–3):161–169
Schöffler H, Braun V (1989) Transport across the outer membrane of Escherichia coli K12 via the FhuA receptor is regulated by the TonB protein of the cytoplasmic membrane. Mol Gen Genet 217(2–3):378–383
Sit CS, Vederas JC (2008) Approaches to the discovery of new antibacterial agents based on bacteriocins. Biochem Cell Biol 86(2):116–123
Smith L, Hasper H, Breukink E, Novak J, Cerkasov J, Hillman JD, Wilson-Stanford S, Orugunty RS (2008) Elucidation of the antimicrobial mechanism of mutacin 1140. Biochemistry 47(10):3308–3314
Stevens KA, Sheldon BW, Klapes NA, Klaenhammer TR (1991) Nisin treatment for inactivation of Salmonella species and other gram-negative bacteria. Appl Environ Microbiol 57(12):3613–3615
Szekat C, Jack RW, Skutlarek D, Färber H, Bierbaum G (2003) Construction of an expression system for site-directed mutagenesis of the lantibiotic mersacidin. Appl Environ Microbiol 69(7):3777–3783
Thumm G, Götz F (1997) Studies on prolysostaphin processing and characterization of the lysostaphin immunity factor (Lif) of Staphylococcus simulans biovar staphylolyticus. Mol Microbiol 23(6):1251–1265
Turner DL, Brennan L, Meyer HE, Lohaus C, Siethoff C, Costa HS, Gonzalez B, Santos H, Suárez JE (1999) Solution structure of plantaricin C, a novel lantibiotic. Eur J Biochem 264(3):833–839
van Heijenoort J (2001) Recent advances in the formation of the bacterial peptidoglycan monomer unit. Nat Prod Rep 18(5):503–519
van Kuijk S, Noll KS, Chikindas ML (2012) The species-specific mode of action of the antimicrobial peptide subtilosin against Listeria monocytogenes Scott A. Lett Appl Microbiol 54(1):52–58
Vollmer W (2008) Structural variation in the glycan strands of bacterial peptidoglycan. FEMS Microbiol Rev 32(2):287–306
Vollmer W, Blanot D, de Pedro MA (2008) Peptidoglycan structure and architecture. FEMS Microbiol Rev 32(2):149–167
Vollmer W, Pilsl H, Hantke K, Höltje JV, Braun V (1997) Pesticin displays muramidase activity. J Bacteriol 179(5):1580–1583
Wheeler R, Mesnage S, Boneca IG, Hobbs JK, Foster SJ (2011) Super-resolution microscopy reveals cell wall dynamics and peptidoglycan architecture in ovococcal bacteria. Mol Microbiol 82(5):1096–1109
Wiedemann I, Böttiger T, Bonelli RR, Schneider T, Sahl HG, Martínez B (2006) Lipid II-based antimicrobial activity of the lantibiotic plantaricin C. Appl Environ Microbiol 72(4):2809–2814
Wiedemann I, Böttiger T, Bonelli RR, Wiese A, Hagge SO, Gutsmann T, Seydel U, Deegan L, Hill C, Ross P, Sahl HG (2006) The mode of action of the lantibiotic lacticin 3147—a complex mechanism involving specific interaction of two peptides and the cell wall precursor lipid II. Mol Microbiol 61(2):285–296
Wiedemann I, Breukink E, van Kraaij C, Kuipers OP, Bierbaum G, de Kruijff B, Sahl HG (2001) Specific binding of nisin to the peptidoglycan precursor lipid II combines pore formation and inhibition of cell wall biosynthesis for potent antibiotic activity. J Biol Chem 276(3):1772–1779
Zasloff M (2002) Antimicrobial peptides of multicellular organisms. Nature 415(6870):389–395
Zeth K, Römer C, Patzer SI, Braun V (2008) Crystal structure of colicin M, a novel phosphatase specifically imported by Escherichia coli. J Biol Chem 283(37):25324–25331
Acknowledgments
Work at DairySafe group has been funded by grants BIO2004-04312, BIO2006-65061, BIO2010-17414 of the Ministerio de Economía y Competitividad (Spain) and grants EQUIP08-01 and COF08-01 from FICYT (Asturias, Spain). C.R. is a recipient of a predoctoral JAE-CSIC fellowship.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Roces, C., Rodríguez, A. & Martínez, B. Cell Wall-active Bacteriocins and Their Applications Beyond Antibiotic Activity. Probiotics & Antimicro. Prot. 4, 259–272 (2012). https://doi.org/10.1007/s12602-012-9116-9
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12602-012-9116-9