Abstract
Genogroup (G) IV norovirus (NoV) has been described in the literature as infectious agents in humans, although there are few reports regarding the frequency and spread of this virus, resulting in insufficient epidemiological data. The aim of this study was to investigate the occurrence of GIV norovirus in the State of Rio de Janeiro, Brazil in order to evaluate frequency, concentration, and genetic diversity using clinical and environmental approaches. For this purpose, 316 stool samples were collected from acute gastroenteritis cases reported over a period of three years. Wastewater samples were also obtained from the main wastewater treatment plant (WWTP) located in Rio de Janeiro throughout one year, totalizing 156 samples. All samples were submitted to quantitative analysis by TaqMan™ real-time PCR for GIV norovirus. Three out of 316 clinical samples were positive (0.9%) for GIV, with viral load ranging from 104 to 106 genome copies (CG) per gram. Regarding wastewater samples, GIV were detected in 52% of raw sewage, with viral load ranging from 104 to 106 CG per liter. Phylogenetic analysis revealed the circulation of a new GIV genotype in both clinical and environmental samples. To our knowledge, this is the first description of GIV norovirus in clinical samples in Brazil. These results demonstrate the importance of performing laboratory surveillance of clinical and environmental samples, assisting the comprehension of the epidemiology pattern of viruses with neglected diagnosis and indefinite impact in the population.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Human norovirus is estimated to be associated with 18% of all diarrheal diseases and is the major causative agent of acute gastroenteritis (AGE) outbreaks worldwide (Ahmed et al. 2014). Genus Norovirus belongs to the Caliciviridae family, which comprises nonenveloped viruses of icosahedral symmetry of approximately 27–30 nm in diameter. The norovirus genome is organized into three open reading frames (ORF). ORF1 encodes six nonstructural proteins, including the RNA-dependent RNA-polymerase; ORF2 encodes the structural protein VP1 that composes the viral capsid, and ORF3 encodes the structural protein VP2 (Hardy 2005). Norovirus is classified into seven genogroups (GI to GVII), and further subdivided into at least 36 genotypes (Kroneman et al. 2013; Vinje 2015; Zheng et al. 2006). GI, GII, and GIV have been described in the literature as infectious agents in humans and outbreaks are mainly caused by GII (Kroneman et al. 2008). While major epidemiological data report the occurrence of GI and GII, few studies have described the occurrence and spread of GIV from clinical and environmental samples worldwide (Di Martino et al. 2014; Kitajima et al. 2009, 2010, 2014, 2016; La Rosa et al. 2008, 2010; Teixeira et al. 2016).
In Brazil, studies regarding epidemiology of norovirus have focused in GI and GII detection and molecular characterization (Aragão et al. 2013; de Andrade et al. 2014; Ferreira et al. 2012; Fioretti et al. 2011, 2014; Portal et al. 2016; Siqueira et al. 2011, 2013; Vicentini et al. 2013; Victoria et al. 2007, 2010), and until now, there is only one study describing detection of GIV norovirus in environmental samples from the Amazon region (Teixeira et al. 2016). Thus, there is a lack of information concerning the prevalence of this genogroup in clinical specimens and its dissemination in environmental waters. Therefore, the aim of this study is to demonstrate the occurrence and impact of GIV infections in AGE cases occurred in the metropolitan area of Rio de Janeiro, evaluating its frequency and distribution in clinical and environmental samples.
Materials and Methods
Clinical Samples
A total of 316 stool samples were obtained from AGE outbreaks or sporadic cases from inpatients and outpatients attended in public health centers including hospitals (n = 192) and central laboratories (n = 124) located in the State of Rio de Janeiro, Brazil. AGE cases included in this study were collected between January/2012 and December/2014 and were previously negative for rotavirus. Samples were obtained from people of all ages, in which 208 were from children <5 years old, 131 from children of 6–12 years old, and 84 from people of 15–81 years old. Stool samples from each individual were collected in plastic bottles and preserved at 4 °C until initial processing.
Wastewater Samples and Virus Concentration Method
Between May/2013 and May/2014, wastewater samples were collected weekly from a wastewater treatment plant (WWTP), located at the metropolitan region of the State of Rio de Janeiro, Brazil. This WWTP is the largest in the state and receives urban sewage from approximately 1,500,000 inhabitants (24.6% of the population), including those residents in the central and peripheral regions of the city. The plant employs a secondary treatment process (an aerobic method using conventional activated sludge) and the final effluents are not chlorinated. Treated effluent from the WWTP is discharged into Guanabara Bay, which is an ecosystem that surrounds a large part of the city, including the ports. Samples were collected along three sites of the WWTP, totaling 156, 52 from each point: influent, which consists in raw sewage; primary effluent, which consists from a preliminary treatment and primary sedimentation; and secondary effluent, which is the final effluent and is obtained after the activated sludge process treatment. For viral concentration, was employed the skimmed-milk flocculation method previously described by Calgua et al. (2013). All samples were spiked with a known concentration of PP7 bacteriophage, using as an internal control.
GIV Norovirus Detection and Quantification
Viral nucleic acid was extracted from 140 μl of a 20% fecal suspensions or wastewater concentrated samples using QIAamp Viral RNA™ Mini Kit (QIAGEN, Valencia, CA, USA), according to the manufacturer’s protocol. cDNA synthesis was carried out using a pd(N)6™ random primer (Amersham Biosciences, UK) and Superscript™ III RNAse H Reverse kit (Invitrogen, USA).
A TaqMan® Real-Time PCR assay was used for quantitative detection of GIV using primers and probe previously described (Trujillo et al. 2006) in both clinical and environmental samples. QPCR reactions were performed using Applied Biosystems® 7500 Real-Time PCR Systems (Applied Biosystems, Foster City, CA, USA). A 10-fold serial plasmids dilution containing ORF1/2 junction of GIV norovirus were used to generate a standard curve for virus quantification. Genome copies (GC) per gram of stool sample (GC/g) or liter (GC/l) of wastewater were determined by adjusting the values according to volumes used for each step of the procedure (fecal suspension/virus concentration, extraction, cDNA synthesis, and qPCR reaction). Undiluted and tenfold dilutions of nucleic acid extract of sewage samples were analyzed in duplicate (4 runs/sample).
Molecular Characterization and Phylogenetic Analysis of GIV Norovirus
For genetic characterization and phylogenetic analysis, a semi-nested-PCR was performed targeting the 5′-end ORF2 region (Kitajima et al. 2010). Amplicons were purified and sequenced using ABI Prism BigDye Terminator Cycle Sequencing Ready Reaction Kit® and the ABI Prism 3130xl DNA Sequencer (Applied Biosystems, Foster City, CA, USA). Following chromatograms analyses, consensual sequences were obtained using BioEdit (Hall 1999). A phylogenetic dendrogram was constructed by the neighbor-joining method using a matrix of genetic distances established under Kimura-two parameter model using MEGA v.7.0.21 (Kumar et al. 2016). The nucleotide sequences obtained in this study were submitted to the National Center for Biotechnology Information (GenBank, http://www.ncbi.nlm.nih.gov/) and received the following accession numbers: KY489824–KY489851.
Statistical Analyses
Statistical analyses were performed using GraphPad Prism v. 5.01 (GraphPad Software, San Diego, CA, USA). Viral load concentration of GIV norovirus in wastewater (influent, primary, and secondary effluent) was analyzed for significant differences using Mann–Whitney U test. Frequencies of GIV detection in wastewater were compared using Fisher`s exact test. A p value inferior to 0.05 was considered statistically significant.
Results
A total of three out of 316 clinical samples were positive (0.9%) for GIV, with viral loads ranging from 1.5 × 104 to 1.2 × 106 GC/g, being two presenting co-infection with GI and GI/GII (data not shown). Positive samples were obtained from outpatients collected in 2013, with ages of 03, 23, and 61 years old.
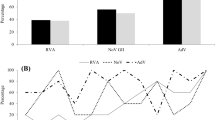
Overall analysis of the 156 wastewater samples detected GIV in 53 (34%). Frequency in influent and primary effluents samples was 52 and 48% [(27/52), (25/52)], respectively. In secondary samples, a clear reduction (p < 0.001) in GIV frequency was observed (1.9%). Quantitative analyses in wastewater samples demonstrated viral load ranging from 4.7 × 104 to 3.5 × 106 GC/l, without statistical differences with the median viral load between influent and primary effluent samples (Fig. 1). The GIV frequency in raw sewage was higher in winter and summer (p < 0.05) (Fig. 2).
All qPCR positive samples (53 from wastewater and 3 from stool) were submitted for semi-nested PCR, being 30 and 2 positive from wastewater and stool samples, respectively. The phylogenetic tree of partial sequence (282 nucleotides) of VP1 gene from 27 samples obtained from stools (2) and sewage (25) demonstrated all samples clustering into a new genotype of GIV, named GIV.3 in this study (Fig. 3). Nucleotide identities among sewage and clinical samples ranged from 99.3 to 100%.
Bacteriophage PP7, used as an internal control of virus concentration method, was detected in 100% of wastewater samples.
Discussion
The present study demonstrates data regarding occurrence of GIV norovirus in clinical and wastewater samples in the great metropolitan region of the State of Rio de Janeiro, Brazil. As our knowledge, no study describing the circulation of this genogroup in clinical samples was reported in Brazil. Recently, GIV norovirus was described for the first time in the country, in samples collected from surface water and untreated sewage in the North region of Brazil (Teixeira et al. 2016).
Out of three clinical samples detected positive for GIV, two presented co-infection with GI and GI/GII. Thus, we could not infer if the AGE case could be associated with GIV infection in affected patients. In Bangladesh, patients with diarrheal disease presented co-infection with the three genogroups (GI, GII, and GIV) in 11% of norovirus positive cases and 2% presented infection with only GIV (Rahman et al. 2016).
Overall, 52% of the wastewater influent and 48% of the primary effluent samples tested positive, while 1.9% of the effluent samples tested positive, demonstrating a sharp reduction of detectable viruses. Nevertheless, viral genome loads below the qPCR assay detection limit could be present in treated effluent samples. Also, because of the sampling procedure used (grab sample), we could not properly infer the real removal efficiency of the WWTP studied. Studies demonstrate positivity rate in untreated wastewater varying from 16.7% in Brazil (North region) (Teixeira et al. 2016), 21.8% in Italy (Muscillo et al. 2013), 50% in Japan (Kitajima et al. 2009), and 67% in the United States (Kitajima et al. 2014).
Concerning frequency distribution in raw sewage during 2013 and 2014, GIV viruses were detected with a higher frequency and viral load during the winter and summer months which differs from data from the US where these viruses were detected year-round (Kitajima et al. 2014).
A higher positivity percentage (34%) of GIV observed in environmental samples when compared to clinical samples (0.9%) was similar to a study performed in Italy, where GIV positivity in untreated sewage samples (21.8%) was seven times higher when compared with diarrhea cases collected from hospitalized patients (3.2%) (Muscillo et al. 2013). These results may suggest association with asymptomatic cases in GIV infection. Nevertheless, it is important to notice that since AGE is not a severe disease in adults, many cases are not notified, which could also explain higher prevalence in untreated wastewater but not in clinical samples.
Recently, Kitajima et al. (2016) described the circulation of a new GIV genotype (GenBank accession number LC150829) based on differences in the clustering pattern among GIV strains. In our study, the phylogenetic analysis revealed the circulation of this new genotype, named GIV.3 in the present study. All sequences from clinical and environmental samples grouped into the same cluster containing Japanese and North American strains. To our knowledge, this would be the first description of the circulation of this novel genotype in Latin America; however, further studies on phylogenetic analysis from complete sequence of VP1 with strains of this proposed novel genotype are needed. Nucleotide identities among sewage and clinical samples varied from 99.3 to 100%, which may suggest circulation of the same strain in humans and in the environment.
Data obtained in this study demonstrate the importance of laboratorial surveillance in AGE cases combined with environmental approaches to understand the impact of a new virus infection in a given population. Continuous monitoring of which norovirus genotypes circulate in a certain geographic area is important when introduction of a norovirus vaccine is being considered.
References
Ahmed, S. M., Hall, A. J., Robinson, A. E., Verhoef, L., Premkumar, P., Parashar, U. D., et al. (2014). Global prevalence of norovirus in cases of gastroenteritis: A systematic review and meta-analysis. The Lancet Infectious Diseases, 14(8), 725–730.
Aragão, G. C., Mascarenhas, J. D., Kaiano, J. H., de Lucena, M. S., Siqueira, J. A., Fumian, T. M., et al. (2013). Norovirus diversity in diarrheic children from na African-descendant settlement in Belém, Northern Brazil. PLoS ONE, 8(2), e56608.
Calgua, B., Rodriguez-Manzano, J., Hundesa, A., Suñen, E., Calvo, M., Bofill-Mas, S., et al. (2013). New methods for the concentration of viruses from urban sewage using quantitative PCR. Journal of Virological Methods, 187(2), 215–221.
de Andrade, J. S., Rocha, M. S., Carvalho-Costa, F. A., Fioretti, J. M., Xavier, M. P., Nunes, Z. M., et al. (2014). Noroviruses associated with outbreaks of acute gastroenteritis in the State of Rio Grande do Sul, Brazil, 2004-2011. Journal of Clinical Virology, 61(3), 345–352.
Di Martino, B., Di Profio, F., Ceci, C., Di Felice, E., Green, K. Y., Bok, K., et al. (2014). Seroprevalence of norovirus genogroup IV antibodies among humans, Italy, 2010-2011. Emerging Infectious Diseases, 20(11), 1828–1832.
Ferreira, M. S., Xavier, M. P., Tinga, A. C., Rose, T. L., Fumian, T. M., Fialho, A. M., et al. (2012). Assessment of gastroenteric viruses frequency in a children’s day care center in Rio De Janeiro, Brazil: A fifteen year study (1994–2008). PLoS ONE, 7(3), e33754.
Fioretti, J. M., Bello, G., Rocha, M. S., Victoria, M., Leite, J. P., & Miagostovich, M. P. (2014). Temporal dynamics of norovirus GII.4 variants in Brazil between 2004 and 2012. PLoS ONE, 9(3), e92988.
Fioretti, J. M., Ferreira, M. S., Victoria, M., Vieira, C. B., Xavier, M. P., Leite, J. P., et al. (2011). Genetic diversity of noroviruses in Brazil. Memorias do Instituto Oswaldo Cruz, 106(8), 942–947.
Hall, T. A. (1999). BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series, 41, 95–98.
Hardy, M. E. (2005). Norovirus protein structure and function. FEMS Microbiology Letters, 253, 1–8.
Kitajima, M., Haramoto, E., Phanuwan, C., Katayama, H., & Ohgaki, S. (2009). Detection of genogroup IV norovirus in wastewater and river water in Japan. Letters in Applied Microbiology, 49(5), 655–658.
Kitajima, M., Iker, B. C., Pepper, I. L., & Gerba, C. P. (2014). Relative abundance and treatment reduction of viruses during wastewater treatment processes–identification of potential viral indicators. Science of the Total Environment, 488–489, 290–296.
Kitajima, M., Oka, T., Haramoto, E., Takeda, N., Katayama, K., & Katayama, H. (2010). Seasonal distribution and genetic diversity of genogroups I, II, and IV noroviruses in the Tamagawa River, Japan. Environmental Science & Technology, 44(18), 7116–7122.
Kitajima, M., Rachmadi, A. T., Iker, B. C., Haramoto, E., & Gerba, C. P. (2016). Genetically distinct genogroup IV norovirus strains identified in wastewater. Archives of Virology, 161(12), 3521–3525.
Kroneman, A., Veja, E., Vennema, H., Vinjé, J., White, P. A., Hansman, G., et al. (2013). Proposal for a unified norovirus nomenclature and genotyping. Archives of Virology, 158(10), 2059–2068.
Kroneman, A., Verhoef, L., Harris, J., Vennema, H., Duizer, E., van Duynhoven, Y., et al. (2008). Analysis of integrated virological and epidemiological reports of norovirus outbreaks collected within the Foodborne Viruses in Europe network from 1 July 2001 to 30 June 2006. Journal of Clinical Microbiology, 46(9), 2959–2965.
Kumar, S., Stecher, G., & Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution, 33, 1870–1874.
La Rosa, G., Iaconelli, M., Pourshaban, M., Fratini, M., & Muscillo, M. (2010). Molecular detection and genetic diversity of norovirus genogroup IV: A yearlong monitoring of sewage throughout Italy. Archives of Virology, 155(4), 589–593.
La Rosa, G., Pourshaban, M., Iaconelli, M., & Muscillo, M. (2008). Detection of genogroup IV noroviruses in environmental and clinical samples and partial sequencing through rapid amplification of cDNA ends. Archives of Virology, 153(11), 2077–2083.
Muscillo, M., Fratini, M., Graffeo, R., Sanguinetti, M., Martella, V., Green, K. Y., et al. (2013). GIV noroviruses in wastewaters and in stool specimens from hospitalized patients. Food and Environmental Virology, 5(4), 194–202.
Portal, T. M., Siqueira, J. A., Costa, L. C., Lima, I. C., Lucena, M. S., Bandeira, R. S., et al. (2016). Caliciviruses in hospitalized children, São Luís, Maranhão, 1997-1999: Detection of norovirus GII.12. Brazilian Journal of Microbiology, 47(3), 724–730.
Rahman, M., Rahman, R., Nahar, S., Hossain, S., Ahmed, S., Golam Faruque, A. S., et al. (2016). Norovirus diarrhea in Bangladesh, 2010-2014: Prevalence, clinical features, and genotypes. Journal of Medical Virology, 88(10), 1742–1750.
Siqueira, J. A., Linhares, A. C., de Carvalho, T. C., Aragão, G. C., Oliveira, D. S., Dos Santos, M. C., et al. (2013). Norovirus infection in children admitted to hospital for acute gastroenteritis in Belém, Pará, Northern Brazil. Journal of Medical Virology, 85(4), 737–744.
Siqueira, J. A., Linhares, A. C., Oliveira, D. S., Soares, L. S., Lucena, M. S., Wanzeller, A. L., et al. (2011). Evaluation of third-generation RIDASCREEN enzyme immunoassay for the detection of norovirus antigens in stool samples of hospitalized children in Belém, Pará, Brazil. Diagnostic Microbiology and Infectious Disease, 71(4), 391–395.
Teixeira, D. M., Hernandez, J. M., Silva, L. D., Oliveira, D. S., Spada, P. K., Gurjão, T. C., et al. (2016). Occurrence of norovirus GIV in environmental water samples from Belém City, Amazon Region, Brazil. Food and Environmental Virology, 8(1), 101–104.
Trujillo, A. A., McCaustland, K. A., Zheng, D. P., Hadley, L. A., Vaughn, G., Adams, S. M., et al. (2006). Use of TaqMan real-time reverse transcription-PCR for rapid detection, quantification, and typing of norovirus. Journal of Clinical Microbiology, 44(4), 1405–1412.
Vicentini, F., Denadai, W., Gomes, Y. M., Rose, T. L., Ferreira, M. S., Le Moullac-Vaidye, B., et al. (2013). Molecular characterization of noroviruses and HBGA from infected Quilombola children in Espirito Santo State, Brazil. PLoS ONE, 8(7), e69348.
Victoria, M., Carvalho-Costa, F. A., Heinemann, M. B., Leite, J. P., & Miagostovich, M. (2007). Prevalence and molecular epidemiology of noroviruses in hospitalized children with acute gastroenteritis in Rio de Janeiro, Brazil, 2004. The Pediatric Infectious Disease Journal, 26(7), 602–606.
Victoria, M., Guimarães, F. R., Fumian, T. M., Ferreira, F. F., Vieira, C. B., Shubo, T., et al. (2010). One year monitoring of norovirus in a sewage treatment plant in Rio de Janeiro, Brazil. Journal of Water and Health, 8(1), 158–165.
Vinje, J. (2015). Advances in laboratory methods for detection and typing of norovirus. Journal of Clinical Microbiology, 53, 373–381.
Zheng, D. P., Ando, T., Fankhauser, R. L., Beard, R. S., Glass, R. I., & Monroe, S. S. (2006). Norovirus classification and proposed strain nomenclature. Virology, 346, 312–323.
Acknowledgement
We kindly thank Peter White, from the School of Biotechnology and Biomolecular Sciences, Faculty of Science, University of New South Wales, Sydney, Australia for the generous provision of plasmids that contained the GIV norovirus genome. The project was financially supported by Fundação de Amparo à pesquisa do Rio de Janeiro (Faperj) (E-26/102.845/2012) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq). This research study is under the scope of the activities of Fiocruz as a Collaborating Center of Pan American Health Organization (PAHO) and World Health Organization (WHO) of Public and Environmental Health.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical Approval
This study was approved by the Ethics Committee of Oswaldo Cruz Foundation (CEP 311/06) and is part of an official Brazilian Ministry of Health’s surveillance. In this ongoing program, diagnosis of gastroenteritis is required to elucidate viral etiology and data are anonymous.
Conflict of Interest
There is no conflict of interest for all the authors.
Rights and permissions
About this article
Cite this article
Fioretti, J.M., Fumian, T.M., Rocha, M.S. et al. Surveillance of Noroviruses in Rio De Janeiro, Brazil: Occurrence of New GIV Genotype in Clinical and Wastewater Samples. Food Environ Virol 10, 1–6 (2018). https://doi.org/10.1007/s12560-017-9308-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12560-017-9308-2