Abstract
Seed osmopriming is a pre-sowing treatment that involves limitation of the seed water imbibition, so that pre-germinative metabolic activities proceed without radicular protrusion. This technique is used for improving germination rate, uniformity of seedling growth and hastening the time to start germination. In Arabidopsis thaliana, seed germination has been associated with the induction of enzymes involved in cell wall modifications, such as expansins. The α-expansins (EXPAs) are involved in cell wall relaxation and extension during seed germination. We used online tools to identify AtEXPA genes with preferential expression during seed germination and RT-qPCR to study the expression of five EXPA genes at different germination stages of non-primed and osmoprimed seeds. In silico promoter analysis of these genes showed that motifs similar to cis-acting elements related to abiotic stress, light and phytohormone responses are the most overrepresented in promoters of these AtEXPA genes, showing that their expression is likely be regulated by intrinsic developmental and environmental signals during Arabidopsis seed germination. The osmopriming conditioning had a decreased time and mean to 50% germination without affecting the percentage of final seed germination. The dried PEG-treated seeds showed noticeable high mRNA levels earlier at the beginning of water imbibition (18 h), showing that transcripts of all five EXPA isoforms were significantly produced during the osmopriming process. The strong up-regulation of these AtEXPA genes, mainly AtEXPA2, were associated with the earlier germination of the osmoprimed seeds, which qualifies them to monitor osmopriming procedures and the advancement of germination.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Germination is a complex phenomenon that can be divided into three stages: Phase I—rapid absorption of water, increase in respiratory intensity and energy production and onset of degradation of the reserve substances; Phase II—active transport of substances, a decrease of respiration and water absorption; Phase III—transported substances are reorganized into more complex ones for the beginning of embryonic axis growth (Bewley et al. 2013).
Of the environmental factors that can influence the germination, the water availability is the most determinant factor as it intensifies the metabolic activities, culminating in the growth of the embryonic axis. However, environmental stresses may also positively influence seed performance as a physiological conditioning (priming) induces improvements in seed quality favoring higher germination rates and more uniform seedling emergence (Chen and Arora 2013; Khadraji et al. 2017; Abid et al. 2018). Seed osmopriming is a pre-sowing treatment which consists of the controlled hydration of the seeds, allowing only the first stages of the germination process (phases I and II) to be completed, with subsequent drying for the seed to return to a quiescent state. In this way, the germination process is initiated without radicular protrusion (phase III) (Bradford 1986). One of the osmopriming methods for controlling the imbibition rate of the seeds uses osmotic solutions of polyethylene glycol (PEG). This solute has desirable characteristics, since it is inert, is not absorbed by the seed and is non-toxic (Heydecker and Gibbins 1978).
There are numerous reports of the positive effect of osmopriming on seed germination ability, uniformity and vigor over non-primed seeds under adverse environmental conditions in different crop species, such as sorghum (Foti et al. 2002) soybean (Zhuo et al. 2009), rice (Sun et al. 2010), chickpeas (Lamichaney and Katiyar 2017), among many others. The beneficial effect of this technique on the germination-related processes has been associated with the accumulation of mRNA and inactive proteins produced during the osmotic conditioning (Bray 1995; Özbingol et al. 1999; Gallardo et al. 2001). For example, Soeda et al. (2005) identified the expression of genes related to stress tolerance during the conditioning of Brassica oleracea seeds, such as those coding for superoxide dismutases (SOD), heat shock proteins (HSP) and late embryogenesis abundant proteins (LEA). These authors also identified a group of genes that were up-regulated during germination in normal conditions, but not during priming. The comparative analysis of proteome changes in Medicago sativa seeds during germination and osmoconditioning showed some proteins specific to osmopriming, some that varied during germination and there were some proteins found to be commonly present in both the conditions, which were mainly involved in cell structure, metabolism and defense (Yacoubi et al. 2011). In spinach, increased germination of osmoprimed seeds was accompanied by the accumulation of dehydrins, proteins that are normally synthesized in maturating seeds during their desiccation (Chen et al. 2012). Priming-induced modulation of genes and enzymes of the proline metabolism, with a consequent improvement of salinity stress tolerance during post-priming germination, was reported in Brassica napus (Kubala et al. 2015).
In Arabidopsis thaliana seeds, the embryo is surrounded by a single layer of cells from the endosperm and a seed coat. The endosperm and its layer cells are responsible for regulating germination, which consists of two sequential events: the testa and endosperm rupture (Holdsworth et al. 2008). One of the main characteristics related to the endosperm rupture by radicle expansion is the weakening of the endosperm, which has been associated with the induction of cell wall loosening by enzymes such as expansins (Kucera et al. 2005). Transcriptomic and proteomic analyses have shown that the expression of several genes and proteins are up- or down-regulated during priming and post-priming germination, including those involved in cell wall loosening and extension (Zhang et al. 2014; Kubala et al. 2015; Sano et al. 2017). However, there is still no specific information on differentially expressed α-expansin (EXPA) isoforms in Arabidopsis seeds subjected to osmoconditioning during subsequent germination under normal conditions.
Expansins are proteins capable of inducing cell extension, acting as mediators in response to acidic pH and catalyzing the cell wall expansion (McQueen-Mason et al. 1992). These proteins are assumed to break non-covalent bonds between cell wall polysaccharides, extending these polysaccharides in an irreversible way without causing structural changes in the cell wall (Cosgrove 1997). They are members of a plant-specific superfamily that comprises two major families: α-expansins (EXPA) and β-expansins (EXPB), and two smaller families named expansin-like A (EXLA) and expansin-like B (EXLB) (Cosgrove 2015). Members of EXPA and EXPB families have already been functionally validated, while the functional aspects for the EXPLA and EXPLB families remain uncertain (Cosgrove 2015). The EXPAs are responsible for controlling cell wall extension and developmental processes, including cell dissociation and separation (McQueen-Mason et al. 1992; Cho and Kende 1997).
The role of expansins in elongation and cell expansion has been observed in different cells and tissues during growth and development, such as the hypocotyls (Caderas et al. 2000), roots and root hairs (Cho and Cosgrove 2002; Lin et al. 2001) and leaves (Muller et al. 2007; Goh et al. 2012). These proteins are also expressed during seed developmental stages (Budzinski et al. 2011; Hussain et al. 2016). As seed germination involves cell elongation and division, it is expected that proteins with such a role would be differentially regulated since the onset of this process. Indeed, transcriptional analyses during the germination of A. thaliana showed that expansin genes were induced in the endosperm and were involved in the processes that lead to testa rupture and weakening of the micropylar endosperm cap (Morris et al. 2011). Additionally, AtEXPA2 has been considered to be a marker for endosperm tissue in non-dormant imbibed or germinating seeds (Penfield et al. 2006; Yan et al. 2014). In rice, various specific expansins were shown to play important roles in coleoptile elongation under submergence stress (Hussain et al. 2016). These authors showed that some α-expansin genes were strongly up-regulated due to chemical and hormonal seed priming treatments compared to non-primed seeds, suggesting that expression of α-expansins might have contributed to priming-induced submergence tolerance in rice.
In this work, we aimed to improve our comprehension of gene expression patterns during A. thaliana seed germination by studying the expression transcription profile of α-expansins (EXPA) genes associated with seed germination. For this, selected EXPA coding sequences and their upstream regulatory elements were analyzed using experimental data available from transcriptome databases and qPCR analyses were performed to verify the expression of EXPA genes at different germination stages of non-primed and osmoprimed seeds. The data presented here demonstrate that osmopriming Arabidopsis seeds with PEG6000 caused differential regulation of α-expansins genes at early stages of germination when compared to non-primed seeds. Understanding the mRNA expression patterns of α-expansins in A. thaliana seeds subjected to physiological conditioning (osmopriming) may provide new insights into the mechanisms involved in seed priming-induced enhancement of seed germination, resulting in genetic manipulation approaches to obtain plants more tolerant to abiotic stresses at their initial development stages.
Materials and methods
Selection of α-expansins expressed during seed germination of A. thaliana
We initially selected the α-expansins from A. thaliana available in the Genevestigator database for heat-map analysis (https://www.genevestigator.com, Zimmermann et al. 2004) (Fig S1). For subsequent analysis, we selected the α-expansins with expression levels greater than or equal to 10 (signal intensity in Affymetrix microarray) and that were expressed preferentially during germination (1–5 days) (Fig S2). Based on these data, only five AtEXPAs genes that are preferentially expressed during germination were selected for further study: AtEXPA2 (AT5G05290), AtEXPA3 (AT2G37640), AtEXPA8 (AT2G40610), AtEXPA9 (AT5G02260) and AtEXPA20 (AT4G38210) (Fig S2). CLC Main Workbench (CLC bio, Qiagen, Denmark) and MEGA 5.1 (Kumar et al. 2016) software was used for deduced amino acid sequence comparison and alignments. The ScanProsite tool (http://prosite.expasy.org/scanprosite/) were applied to predict the putative motifs and domains.
For the sake of comparison with later RT-qPCR results, we have also analyzed the expression profile of the selected α-expansins in non-primed seeds subjected to various water soaking periods (1, 3, 6, 12 and 24 h at 4 °C) available at the BAR eFP database (Winter et al. 2007; http://bar.utoronto.ca/welcome.htm) (Fig S3).
In silico analysis of promoter sequences
A 1000 bp region upstream from translation start codon of AtEXPA genes was selected and analyzed using different databases: RegSite (http://softberry.com), PlantCare (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Lescot et al. 2002), cis-regulatory DNA Elements PLACE (http://www.dna.affrc.go.jp/PLACE/signalscan.html) (Higo et al. 1999) in order to find regulatory motifs on the putative promoter region. The motifs reported previously in the literature as related to seed germination were selected for further analyses. Pattern Matching in RSA-Tools (http://rsat.scmbb.ulb.ac.be/rsat/) (Thomas-Chollier et al. 2011) was used to map occurrences and frequency of the selected cis-elements onto the five ProAtEXPA sequences. Pattern matching and image generation were conducted using the default settings.
Plant material and growth conditions
Arabidopsis thaliana ecotype Columbia-0 seeds were germinated in Petri dishes containing moistened qualitative filter paper (Qualy™—J. Prolab, Brazil) at 15 °C in the dark. The seedlings were then transplanted into 5 cm diameter pots containing 80% Bioplant™ substrate with coconut fiber and 20% sphagnum, and fertilized with 1 g/L (w/v) of Osmocote™ and maintained in a growth room under a photoperiod of 8 h of light (120 μE m2 s1) at 20 °C. The plants were then allowed to grow until the siliques matured, and then the seeds were collected, dried and stored for the priming treatments.
Osmopriming and germination curve
Priming was carried out for 7 days at 15 °C by incubating seeds on germination paper moistened with a solution of polyethylene glycol 6000 (PEG 6000; Sigma-Aldrich, St. Louis, MO, US) equivalent to osmotic potential of − 0.75 MPa (Gallardo et al. 2001). Subsequently, the seeds were washed to remove the PEG and dried in an airtight mini chamber containing silica gel at 20 °C.
Primed (PEG) and non-primed seeds (control) were placed in 10-cm Petri dishes and distilled water equivalent to 2.5 times the dry weight of the germination paper was added to each Petri dish. The samples were kept in a germination chamber at a constant temperature of 15 °C ± 1 under 8 h of light (Baskin and Baskin 1972) and the germination rates were scored every 6 h during 120 h based on the number of seeds with primary root emission. The germination experiment consisted of a completely randomized design with 5 replicates with 50 seeds per replicate. The germination indices as maximum germination (GMax, as a percentage), germination times (T10, T50 and mean germination, in hours), uniformity of germination (U75–25: time interval between 25 and 75% of viable seeds to germinate) and area under the curve (AUC) were calculated using the GERMINATOR software (Joosen et al. 2010) for a 120 h period using the mathematical approach described by El-Kassaby et al. (2008). The percentage data, such as MaxG, were transformed to arcsine √x/100 (untransformed values are shown in the table to enable comparison).
RNA extraction, primers and RT-qPCR analysis
One gram of non-primed (control treatment) and osmoprimed seeds were sown on distilled water wetted filter paper in Petri dishes at 15 °C in the dark. Samples were collected at 0, 6, 18, 30, 42 and 54 h (20 mg per treatment, approximately 400 seeds) for total RNA extraction using the SV Total RNA Isolation System (Promega, Madison, WI, USA). The extracted RNAs were quantified by the spectrophotometry at A260 nm and purity evaluated via the absorbance ratios at A260/A280 nm. RNA samples were treated with Turbo DNAse I (Ambion, Austin, TX, USA to eliminate DNA contamination and the quality and integrity of the samples were checked at 1% (w/v) electrophoresis agarose gel dyed with ethidium bromide. The cDNA synthesis was made using SuperScript® IV First-Strand Synthesis SuperMix kit (Invitrogen, Carlsbad, CA, USA) starting from 2 μg of total RNA. cDNAs were subjected to a PCR reaction with the specific primers of each gene to verify whether the primers amplify only the targeted 80–100 base pairs sequence.
Specific primers were designed using Primer Express v. 3.0 software (Applied Biosystems, Foster City, CA, USA) in order to obtain amplicons of 80–100 base pairs in the regions of greatest dissimilarity, mainly from the 3′-untranslated region (3′-UTR). The amplification efficiencies for all primers were calculated using the LinRegPCR software (Ramakers et al. 2003; Table 1). For preventing non-specific annealing, all primers and amplicons were confronted with GenBank sequences using Primer BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) to confirm their specificity. In addition, the primers were evaluated with Primer3Plus (https://primer3plus.com/cgi-bin/dev/primer3plus.cgi) to check for primer dimers and hairpins.
The RT-qPCR was performed on the StepOnePlus™ Real-Time PCR (Thermo Fisher Scientific, Waltham, MA, USA) apparatus using the SYBR Green detection method. The final cDNA products were diluted 10-fold before being used in RT-qPCR. Each reaction contained 5 μl 2× SYBR Green/ROX qPCR Master Mix (Invitrogen, Carlsbad, CA, USA), 0.4 μl of each primer (5 µM), 1 μl of 1:10 diluted cDNA in milli-Q water, with a final volume of 10 μl. The reaction conditions were 2 min at 95 °C, followed by 40 cycles of amplification at 95 °C for 30 s and 60 °C for 60 s. The reactions were performed in triplicate for each of the three biological replicates per sampling time-point of each treatment. In all analyses, transcript levels of the target genes were normalized against the transcriptional profile of the reference gene (At3g25800) reported by Dekkers et al. (2012). The relative expression of the genes was calculated as the expression levels of the target genes minus the At3g2580 gene as the internal reference (ΔCt). Statistical analyses among the different time-points were performed using one-way ANOVA followed by the Tukey test (p value < 0.05). Comparison between non-primed and osmoprimed seeds at each time-point was performed by paired t test for means (*p < 0.05).
The RT-qPCR experiment followed the “Minimum Information for Publication of Quantitative Real-Time PCR Experiments” MIQE (Bustin et al. 2009).
Results
In silico analysis of cis-elements in AtEXPAs gene promoters
From all α-expansins in A. thaliana, we selected those whose expression levels were greater than or equal to 10 (signal intensity in Affymetrix microarray) in the Genevestigator database (Fig S1 and Fig S2). Although AtEXPA10 presented high expression during the germination, it was excluded from analysis due to its similar and constant expression profile in the remaining stages of plant development (Fig S2). AtEXPA11 and AtEXPA13 were also excluded from analysis because their higher expression peaks were in other developmental stage than seed germination, such as in rosette phase for AtEXPA11, and mature seed and for AtEXPA13 (Fig S2). Therefore, five AtEXPA that were preferentially expressed during germination (AtEXPA2, AtEXPA3, AtEXPA8, AtEXPA9 and AtEXPA20) were chosen for further analysis.
In silico promoter analyses of the 1 kb region upstream from the start codon of these five α-expansin genes (ProAtEXPAs), which are located at different positions on chromosomes 2, 4 and 5 of A. thaliana (Fig S4) were performed to support the choice of these isoforms for the mRNA expression experiment. More than 300 cis-element motifs were identified in the putative regulatory regions. Based on this initial analysis, we selected the cis-elements that have been previously reported in literature to be related to seed germination. Most of the motifs found are related to the response to GA, ABA, light and seed specific (Table 2). We mapped the frequencies of identified putative cis-elements related to seed germination onto the selected ProAtEXPAs using Pattern Matching in RSA-Tools (Fig. S5). The most frequent seed specific elements were DOF in ProAtEXPA9 (21 copies), CAATBOX1 in ProAtEXPA20 (11 copies), POLASIG1 in ProAtEXPA9 (7 copies) and EBOXBNNAPA in ProAtEXPA2 (7 copies), even though being found in all ProAtEXPAs studied in this work.
AtEXPAs gene expression patterns in non-primed and osmoprimed seeds during germination
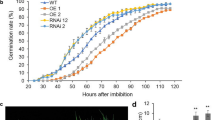
Our results are in agreement to what is usually expected for seeds of various plant species after osmoconditioning (Fig. 1). Time needed for the primary root emission decreased from 36 to 6 h in osmoprimed seeds as compared to non-primed seeds (Fig. 1). From day 6, the germination of primed seeds progressed gradually and linearly for 54 h and plateau at approximately 66 h after the seeds were placed on wetted filter paper. Moreover, ~ 14-fold more primed seeds than non-primed ones were germinated after 42 h of imbibition (Fig. 1). On the other hand, the radical protrusion of the non-primed seeds was only detected after 36 h, followed by a rapid increase in the germination rate, almost leveling at approximately 72 h after the beginning of the experiment (Fig. 1).
The results presented in Table 3 show that the osmopriming treatment had no effect on the maximum percentage germination as the values of this parameter were similar in seeds subjected to the PEG treatment and dry non-primed seeds (control). However, the time required for germination of 10% and 50% of seeds was ~ 3.0 and 1.7 times shorter in PEG treated seeds compared with the non-primed seeds and the mean germination time was ~ 50% higher in response to osmopriming (Table 3). The time interval between 25 and 75% of viable seeds to germinate (U75–25) was 2.5-fold longer for osmoprimed seeds compared to the control seeds, showing that osmopriming caused less uniform seed germination (Table 3). Overall, the AUC measurements, which integrates the parameters Gmax, t50 and U7525, demonstrated that PEG-treated seeds had better performance (~ 23% higher AUC values) than non-primed seeds.
We next examined the effect of osmopriming on the expression pattern of α-expansins, RT-qPCR analyses were performed using germinating osmoprimed and non-primed A. thaliana seeds collected at the same germination time-points. We first analyzed the expression profile of the five selected α-expansins in seeds subjected to the germination process following different water soaking periods using the in silico data available in the BAR eFP database (Fig S3). The peak of transcript accumulation for the five selected AtEXPAs was observed when the seeds were soaked in water for at least 12 h. Among all α-expansins studied, AtEXPA8 and AtEXPA9 showed the highest mRNA expression level at 12 and 24 h after imbibition, respectively (Fig S3).
Corroborating the data obtained from in silico analysis, the RT-qPCR analysis in different time-points post-germination demonstrated that the AtEXPAs displayed high levels of expression during germination of dry non-primed seeds. Here, the peak of transcription activity for the majority of the AtEXPAs was observed at later time (30 h) than reported by microarray analysis (Fig S3). This difference is probably due to the different temperatures used during seed imbibition (in our case 15 °C). From the five AtEXPA selected in our study, the AtEXPA2 reached the highest expression at 30 h, exhibiting a fold change of ~ 690 compared with 0 h time point (Fig. 2a). After this sampling-point, all AtEXPAs were progressively down-regulated, with exception of AtEXPA20 (Fig. 2d). AtEXPA3 showed the highest decline after 30 h of water imbibition (Fig. 2b).
Relative expression of AtEXPA genes in non-primed and osmoprimed seeds of A. thaliana collected at different germination time-points. The results are expressed as relative expression levels of the target genes with At3g2580 as the internal reference (ΔCt). Values represent mean (± SD) calculated from three biological replicates per sampling time-point. Statistical analysis among the different time-points was performed using one-way ANOVA followed by the Tukey test (p < 0.05). Lower- and upper-case letters indicate comparisons at different time-points in non-primed and osmoprimed seeds, respectively. Comparison between non-primed and osmoprimed seeds at each time-point was performed by paired t test for means (*p < 0.05)
As observed in Fig. 2, the most striking difference between osmoprimed and non-primed seeds was the level of the AtEXPAs transcripts at the 18-h time point of water soaking initiation. At this point, a significant amount of AtEXPA mRNAs were already detected in the PEG-treated seeds. In the case of the AtEXPA2, the highest-expressed isoform, a further increase in the number of transcripts was observed at 30 h post-imbibition, with expression levels rapidly decreasing in subsequent time points. Similar to the non-primed seeds, the mRNA levels of genes AtEXPA8 and AtEXPA9 were gradually decreased throughout the germination period, while the AtEXPA20 was the least expressed gene in all sampling points.
Discussion
Analysis of cis-elements in AtEXPA promoters
The regulation of gene expression of various biological processes, including germination, usually depends on the type, number, position and combination of regulatory elements present in and around the promoter (Hernandez-Garcia and Finer 2014). Therefore, the identification of such cis-elements in EXPA genes help us to assess the specificity of the selected isoforms prior to experimental verification.
Among the cis-elements implicated in control of seed germination found in the ProAtEXPAs, EBOXBNNAPA (Stålberg et al. 1996), DOF (Yanagisawa and Schmidt 1999), POLASIG1 (O’Neill et al. 1990), A-box motif (ACGTABOX) (Toyofuku et al. 1998), CAATBOX1 (Wenkel et al. 2006) and − 300 element (Colot et al. 1987) were present in all sequences analyzed. DOF (AAAG) was the most frequent motif in the α-expansin promoters. A strong enrichment of motifs associated with DOF-type transcription factors was observed in the promoters coding for the DELLA protein RGL2 (one of the major negative regulators of gibberellin signaling), which inhibits seed germination by hindering the transcription of EXPA3 and EXPA8 (Stamm et al. 2012).
Several light-responsive cis-elements are present in the AtEXPA promoters analyzed. Among them, the elements GATA (Gilmartin et al. 1990), GT1 (Terzaghi and Cashmore 1995) and SORLIP1AT (Jiao et al. 2005) were found in all five ProAtEXPAs. These results can be explained by the fact that the luminosity acidifies the cell wall (Elzenga et al. 2000) and that cellular acidification is a pre-requisite for the activity of the expansins (Cosgrove 1997).
Expansin isoforms exhibit different expression patterns according to external regulatory stimuli and response to plant growth regulators (Karaaslan and Hrazdina 2010; Cosgrove 2015; Lamichaney and Katiyar 2017). Motifs related to gibberellin were identified in all analyzed EXPA promoters. For example, GAREOSREP1, which is expressed in the aleurone cells of rice seeds (Sutoh and Yamauchi 2003), was found in ProAtEXPA9. The GARE motif (TAACAGA) (Ogawa et al. 2003) was present in ProAtEXPA2 and 3, and the element MYBGAHV in ProAtEXPA2 which is a central component in the gibberellin response complex (GARC) (Olszewski et al. 2002).
The presence of several ABA motifs in all the AtEXPAs promoters analyzed might be related to the secondary responses of the seeds to light and desiccation stress tolerance. The cis-elements W box (Xie et al. 2005) and PYRIMIDINE box (GA repressor) (Sutoh and Yamauchi 2003) were identified in all five ProAtEXPAs analyzed. Various motifs related to this phytoregulator were also present in ProAtEXPAs, as for example, the CBFHV (Xue 2002) and YACGTGGC (Hattori et al. 2002), related to dehydration and the ABA response, the DPBFCOREDCDC3, usually embryo-specific (Finkelstein and Lynch 2000) and the binding site for the transcription factor MYB (CNGTTR) (Abe et al. 2003).
ABA synthesis in imbibed seeds is required for maintaining seed dormancy and preventing precocious germination (Nambara et al. 2010). On the other hand, during germination, GA acts as an inducer of transcription of cell wall loosening protein genes, such as expansins and pectin methyl esterases (PMEs) (Ogawa et al. 2003). Thus, the balance of the antagonistic effects of GA/ABA might be involved in the expression patterns of the α-expansins during the osmopriming treatment. Indeed, Seo et al. (2006) showed that the balance of these two hormones affects the expression of AtEXPA2.
Expression of EXPAs genes in seeds A. thaliana during post-priming
Seed priming often involves restriction of the imbibition time and is used to improve seed performance including germinability, vigor and stress tolerance. In this study, osmopriming induced earlier (without an apparent lag phase) and faster germination, but less uniform, compared with the non-primed seeds at 15 °C (Fig. 1; Table 3). The higher values for AUC of the PEG-treated seeds indicates that the earlier germination and higher germination rate compensate for the asynchronous germination, which can be associated with changes in thermal time requirement and length of the priming treatment (Elkoca et al. 2007). As it has been pointed out, modifications of mRNA levels and protein accumulation, occur during osmopriming and post-priming germination (Gallardo et al. 2001; Soeda et al. 2005; Galland et al. 2014; Kubala et al. 2015). Therefore, it is expected that mRNA levels of several genes, including those involved in the relaxation of primary cell walls (such as the EXPAs), should be distinctively modulated in primed seeds during the germination. Previous studies have demonstrated that EXPAs play distinct tissue-specific and temporal functions during seed germination (Chen and Bradford 2000; Chen et al. 2001). The in silico expression analysis of the five selected AtEXPAs gene using the Arabidopsis eFP browser (Winter et al. 2007; Fig S3) suggests enhanced expression in seeds after water imbibition. Here, we were able to compare the transcript levels of AtEXPAs in dried osmoprimed seeds with non-primed seeds during the initial stages of germination by RT-qPCR. This analysis confirmed the increased expression levels of all five AtEXPAs during the early phases of seed germination and, in general, that the transcripts abundance mostly increased until circa 30 h of water imbibition for both non-primed and osmoprimed seeds (Fig. 2). Consistent with the microarray data (Winter et al. 2007; Fig S3), AtEXPA 2, 3, 8 and 9, with emphasis for the first isoforms, were the most expressed genes in non-primed germinating seeds (Fig. 2). In fact, based on gene expression studies, it has been proposed the use of AtEXPA2 as a molecular marker for specific tissues of non-dormant or germinating A. thaliana seeds (Penfield et al. 2006; Yan et al. 2014).
The primed seeds presented high AtEXPAs mRNA levels at 18 h of water imbibition for all AtEXPAs but AtEXP20, showing that transcripts of all five EXPA isoforms are produced in more quantities earlier than non-primed seeds due to the osmopriming treatment. Specifically, the higher fold changes comparatively to non-primed seeds were observed for AtEXPA8 and 9 at 18 h when elevated levels of transcripts (almost 2.7-fold) were already detected (Fig. 2). Together with AtEXPA2, the mRNAs of these EXPA were highly detected in the endosperm cap prior to the onset of endosperm weakening process that leads to testa rupture (Penfield et al. 2006, Holdsworth et al. 2008; Morris et al. 2011). The AtEXPA3, reported to be specifically expressed in the radicle (Morris et al. 2011), showed a great decline in expression after 30 h imbibition (~ 60-fold) with progression of the germination process (late germination phase and endosperm rupture) (Fig. 2b).
Despite the great difference in mRNA abundance at the 18 h time point, the osmoprimed and non-primed seeds presented similar gene expression levels of AtEXPAs at 0 and 6 h post-germination (Fig. 2). This equalization in expression indicates that the de novo transcription of the studied EXPAs at the very beginning of water imbibition does not reflect in a faster germination rate of the primed seeds (Fig. 1). In addition to the AtEXPA mRNAs triggered by the applied osmopriming procedure (PEG treatment and post-priming drying), we cannot discard the possibility that active α-expansins accumulated during the 7 days of osmopriming treatment may have had some effect on the earlier germination times observed in the osmoprimed seeds (Fig. 1). Seed treatment with a low osmotic potential PEG-solution during several days, as our case, can be considered a water stress situation. It is well known that EXPAs are up-regulated in response to water deficit condition (Cosgrove 2015; Marowa et al. 2016); hence, the accumulation of AtEXPAs may have occurred under water restriction conditions caused by the PEG treatment during the osmotic stress or even during the post-priming drying. In cucumber hypocotyl cell walls, expansins were detected at very low concentrations under normal conditions (McQueen-Mason and Cosgrove 1995). It is well known that transcriptional regulation of gene expression is a major mechanism used by plants for adaptation in changing environments. However, complex processes exert profound influence on the modulation of the final protein level. As such, further detailed studies on post-transcriptional and post-transductional regulation from a more specific perspective are necessary to fully assess the role of EXPAs during short or long treatments with different PEG concentration, either during and after water imbibition in osmoprimed seeds compared to the non-primed ones. Finally, our results are consistent with the knowledge that the osmopriming process (PEG treatment and post-priming drying) acts as a strong activator in the germination process (Heydecker and Gibbins 1978; Bray 1995) evoking stronger and earlier expression of the AtEXPA genes associated with the acquisition of germination capacity. Given the high mRNA levels of the α-expansin isoforms (AtEXPA2, 3, 8 and 9), this observation suggests that these genes may serve as markers of the earlier stages of germination and priming efficiency in Arabidopsis. Also, as plant promoters tend to respond to several transcription factors and are consequently activated by more than one pathway, the biological significance of the identified cis-acting elements occurring onto the promoters of up-regulated AtEXPAs and their effective involvement in different stages and tissues should be further investigated in non-primed and osmoprimed seeds.
References
Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Physiol 15:63–78
Abid M, Hakeem A, Shao Y, Liu Y, Zahoor R, Fan Y, Suyu J, Ata-Ul-Karim ST, Tian Z, Jiang D, Snider JL, Dai T (2018) Seed osmopriming invokes stress memory against post-germinative drought stress in wheat (Triticum aestivum L.). Environ Exp Bot 145:12–20
Baskin JM, Baskin CC (1972) Ecological life cycle and physiological ecology of seed germination of Arabidopsis thaliana. Can J Bot 50:353–360
Bewley JD, Bradford K, Hilhorst H, Nonogaki H (2013) Seeds: physiology of development, germination and dormancy. Springer, Berlin
Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Görlach J (2001) Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell 13:1499–1510
Bradford KJ (1986) Manipulation of seed water relations via osmotic priming to improve germination under stress conditions. HortScience 21:1105–1112
Bray CM (1995) Biochemical processes during the osmopriming of seeds. In: Kigel J, Galili G (eds) Seed development and germination. Marcel Dekker, New York, pp 767–789
Budzinski IGF, dos Santos TB, Sera T, Pot D, Vieira LGE, Pereira LFP (2011) Expression patterns of three α-expansin isoforms in Coffea arabica during fruit development. Plant Biol 13:462–471
Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT (2009) The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55:611–622
Caderas D, Muster M, Vogler H, Mandel T, Rose JKC, McQueen-Manson S, Kuhlemeier C (2000) Limited correlation between expansin gene expression and elongation growth rate. Plant Physiol 123:1399–1414
Chen K, Arora R (2013) Priming memory invokes seed stress-tolerance. Environ Exp Bot 94:33–45
Chen F, Bradford KJ (2000) Expression of an expansin is associated with endosperm weakening during tomato seed germination. Plant Physiol 124:1265–1274
Chen F, Dahal P, Bradford K (2001) Two tomato expansin genes show divergent expression and localization in embryos during seed development and germination. Plant Physiol 127:928–936
Chen K, Fessehaie A, Arora R (2012) Dehydrin metabolism is altered during seed osmopriming and subsequent germination under chilling and desiccation in Spinacia oleracea L. cv. Bloomsdale: possible role in stress tolerance. Plant Sci 183:27–36
Cho H, Cosgrove DJ (2002) Regulation of root hair initiation and expansin gene expression in Arabidopsis. Plant Cell 14:3237–3253
Cho H, Kende H (1997) Expansins and internodal growth of deepwater rice. Plant Physiol 113:1145–1151
Colot V, Robert LS, Kavanagh TA, Bevan MM, Thompson RD (1987) Localization of sequences in wheat endosperm protein genes which confer tissue-specific expression in tobacco. EMBO J 6:3559–3564
Cosgrove DJ (1997) Relaxation in a high-stress environment: the molecular bases of extensible cell walls and cell enlargement. Plant Cell 9:1031–1041
Cosgrove DJ (2015) Plant expansins: diversity and interactions with plant cell walls. Curr Opin Plant Biol 25:162–172
Dekkers BJW, Willems L, Bassel GW, van Bolderen-Veldkamp RPM, Ligterink W, Hilhorst HWM, Bentsink L (2012) Identification of reference genes for RT-qPCR expression analysis in Arabidopsis and tomato seeds. Plant Cell Physiol 53:28–37
El-Kassaby YA, Moss I, Kolotelo D, Stoehr M (2008) Seed germination: mathematical representation and parameters extraction. For Sci 54:220–222
Elkoca E, Haliloglu K, Esitken A, Ercisli S (2007) Hydro- and osmopriming improve chickpea germination. Acta Agric Scan Sect B Soil Plant Sci 57:193–200
Elzenga JTM, Staal M, Prins HBA (2000) Modulation by phytochrome of the blue light-induced extracellular acidification by leaf epidermal cells of pea (Pisum sativum L.): a kinetic analysis. Plant J 22:377–389
Finkelstein R, Lynch T (2000) The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12:599–609
Foti S, Cosentino SL, Patanè C, D’Agosta GM (2002) Effects of osmoconditioning upon seed germination of sorghum (Sorghum bicolor (L.) Moench) under low temperatures. Seed Sci Technol 30:521–533
Galland M, Huguet R, Arc E, Cueff G, Job D, Rajjou L (2014) Dynamic proteomics emphasizes the importance of selective mRNA translation and protein turnover during Arabidopsis seed germination. Mol Cell Proteomics 13:252–268
Gallardo K, Job C, Groot SPC, Puype M, Demol H, Vandekerckhove J, Job D (2001) Proteomic analysis of Arabidopsis seed germination and priming. Plant Physiol 126:835–848
Gilmartin PM, Sarokin L, Memelink J, Chua NH (1990) Molecular light switches for plant genes. Plant Cell 2:369–378
Goh HH, Sloan J, Dorca-Fornell C, Fleming A (2012) Inducible repression of multiple expansin genes leads to growth suppression during leaf development. Plant Physiol 159:1759–1770
Hattori T, Totsuka M, Hobo T, Kagaya Y, Yamamoto-Toyoda A (2002) Experimentally determined sequence requirement of ACGT-containing abscisic acid response element. Plant Cell Physiol 43:136–140
Hernandez-Garcia CM, Finer JJ (2014) Identification and validation of promoters and cis-acting regulatory elements. Plant Sci 217:109–119
Heydecker W, Gibbins BM (1978) The priming of seeds. Acta Hortic 83:213–223
Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database. Nucleic Acids Res 27:297–300
Holdsworth MJ, Bentsink L, Soppe WJJ (2008) Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol 179:33–54
Hussain S, Yin H, Peng S, Khan FA, Khan F, Sameeullah M, Hussain HA, Huang J, Cui K, Ni L (2016) Comparative transcriptional profiling of primed and non-primed rice seedlings under submergence. Front Plant Sci 7:1125
Jiao Y, Strickland E, Deng XW (2005) Conservation and divergence of light-regulated genome expression patterns during seedling development in rice and arabidopsis. Plant Cell 17:3239–3256
Joosen RV, Kodde J, Willems LA, Ligterink W, van der Plas LH, Hilhorst HW (2010) GERMINATOR: a software package for high-throughput scoring and curve fitting of Arabidopsis seed germination. Plant J 62:148–159
Karaaslan M, Hrazdina G (2010) Characterization of an expansin gene and its ripening-specific promoter fragments from sour cherry (Prunus cerasus L.) cultivars. Acta Physiol Plant 32:1073–1084
Khadraji A, Mouradi M, Houasli C, Qaddoury A, Ghoulam C (2017) Growth and antioxidant responses during early growth of winter and spring chickpea (Cicer arietinum) under water deficit as affected by osmopriming. Seed Sci Technol 45:198–211
Kubala S, Wojtyla Ł, Quinet M, Lechowska K, Lutts S, Garnczarska M (2015) Enhanced expression of the proline synthesis gene P5CSA in relation to seed osmopriming improvement of Brassica napus germination under salinity stress. J Plant Physiol 183:1–12
Kucera B, Cohn MA, Leubner-Metzger G (2005) Plant hormone interactions during seed dormancy release and germination. Seed Sci Res 15:281–307
Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1874
Lamichaney A, Katiyar PK (2017) Plant emergence and t50 responses of two chickpea cultivar differing in seed coat colour to PEG-osmopriming at sub-optimal temperature. Natl Acad Sci Lett 40:399–403
Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30:325–327
Lin C, Choi HS, Cho HT (2001) Root hair-specific EXPANSIN A7 is required for root hair elongation in Arabidopsis. Mol Cell 31:393–397
Marowa P, Ding A, Kong Y (2016) Expansins: roles in plant growth and potential applications in crop improvement. Plant Cell Rep 35:949–965
McQueen-Mason SJ, Cosgrove DJ (1995) Expansin mode of action on cell walls: analysis of wall hydrolysis, stress relaxation and binding. Plant Physiol 107:87–100
McQueen-Mason S, Durachko DM, Cosgrove DJ (1992) Two endogenous proteins that induce cell wall extension in plants. Plant Cell 4:1425–1433
Morris K, Linkies A, Müller K, Oracz K, Wang X, Lynn JR, Leubner-Metzger G, Finch-Savage WE (2011) Regulation of seed germination in the close Arabidopsis relative Lepidium sativum: a global tissue-specific transcript analysis. Plant Physiol 155:1851–1870
Muller B, Bourdais G, Reidy B, Bencivenni C, Massonneau A, Condamine P, Rolland G, Conéjéro G, Rogowsky P, Tardieu F (2007) Association of specific expansins with growth in maize leaves is maintained under environmental, genetic, and developmental sources of variation. Plant Physiol 143:278–290
Nakabayashi K, Okamoto M, Koshiba TT, Kamiya Y, Nambara E (2005) Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: epigenetic and genetic regulation of transcription in seed. Plant J 41:697–709
Nambara E, Okamoto M, Tatematsu K, Yano R, Seo M, Kamiya Y (2010) Abscisic acid and the control of seed dormancy and germination. Seed Sci Res 20:55–67
O’Neill SD, Kumagai MH, Majumdar A, Huang N, Sutliff TD, Rodriguez RL (1990) The alpha-amylase genes in Oryza sativa: characterization of cDNA clones and mRNA expression during seed germination. Mol Gen Genet 221:235–244
Ogawa M, Hanada A, Yamauchi Y, Kuwalhara A, Kamiya Y, Yamaguchi S (2003) Gibberellin biosynthesis and response during arabidopsis seed germination. Plant Cell 15:1591–1604
Olszewski N, Sun T, Gubler F (2002) Gibberellin signaling: biosynthesis, catabolism, and response pathways. Plant Cell 14:61–80
Özbingol N, Corbineau F, Groot SPC, Bino RJ, Côme D (1999) Activation of the cell cycle in tomato (Lycopersicon esculentum Mill.) seeds during osmoconditioning as related to temperature and oxygen. Ann Bot 84:245–251
Penfield S, Li Y, Gilday AD, Graham S, Graham IA (2006) Arabidopsis ABA INSENSITIVE4 regulates lipid mobilization in the embryo and reveals repression of seed germination by the endosperm. Plant Cell 18:1887–1899
Ramakers C, Ruijter JM, Deprez RH, Moorman AF (2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 339:62–66
Sano N, Kim J-S, Onda Y, Nomura T, Mochida K, Okamoto M, Seo M (2017) RNA-Seq using bulked recombinant inbred line populations uncovers the importance of brassinosteroid for seed longevity after priming treatments. Sci Rep 7:8095
Seo M, Hanada A, Kuwahara A, Endo A, Okamoto M, Yamauchi Y (2006) Regulation of hormone metabolism in Arabidopsis seeds: phytochrome regulation of abscisic acid metabolism and abscisic acid regulation of gibberellin metabolism. Plant J 48:354–366
Soeda Y, Konings MCJM, Vorst O, Van AMML, Houwelingen GM, Stiipen CA, Maliepaard J, Kodde RJ, Bino SPC (2005) Gene expression programs during Brassica oleracea seed maturation, osmopriming, and germination are indicators of progression of the germination process and the stress tolerance level. Plant Physiol 137:354–368
Stålberg K, Ellerstöm M, Ezcurra I, Ablov S, Rask L (1996) Disruption of an overlapping E-box/ABRE motif abolished high transcription of the napA storage-protein promoter in transgenic Brassica napus seeds. Planta 199:515–2199
Stamm P, Ravindran P, Mohanty B, Tan EL, Yu H, Kumar PP (2012) Insights into the molecular mechanism of RGL2-mediated inhibition of seed germination in Arabidopsis thaliana. BMC Plant Biol 12:179
Sun YY, Sun YJ, Wang MT, Li XY, Guo X, Hu R, Ma J (2010) Effects of seed priming on germination and seedling growth under water stress in rice. Acta Agron Sin 36:1931–1940
Sutoh K, Yamauchi D (2003) Two cis-acting elements necessary and sufficient for gibberellin-upregulated proteinase expression in rice seeds. Plant J 34:635–645
Terzaghi WB, Cashmore AR (1995) Light-regulated transcription. Annu Rev Plant Physiol 46:445–474
Thomas-Chollier M, Defrance M, Medina-Rivera A, Sand O, Hermann C, Thieffry D, Van Helden J (2011) RSAT 2011: regulatory sequence analysis tools. Nucleic Acids Res 39:W86–W91
Toyofuku K, Umemura T, Yamaguchi J (1998) Promoter elements required for sugar-repression of the RAmy3D gene for alpha-amylase in rice. FEBS Lett 428:275–280
Wenkel S, Turck F, Singer K, Gissot L, Le Gourrierec J, Samach A, Coupland G (2006) CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell 18:2971–2984
Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ (2007) An “electronic fluorescent pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2:e718
Xie Z, Zhang Z, Zou X, Huang J (2005) Annotations and functional analyses of the rice WRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells. Plant Physiol 137:176–189
Xue GP (2002) Characterisation of the DNA-binding profile of barley HvCBF1 using an enzymatic method for rapid, quantitative and high-throughput analysis of the DNA-binding activity. Nucleic Acids Res 30:e77
Yacoubi R, Job C, Belghazi M, Chaibi W, Job D (2011) Toward characterizing seed vigor in alfalfa through proteomic analysis of germination and priming. J Protome Res 10:3891–3903
Yamauchi Y, Ogawa M, Kuwahara A, Hanada A, Kamiya Y, Yamaguchi S (2004) Activation of gibberellin biosynthesis and response pathways by low temperature during imbibition of Arabidopsis thaliana seeds. Plant Cell 16:367–378
Yan A, Wu M, Yan L, Hu R, Ali I, Gan Y (2014) AtEXP2 is involved in seed germination and abiotic stress response in Arabidopsis. PLoS ONE 9:e85208
Yanagisawa S, Schmidt RJ (1999) Diversity and similarity among recognition sequences of Dof transcription factors. Plant J 17:209–214
Zhang W, Yan H, Chen W, Liu J, Jiang C, Jiang H, Zhu S, Cheng B (2014) Genome-wide identification and characterization of maize expansin genes expressed in endosperm. Mol Genet Genomics 289:1061–1074
Zhuo J, Wang W, Lu Y, Sen W, Wang X (2009) Osmopriming-regulated changes of plasma membrane composition and function were inhibited by phenylarsine oxide in soybean seeds. J Integr Plant Biol 51:858–867
Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol 136:2621–2632
Acknowledgements
We are grateful by the suggestions provided by Dr. Peter E. Toorop. All authors read and approved the final manuscript.
Author information
Authors and Affiliations
Contributions
LGEV and AFR conceived and designed the experiments. FLA, CCC and NBMN performed the germination and priming experiments. AFR and NVS carried out the bioinformatics analysis. AFR and TBS carried out the RT-qPCR assays and analyzed the data. LGEV, AFR and NVC wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
NVS has received a grant from São Paulo Research Foundation (FAPESP) and Coordination for the Improvement of Higher Education Personnel (CAPES) for the fellowship during her doctor degree and National Council for Scientific and Technological Development—CNPq for the fellowship during her master degree. TBS has received a grant from Coordination for the Improvement of Higher Education Personnel (CAPES) for postdoctoral fellowship. LGEV has received a grant from National Council for Scientific and Technological Development—CNPq for the research fellowship. AFR, FLA, CCC, NBM declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1
Heatmap produced by GENEVESTIGATOR (https://www.genevestigator.com) showing the expression pattern of 23 α-expansins during developmental stages of A. thaliana. The stages of development from seed germination to senescence are grouped arbitrarily based on Boyes et al. (2001). Data were retrieved from GENEVESTIGATOR (Zimmermann et al. 2004) (TIFF 1478 kb)
Fig. S2
Expression pattern of eight α-expansins during developmental stages of A. thaliana produced by GENEVESTIGATOR (https://www.genevestigator.com). The stages of development from seed germination to senescence are grouped arbitrarily based on Boyes et al. (2001). Data were retrieved from GENEVESTIGATOR (Zimmermann et al. 2004) (TIFF 1415 kb)
Fig. S3
Transcriptional profiles of five AtEXPAs during seed germination. The data were generated in the database BAR eFP Browser - http://bar.utoronto.ca/efp_arabidopsis/cgi-bin/efpWeb.cgi (Winter et al. 2007) - based on experiments for (A) non-dormant, non-stratified after-ripened wild-type seeds (Nakabayashi et al. 2005), (B) cold-stratified wild-type seeds (Yamauchi et al. 2004) (TIFF 586 kb)
Fig. S4
Distribution of the identified α-expansins genes on five chromosomes of A. thaliana. The chromosomal position of each gene was mapped according to the A. thaliana genome. The location of each gene was indicated by a line (TIFF 1576 kb)
Fig. S5
Putative cis-regulatory elements mapped onto the promoters of five EXPAs of A. thaliana up-regulated during seed germination. A Gibberellic acid motifs; B Abscisic acid/Abiotic stress motifs; C Light related motifs and D Seed related motifs. The size of each promoter region was one kb in relation to the translation start codon (ATG). A color key of cis-elements is given at the left of the image. Horizontal black lines in each block represent the promoter sequences and colored squares show the position of the mapped cis-element occurrences found in sense (above line) or anti-sense orientation (below line) (TIFF 1871 kb)
Rights and permissions
About this article
Cite this article
Ferreira Ribas, A., Volpi e Silva, N., dos Santos, T.B. et al. Regulation of α-expansins genes in Arabidopsis thaliana seeds during post-osmopriming germination. Physiol Mol Biol Plants 25, 511–522 (2019). https://doi.org/10.1007/s12298-018-0620-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12298-018-0620-6