Abstract
By promoting cell wall loosening, expansins contribute to cell enlargement during various developmental processes. Nevertheless, the role of expansins in the expansion and development of endosperm—a major seed component whose cell size is significantly associated with grain yield—is poorly understood. To explore associated biological processes and the evolution of expansins in maize, we performed a systematic analysis of the expansin gene family encompassing gene structure, phylogeny, chromosomal location, gene duplication, and gene ontology. A total of 88 maize expansin genes (ZmEXPs) were identified and categorized into three subfamilies according to their phylogenetic relationships. Expression patterns of ZmEXPs were also investigated in nine different tissues by semi-quantitative RT-PCR. The expression of eight ZmEXPs was detected in endosperm, with five showing endosperm-specific expression. Quantitative RT-PCR was used to analyze expression patterns of the eight ZmEXPs in endosperm (10 days after pollination) under abscisic acid (ABA) and gibberellic acid (GA3) treatments. All eight ZmEXPs were found to be significantly regulated by ABA and GA3 in endosperm, suggesting important roles for these hormones in the regulation of ZmEXPs during endosperm development. Our results provide essential information for ZmEXPs cloning and functional exploration, which will assist research on expansin-related mechanisms and contribute to future enhancement of maize grain yield.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Maize is one of the most important cereal plant crops, providing food for humans and animals as well as raw material for industry. A better understanding of endosperm development is vital for increased maize production, as endosperm is an important seed component (Guo et al. 2013) occupying 85 % of seed mass or volume (Lopes and Larkins 1993). Numerous studies have shown that expansin proteins have essential functions in plant developmental processes such as root growth, pollen tube elongation, and leaf expansion. Interestingly, several investigations have revealed that expression of expansins is associated with critical periods of seed (perisperm, endosperm and pericarp) development (Budzinski et al. 2011; Chen et al. 2001). For example, two expansins (CaEXPA1 and CaEXP3) in different isoforms (IAPAR-59 and IAPAR-59 Graúdo) participate in grain size regulation in Coffea arabica (Budzinski et al. 2011). Expansins may thus be important factors in the final determination of grain size (Lizana et al. 2010). To date, the roles and expression patterns of expansins in maize endosperm remain unclear. A clearer understanding of maize expansin gene family systematics and the roles of expansins in endosperm development would provide potentially beneficial insights for maize breeding.
Expansins are involved in plant cell enlargement and other developmental processes, such as endosperm expansion (Lizana et al. 2010), fruit softening, pollen tube growth, and root hair growth (Lee et al. 2001). The regulatory role of expansins in plant cell enlargement has been investigated in a number of plant species. In wheat, RNA in situ hybridization revealed that the TaExpA6 transcript was principally found in the endosperm and pericarp during early grain development. Expansin gene expression profiles were well correlated with the critical period of early grain expansion, suggesting that expansins might be a factor in the final determination of grain size in wheat (Lizana et al. 2010). Transgenic plants overexpressing OsEXPA8, a root-specific α-expansin gene in rice, showed pleiotropic phenotypes: improved root system architecture, increased plant height, and increased leaf number and size. Expansins were thus proposed to be required for cell enlargement and cell wall loosening in rice (Ma et al. 2013). In another study, ectopic expression of the expansin gene NtEXPA5 (Kuluev et al. 2013) increased leaf and stem sizes of transgenic tobacco plants. Further analysis revealed that the phenotypic changes were induced by cell enlargement (Kuluev et al. 2013). An increasing number of studies have demonstrated the involvement of expansin genes in cell growth of various plant species, including Arabidopsis (Goh et al. 2014), tobacco (Kuluev et al. 2013), rice (Ma et al. 2013), wheat (Zhao et al. 2012), and maize (Kapu and Cosgrove 2010; Valdivia et al. 2009).
Expansins were first identified through their involvement in the pH-associated mechanism of cell wall enlargement (Cosgrove 2000a; McQueen-Mason et al. 1992). These proteins are ubiquitous in cells throughout the plant world, ranging from green algae to land plants (Cosgrove 2000b). Expansin proteins include signal sequences and comprise two conserved domains (DPPB_1 and Pollen_allerg_1) (Yennawar et al. 2006) that cooperate during cell wall loosening (Sampedro and Cosgrove 2005; Yennawar et al. 2006). DPPB_1 domains share high homology with glycoside hydrolase family 45 (GH45) proteins (Sampedro and Cosgrove 2005), but do not possess enzymatic activity (Lee et al. 2001; Sampedro and Cosgrove 2005), presumably because of the highly specific and transient interaction between expansin and its substrate (McQueen-Mason and Cosgrove 1995). Pollen_allerg_1 domains share homology with group 2 grass pollen allergens, whose physiological roles remain unknown (Cosgrove 2005). No significant sequence similarity exists with any other proteins currently in GenBank. On the basis of conserved aromatic and polar residues on the protein surface, Pollen_allerg_1 domains are thought to bind polysaccharides (Cosgrove et al. 1997).
Low extracellular pH (4.5–6) (Cosgrove 2005) has long been known to activate expansin proteins, which then regulate plant cell enlargement by irreversibly loosening the cell wall (Sampedro and Cosgrove 2005). A hypothesized mechanism for this cell wall loosening under acidic conditions involves transient disruption of the non-covalent bond between hemi-cellulose and cellulose microfibrils by the activated expansins (Cosgrove 2005).
In the cell wall loosening process, expression of expansin genes is regulated by different plant hormones (Lee et al. 2001). These plant hormones exert their influence by stimulating the cell to modify the cell wall pH through control of the plasma membrane proton pump (H+-ATPase) (Cosgrove 2005) or by other mechanisms (Zhao et al. 2012). In Arabidopsis, AtEXPA2 is exclusively expressed in germinating seeds, suggesting its involvement in control of seed germination (Yan et al. 2014). During seed germination, AtEXP2 expression increases when seeds are treated with exogenous gibberellin (GA) (Yan et al. 2014). In wheat, treatment of coleoptiles with exogenous abscisic acid (ABA) enhances expansin expression levels, as ABA induces expansin activity (Zhao et al. 2012). Expansin genes have also been found to be responsive to auxin and ethylene (Cho and Cosgrove 2002; Ding et al. 2008; Kwon et al. 2008). These plant hormones (GA and ABA) have been shown to regulate key processes during seed development (Chen et al. 2014).
By suggesting a probable role for expansins in seed development, previous studies of expansin gene expression have set the stage for further research (Wu et al. 2001). The B73 maize genome sequencing project was completed in 2009 (Schnable et al. 2009), thereby providing an important resource for whole-genome annotation and classification and comparative genomics research. Although genome-wide studies have been reported on expansins in Arabidopsis, rice (Lee et al. 2001; Lee and Kende 2002), and some other species (Dal Santo et al. 2013), a systematic analysis of gene family members in maize has not been published to date. Furthermore, the function and expression pattern of expansins in maize endosperm development remains unclear. To detect expressed expansin genes, including those specifically expressed in endosperm, we consequently explored the expression pattern of maize expansin genes in different tissues. Expansin genes whose expression was affected by ABA and gibberellic acid (GA3) during endosperm development were identified, and their spatial expression patterns in endosperm were characterized. Our results should lead to further studies on expansin function in endosperm and should also be applicable to future efforts to improve maize yield.
Materials and methods
Identification and sequence analysis of expansin genes and proteins
Maize, rice, and Arabidopsis genome sequences were downloaded from the following databases: http://ftp.maizesequence.org/current/ (maize), http://rapdb.dna.affrc.go.jp/download/irgsp1.html (rice), and http://www.arabidopsis.org/browse/genefamily/index.jsp (Arabidopsis). Local genome and protein databases were constructed from these databases using the DNATOOLS program (Zhao et al. 2011b). To identify expansin proteins in these databases, a hidden Markov model (HMM) of the DPBB_1 domain (CL0199), based on the Arabidopsis AtEXP1 sequence, was used as a query in the Pfam database (http://pfam.sanger.ac.uk/). The maize genome was searched for sequences containing the DPBB_1 HMM using the BLASTp program (p = 0.001) (Lin et al. 2011). Pfam was then used to find additional genes encoding DPBB_1 and Pollen_allerg_1 domains, and the SMART program (http://smart.embl-heidelberg.de/) (Schultz et al. 2000) was used to confirm the presence of DPBB_1 domains. At this stage, all expansin gene family members were compiled for further analyses. Existing names of ZmEXPs were used (Table S4), and newly identified members were named according to Kende et al. (2004) (Kende et al. 2004).
Molecular weight and isoelectric point (PI) were calculated for all ZmEXPs using ExPASy (http://web.expasy.org/compute_pi/) (Gasteiger et al. 2005). The SignalP 4.1 server (http://www.cbs.dtu.dk/services/SignalP/) (Petersen et al. 2011) was used to predict the presence and location of signal peptide cleavage sites (Gan et al. 2011). Cellular roles and gene ontology (GO) categories were predicted using the ProtFun2.2 server (http://www.cbs.dtu.dk/services/ProtFun/) (Jensen et al. 2002, 2003), which relies solely on the protein sequence as input (Jain et al. 2008). A phylogenetic tree of the expansin family was constructed using the neighbor-joining method in MEGA 4.0 software (Tamura et al. 2007) with 1,000 bootstrap replicates. Subgroups were divided into classes based on bootstrap values ≥50 % (Peng et al. 2012). ClustalW was used to align gene and protein sequences (Thompson et al. 1994).
Using gene and cDNA sequences from the B73 genome database as inputs, GSDS (http://gsds.cbi.pku.edu.cn/) (Guo et al. 2007) was employed to predict gene structure. To analyze chromosomal locations, the start and end positions of each open reading frame were obtained from the genome database, with the MapInspect program (http://mapinspect.software.informer.com/) used to visualize this information (Cheng et al. 2012; Peng et al. 2012; Zhao et al. 2011b). Based on chromosomal location, gene clusters were identified that met specific clustering criteria, namely chromosome regions containing two or more homologous genes within 200 kb (Cheng et al. 2012; Holub 2001).
Phylogenetic and chromosomal location analyses were used to identify duplicated genes. For detection of tandem and segmental duplications, paralogs were regarded as tandemly duplicated genes separated by five or fewer gene loci according to the maize B73 genome annotation. Paralogs were designated as segmental duplicated genes if they were placed on duplicated chromosomal blocks as previously proposed by Wei et al. (2007) (Zhang et al. 2011; Zhao et al. 2011b). The number of nonsynonymous substitutions per nonsynonymous site (Ka) and synonymous substitutions per synonymous site (Ks) were calculated by DnaSPv5.0 (Librado and Rozas 2009; Rozas et al. 2003). To explore different selective constraints, Ks and Ka/Ks ratios for each pair of duplicated ZmEXP genes were calculated (Peng et al. 2012). Ratios of nonsynonymous to synonymous nucleotide substitutions (Ka/Ks) between paralogs were analyzed to determine the mode of selection (Peng et al. 2012).
To identify conserved motifs, all expansin protein sequences were submitted to the MEME program (http://meme.nbcr.net/meme/cgi-bin/meme.cgi) (Bailey et al. 2009) using the following parameters: optimum motif width ≥6 and ≤200, and maximum number of motifs = 20 (Lin et al. 2011). To predict hormone-responsive elements in promoter regions, 2-kb genomic sequences upstream of the transcription start site (ATG) were submitted to PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Lescot et al. 2002; Rombauts et al. 1999) and PLACE (http://www.dna.affrc.go.jp/PLACE/signalscan.html) (Higo et al. 1999; Peng et al. 2012). Four putative stress-related cis-elements were identified: ABA-responsive, GA-responsive, auxin-responsive, and ethylene-responsive elements.
Plant materials and treatments
The B73 maize line was used to investigate expression profiles. B73 plants were grown in the greenhouse at 28 ± 2 °C under a 14-h light/10-h dark cycle (Zhao et al. 2011b), and nine types of tissues were harvested at the following ages or stages: shoot (7 days), root (10 days), leaf (3 weeks), stem (6 weeks), ear (5–8 cm), silk (6–10 cm), tassel (8–12 cm), endosperm, and embryo (3 weeks after pollination). Tissues were frozen immediately in liquid nitrogen for at least 10 min and stored at −80 °C to preserve RNA for semi-quantitative reverse-transcription PCR (semi-RT-PCR). Plants were treated by spraying 100 nM ABA or GA3 (Fu et al. 2012; Guan et al. 1996; Kaya et al. 2006) on the leaf surfaces 10 days after pollination (Jin et al. 2013; Lu et al. 2013b), and endosperm tissue was harvested 0, 3, 6, 12, 24, and 48 h after treatment for quantitative real-time PCR (qRT-PCR).
RNA isolation and semi- and qRT-PCR
Total RNA was isolated from the stored samples using Trizol reagent (Invitrogen) and treated with DNase I (Takara). The first cDNA strand was synthesized using AMV reverse transcriptase (Promega), and the rTaq PCR system (Takara) was used for semi-RT-PCR. Primers were analyzed by Oligo 6 (Table S3A). Expression was normalized against the maize GAPDH housekeeping gene for semi-RT-PCR (Chen et al. 2012). Three biological and technical replicates were used for detection of expression patterns by semi-RT-PCR and subsequent qRT-PCR.
An ABI 7300 Real-Time System (Applied Biosystems) was used for qRT-PCR experiments. Primers were analyzed by Primer Express 3.0 (Applied Biosystems) (Table S3B). The first cDNA strand was synthesized using an M-MLV First Strand kit (Applied Biosystems). Reactions contained 2 µl of cDNA sample, 400 nM forward and reverse primers, 10 µl SYBR Select master mix, and sterile water to 20 µl. Thermal cycling was as follows: 50 °C for 2 min, 95 °C for 2 min, 40 cycles of 95 °C for 15 s, and 60 °C for 1 min. To analyze the specificity of each gene, melting curves were constructed by increasing the final temperature from 60 to 95 °C. The maize 18S rRNA gene was used as an internal control in the qRT-PCR experiments. Relative expression levels were calculated as 2 −ΔΔCT [where ΔC T = C T, target − C T, 18S and ΔΔC T = ΔC T, treatment − ΔC T, CK (0 h)]. Relative expression levels (2 −ΔΔC, CK (0 h)T ) in untreated control plants were normalized to 1 as described previously (Livak and Schmittgen 2001; Peng et al. 2012). Statistical analyses were performed using SDS 1.3.1 (Applied Biosystems).
Results
Identification of expansin genes in maize
Using the conserved amino acid sequences of DPBB_1 HMM (Pfam: CL0199) and Pollen_allerg_1 domains as queries in the BLASTp program, we identified 121 candidate expansin family members in maize. Pfam and SMART analyses were used to confirm the presence of the two domains in all putative expansins, as both domains are required for expansin activity (Yennawar et al. 2006). As a result, 88 expansin genes were confirmed (Table 1). The maize genome was also found to contain a greater number of expansin genes than those present in rice and Arabidopsis genomes (Table 1). Maize expansin genes were named consistently and compiled, and their sequence characteristics were analyzed (including sequence ID, number of residues, molecular weight, PI, signal sequence position, and cleavage site) (Table 2). Lengths of the 88 ZmEXPs ranged from 175 to 717 bp, with 61 ranging from 250 to 290 bp (Fig. S1a). Lengths of their signal sequences varied between 20 and 30 bp (Fig. S1b).
Phylogenetic analysis and evaluation of gene structure and conserved motifs
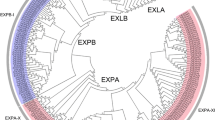
Based on the above analysis, full-length expansins sequences from maize and Arabidopsis were aligned to generate a phylogenetic tree (Fig. S2) for classification of maize expansin genes (Zhao et al. 2011a). According to the evolutionary relationships in this unrooted tree, the 88 ZmEXPs were divided into three subfamilies (Table 1): α-expansin (EXPA), β-expansin (EXPB), and expansin-like A (EXLA). To investigate evolutionary relationships between maize expansin protein family members, an unrooted phylogenetic tree was generated from aligned full-length sequences of all 88 ZmEXPs (Fig. 1a). In the resulting tree, EXPA and EXPB subfamilies contained 11 and 7 subgroups, respectively, with bootstrap support values >50 %. Eleven gene pairs were supported by bootstrap values >95 % (Fig. 1a). Comparative phylogenetic and exon–intron structural analysis (Fig. 1b) revealed that ZmEXP genes within each subgroup possessed similar gene structures. Most genes containing at least one intron were relatively short. Of the 88 ZmEXPs, 85 contained fewer than four introns (Fig. S3).
Besides containing two conserved domains (DPPB_1 and Pollen_allerg_1), expansin proteins carry several other functional domains. Twenty conserved motifs were identified in the identified ZmEXPs using the MEME program (Fig. S4, Table S1). All 88 ZmEXPs necessarily included some conserved motifs, although their types and locations varied. All proteins contained five motifs (motifs 1–5). Pfam analysis revealed that the DPPB_1 domain included motifs 1 and 3, while the Pollen_allerg_1 domain contained motifs 4 and 5. An additional motif, motif 2, was sited between the two domains DPPB_1 and Pollen_allerg_1.
Chromosomal locations and gene duplications
On the basis of gene start positions on each chromosome, the 88 maize expansin genes were found to be unevenly distributed on all 10 chromosomes except for chromosome 8 (Fig. 2). Chromosome 1 contained the largest number of maize expansin genes (23), whereas chromosome 4 contained the fewest (1). In addition, 18 gene clusters including 63 genes were detected on these chromosomes. We searched for evidence of past gene duplication events (tandem and segmental) to elucidate the mechanism(s) responsible for the ZmEXP gene family expansion that is believed to have occurred during the process of evolution. According to the phylogenetic analysis and the chromosomal distribution of the ZmEXP genes, five gene pairs involved in segmental duplication and five pairs involved in tandem duplication were identified (Table 3).
Chromosomal locations of ZmEXPs on nine chromosomes. Different markers represent different subfamilies. Chromosome numbers are shown on top of each vertical bar. Names on the left side of each chromosome correspond to the approximate location of each expansin gene. Genes involved in segmental duplication are joined by dashed lines, and the red rectangle indicates the gene cluster on each chromosome. The scale is in megabases (Mb) (color figure online)
To explore different selective constraints on the duplicated ZmEXP genes, we calculated Ks and Ka/Ks ratios for each duplicated pair. A Ka/Ks ratio less than 1 generally indicates accelerated evolution with positive selection, a ratio equal to 1 corresponds to neutral selection, and a ratio less than 1 indicates negative or purifying selection. Ka/Ks ratios of all 10 duplicated pairs were less than 1. Except for the duplicated pair ZmEXPL2–ZmEXPL4, ratios were less than 0.4, implying strong purifying selection (Table 3). These results suggest that functions of the duplicated genes have not diverged drastically over the course of genome evolution following the duplication events.
GO analysis
To predict ZmEXP biological functions, cellular roles and GO category annotations were explored using the ProtFun server. The majority of genes fell into the cell envelope functional category and GO categories of stress response and immune response (Fig. 3, Table S2), indicating their possible roles in these processes. In particular, 86 % of ZmEXPs were classified into the cell envelope functional category, and more than 43 % of ZmEXPs belonged to the stress response category.
Detected ZmEXPs expressed in endosperm
Expression patterns of the 88 ZmEXPs in nine different tissues were studied using semi-RT-PCR (Fig. 4). Of the 88 ZmEXPs, only 33 were expressed in any of the nine tissues; eight were expressed in endosperm. We observed that 25 genes were expressed in specific tissues (16 in tassels, 5 in endosperm, 2 in shoots, 1 in ears, and 1 in embryos), but no single gene was expressed in all nine tissues. Some genes were expressed in more than one tissue, such as ZmEXPA6 expressed in endosperm and embryos and ZmEXPA4 and ZmEXPA9 expressed in endosperm, embryos, and roots.
To explore the roles of ZmEXPs in endosperm development, qRT-PCR was used to further analyze expression patterns of the eight endosperm-expressed genes (ZmEXPA4, A5, A6, A9, A23, A26, ZmEXPB12, and ZmEXPB13) (Fig. 4). Many studies have revealed that expansin gene expression is regulated by GA3 and ABA treatment (Vogler et al. 2003; Wu et al. 2001). For example, increased expression of five α-expansin genes has been observed in rice internodes under GA treatment (Lee and Kende 2002). Expansin expression increases in plants under various abiotic stresses, as exemplified by the induction of RhEXPA4 promoter activity by ABA (Lu et al. 2013a). We accordingly harvested different stages of endosperm tissue 0, 3, 6, 12, 24, and 48 h after treatment (10 days post-pollination) with ABA or GA3. Although ZmEXPA9, ZmEXPA23, and ZmEXPA26 were not expressed in the untreated (0 h) endosperm sample, expression levels of all eight genes increased strongly after ABA or GA3 treatment (Fig. 5). Three hours after treatment with ABA or GA3, expression levels of all eight genes were noticeably higher than in the control. Expressions were much higher following treatment with ABA than with GA3, except that the 24-h expression level of ZmEXPB13 was higher after GA3 exposure. Most notably, ZmEXPA4 and ZmEXPA6 were strongly up-regulated (almost 100- and 200-fold, respectively) 3 h after ABA or GA3 treatment. These results demonstrate that expression levels of all eight ZmEXPs were modified by ABA or GA3 treatment.
Expression pattern analysis of eight endosperm-expressed ZmEXPs by qRT-PCR following ABA and GA3 treatments (normalized against maize 18S rRNA). Endosperm was sampled at 0 (no treatment), 3, 6, 12, 24, and 48 h following ABA or GA3 treatments 10 days after pollination. Black bar ABA treatment; gray bar GA3 treatment. Error bars show ±SE
Discussion
Maize is one of the world’s most important crop plants, and an increasing body of research has focused on improvement of maize yield. Because endosperm is a major seed component (Guo et al. 2013), regulation of endosperm cell size and volume is a key method for increasing crop yield. On the basis of previous studies, we conclude that expansins play crucial roles in endosperm development by stimulating the loosening of plant cell walls and thereby promoting plant cell enlargement (Cosgrove et al. 2002). The association of expansin expression with grain size dynamics has been reported in wheat (Lizana et al. 2010). In contrast to Arabidopsis and other species, however, a genome-wide analysis of the maize expansin gene family has not been reported, and the sequence characteristics and function of maize expansins remain unknown. A search for ZmEXPs possibly involved in endosperm development would provide essential information for cloning and functional research of candidate ZmEXPs that can be adopted in maize breeding programs to increase yield. In this study, we identified 88 expansins (ZmEXPs) in the maize genome using bioinformatics methods and investigated their sequence characteristics and evolutionary relationships. Importantly, expression profiles of ZmEXPs in nine different tissues were detected by semi-RT-PCR, allowing us to explore their functions in endosperm development. Previous research has shown that the expression of expansins is regulated by plant hormones such as ABA and GA3 (Nakaune et al. 2012; Oka et al. 2001; Yan et al. 2014). We therefore used qRT-PCR to investigate expression patterns of ZmEXPs showing endosperm-specific expression under ABA or GA3 treatment at 10 days after pollination (Xin et al. 2013). The qRT-PCR analysis not only confirmed some previously described features of this gene family but also identified some novel characteristics not reported in earlier studies.
To reconstruct the evolutionary history of maize expansins, we generated an unrooted phylogenetic tree of maize, Arabidopsis, and rice expansins (Fig. S5). These expansins were divided into four groups (EXPA, EXPB, EXPLA, and EXPLB) sharing a common origin in ancient organisms (Lee et al. 2001; Liu et al. 2011). None of the maize expansins were placed in the well-supported (>90 % bootstrap value) EXPLB subfamily in the phylogenetic tree, indicating that expansin-like A subfamily proteins have evolved more slowly in maize than in the other species. Although genetic relationships between proteins of the three species are very close, maize expansin genes are more closely related to those of rice than of Arabidopsis based on the prevalence of ZmEXP ortholog pairs in the phylogenetic tree. For example, ZmEXPs were more closely grouped with OsEXPs than with AtEXPs. This situation is likely due to the diversification of some clades of expansins that accompanied genome evolution following the monocot-dicot split, leading to sequence variation between their members (Peng et al. 2012; Zhao et al. 2011b).
Previous studies have demonstrated that gene duplication has been largely responsible for the expansion of gene families such as CCCH and HD-zip (Peng et al. 2012; Zhao et al. 2011b). In our study, five sister gene pairs of ZmEXPs were determined to be involved in segmental duplications, as deduced by shared phylogenetic clade combinations within the same groups and by their locations within segmentally duplicated blocks. An additional five sister gene pairs on the 10 chromosomes were determined to be involved in tandem duplications. We therefore conclude that segmental and tandem duplications are the main contributors to the diversification of the maize expansin gene family. Ka/Ks ratios calculated for the duplicated gene pairs were all less than 1, indicating that purifying selection may have played an important role in maize expansin gene family evolution (Peng et al. 2012).
Expansin-associated regulation of cell enlargement has been linked to a variety of developmental processes (Cosgrove 2005; Lee et al. 2001; Sampedro and Cosgrove 2005). Although many studies have addressed the function of expansins in numerous plant species, the involvement of ZmEXPs in endosperm development has not been reported. We therefore initiated this study to investigate the functions of maize expansins in future study. Information regarding their functions was gleaned from known functions of rice proteins (Jain et al. 2008) and by comparative analyses using the ProtFun program (Table S2). The majority of maize and rice expansins fell into the cell envelope functional category, although the exact percentages varied between species and proteins.
To further confirm the involvement of expansin genes in seed development (Wu et al. 2001), we analyzed their expression patterns. Fifty-five of the 88 ZmEXPs were not expressed in any of the nine tissues studied. The low proportion of expressed genes may indicate that the expression of some ZmEXPs is highly specific and limited to a single organ or cell type (Wu et al. 2001). Alternatively, expression levels of some genes may have been lower than the detection limit of this study. Nevertheless, eight ZmEXPs were observed to be expressed in endosperm, with five of them exhibiting endosperm-specific expression.
The above results provide important information for future research on the roles of expansins in endosperm. Although regulation of expansin expression by auxin, GA3, ABA, ethylene, and cytokine has been reported (Lee et al. 2001), no analyses have been conducted on hormone-responsive cis-elements of expansin gene families of any plant species. Our analysis of promoter regions revealed the presence of known ABA-, auxin-, GA-, and ethylene-responsive elements. Even more exciting, almost all of the 2-kb promoter sequences (Peng et al. 2012) of the 88 ZmEXPs contained GA3- or ABA-responsive elements (Fig. 6, Table S5). This result indicates that ABA and GA3 are the main hormone-responsive elements involved in ZmEXP expression. The eight ZmEXPs expressed in endosperm contained at least one GA3- or ABA-responsive cis-element in their promoter regions, strongly suggesting their involvement in two hormone-responsive regulatory pathways (Lee et al. 2001) and the two signaling pathways that regulate development. On the basis on these analyses, we postulated that the two hormones upregulate the genes via pH modification (Cosgrove 2005; Zhao et al. 2012). Both ABA and GA3 have been previously linked to pH regulation of seed development (Sampedro and Cosgrove 2005). To test this hypothesis, expressions of the eight ZmEXPs in endosperm under ABA and GA3 treatment were studied using qRT-PCR. All eight ZmEXPs were sharply upregulated after ABA or GA3 treatment. The sensitivity of these ZmEXP genes to ABA and GA3 treatment suggests important roles for ABA and GA3 in the regulation of the eight ZmEXPs during endosperm development and also indicates a potential role in the control of adaptive cell wall modification under treatment (Dal Santo et al. 2013). We plan to experimentally examine the biological functions of ZmEXPs in future studies (Fig. 6).
ZmEXP promoter region hormone-responsive elements. Cis-elements on sense and reverse strands are shown above and below black lines, respectively. ABA-responsive elements (ABRE), GA-responsive elements (GARE, P), auxin-responsive elements (TGA), and ethylene-responsive elements (ERE) are indicated by different markers
In conclusion, we identified and characterized expansin genes in maize by analyzing their structural diversity, chromosomal distribution, GO annotations, and phylogenetic relationships, and subsequently investigated their expression patterns. Eight ZmEXPs were observed to be expressed in endosperm, with five exhibiting endosperm-specific expression. Notably, almost all promoter regions of the eight ZmEXPs contained one or more GA3- or ABA-responsive elements. This finding, which was subsequently verified by qRT-PCR expression analysis of these genes under ABA or GA3 treatment in endosperm, suggests important roles for ABA and GA3 in the regulation of ZmEXPs during endosperm development. Our comprehensive and systematic analysis of the maize expansin gene family and expression patterns of ZmEXPs in endosperm development has provided essential information for further research on the functions of ZmEXPs in the endosperm development process. These findings should assist research into expansin-related mechanisms and aid efforts to improve maize yield.
References
Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37:W202–W208
Budzinski I, Santos T, Sera T, Pot D, Vieira L, Pereira L (2011) Expression patterns of three α-expansin isoforms in Coffea arabica during fruit development. Plant Biol 13:462–471
Chen F, Dahal P, Bradford KJ (2001) Two tomato expansin genes show divergent expression and localization in embryos during seed development and germination. Plant Physiol 127:928–936
Chen KT, Fessehaie A, Arora R (2012) Selection of Reference Genes for Normalizing Gene Expression During Seed Priming and Germination Using qPCR in Zea mays and Spinacia oleracea. Plant Mol Biol Rep 30:478–487. doi:10.1007/s11105-011-0354-x
Chen J, Lausser A, Dresselhaus T (2014) Hormonal responses during early embryogenesis in maize. Biochem Soc Trans 42:325–331
Cheng Y, Li X, Jiang H, Ma W, Miao W, Yamada T, Zhang M (2012) Systematic analysis and comparison of nucleotide-binding site disease resistance genes in maize. FEBS J 279:2431–2443
Cho HT, Cosgrove DJ (2002) Regulation of root hair initiation and expansin gene expression in Arabidopsis. Plant Cell 14:3237–3253. doi:10.1105/Tpc.006437
Cosgrove DJ (2000a) Loosening of plant cell walls by expansins. Nature 407:321–326
Cosgrove DJ (2000b) New genes and new biological roles for expansins Curr Opin Plant Biol 3:73-78
Cosgrove DJ (2005) Growth of the plant cell wall. Nat Rev Mol Cell Biol 6:850–861
Cosgrove DJ, Bedinger P, Durachko DM (1997) Group I allergens of grass pollen as cell wall-loosening agents. Proc Natl Acad Sci USA 94:6559–6564
Cosgrove DJ, Li LC, Cho H-T, Hoffmann-Benning S, Moore RC, Blecker D (2002) The growing world of expansins. Plant Cell Physiol 43:1436–1444
Dal Santo S, Vannozzi A, Tornielli GB, Fasoli M, Venturini L, Pezzotti M, Zenoni S (2013) Genome-wide analysis of the expansin gene superfamily reveals grapevine-specific structural and functional characteristics. PloS One 8:e62206. doi:10.1371/journal.pone.0062206
Ding X, Cao Y, Huang L, Zhao J, Xu C, Li X, Wang S (2008) Activation of the indole-3-acetic acid–amido synthetase GH3-8 suppresses expansin expression and promotes salicylate-and jasmonate-independent basal immunity in rice. Plant Cell 20:228–240
Fu J, Zhang DF, Liu YH, Ying S, Shi YS, Song YC, Li Y, Wang TY (2012) Isolation and characterization of maize PMP3 genes involved in salt stress tolerance. PLoS ONE 7:e31101
Gan D, Jiang H, Zhang J, Zhao Y, Zhu S, Cheng B (2011) Genome-wide analysis of BURP domain-containing genes in Maize and Sorghum. Mol Biol Rep 38:4553–4563
Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A (2005) Protein identification and analysis tools on the ExPASy server. In: Walker JM (ed) The proteomics protocols handbook. Humana Press, pp 571–607
Goh H-H, Sloan J, Malinowski R, Fleming A (2014) Variable expansin expression in Arabidopsis leads to different growth responses. J Plant Physiol 171:329–339. doi:10.1016/j.jblph.2013.09.009
Guan L, Polidoros AN, Scandalios JG (1996) Isolation, characterization and expression of the maize Cat2 catalase gene. Plant Mol Biol 30:913–924
Guo A-Y, Zhu Q-H, Chen X, Luo J-C (2007) GSDS: a gene structure display server. Yi chuan=Hereditas/Zhongguo yi chuan xue hui bian ji 29:1023
Guo X, Yuan L, Chen H, Sato SJ, Clemente TE, Holding DR (2013) Non-redundant function of zeins and their correct stoichiometric ratio drive protein body formation in maize endosperm. Plant Physiol 162:1359–1369
Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database. Nucleic Acids Res 27:297–300
Holub EB (2001) The arms race is ancient history in Arabidopsis, the wildflower. Nat Rev Genet 2:516–527. doi:10.1038/35080508
Jain M, Khurana P, Tyagi AK, Khurana JP (2008) Genome-wide analysis of intronless genes in rice and Arabidopsis. Funct Integr Genomics 8:69–78
Jensen LJ, Gupta R, Blom N, Devos D, Tamames J, Kesmir C, Nielsen H, Staerfeldt HH, Rapacki K, Workman C, Andersen CA, Knudsen S, Krogh A, Valencia A, Brunak S (2002) Prediction of human protein function from post-translational modifications and localization features. J Mol Biol 319:1257–1265. doi:10.1016/s0022-2836(02)00379-0
Jensen LJ, Gupta R, Staerfeldt HH, Brunak S (2003) Prediction of human protein function according to Gene Ontology categories. Bioinformatics 19:635–642. doi:10.1093/bioinformatics/btg036
Jin X, Fu Z, Ding D, Li W, Liu Z, Tang J (2013) Proteomic identification of genes associated with maize grain-filling rate. PloS One 8:e59353
Kapu NUS, Cosgrove DJ (2010) Changes in growth and cell wall extensibility of maize silks following pollination. J Exp Bot 61:4097–4107
Kaya C, Tuna AL, Alfredo AA (2006) Gibberellic acid improves water deficit tolerance in maize plants. Acta Physiol Plant 28:331–337
Kende H, Bradford K, Brummell D, Cho HT, Cosgrove D, Fleming A, Gehring C, Lee Y, McQueen-Mason S, Rose J, Voesenek LA (2004) Nomenclature for members of the expansin superfamily of genes and proteins. Plant Mol Biol 55:311–314. doi:10.1007/s11103-004-0158-6
Kuluev B, Safiullina M, Knyazev A, Chemeris A (2013) Effect of ectopic expression of NtEXPA5 gene on cell size and growth of organs of transgenic tobacco plants. Russ J Dev Biol 44:28–34
Kwon YR, Lee HJ, Kim KH, Hong SW, Lee H (2008) Ectopic expression of Expansin3 or Expansin beta 1 causes enhanced hormone and salt stress sensitivity in Arabidopsis. Biotechnol Lett 30:1281–1288. doi:10.1007/s10529-008-9678-5
Lee Y, Kende H (2002) Expression of alpha-expansin and expansin-like genes in deepwater rice. Plant Physiol 130:1396–1405. doi:10.1104/Pp.008888
Lee Y, Choi D, Kende H (2001) Expansins: ever-expanding numbers and functions. Curr Opin Plant Biol 4:527–532
Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30:325–327
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452
Lin Y-X, Jiang H-Y, Chu Z-X, Tang X-L, Zhu S-W, Cheng B-J (2011) Genome-wide identification, classification and analysis of heat shock transcription factor family in maize. BMC Genomics 12:76
Liu Y, Jiang H, Chen W, Qian Y, Ma Q, Cheng B, Zhu S (2011) Genome-wide analysis of the auxin response factor (ARF) gene family in maize (Zea mays). Plant Growth Regul 63:225–234
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408
Lizana XC, Riegel R, Gomez LD, Herrera J, Isla A, McQueen-Mason SJ, Calderini DF (2010) Expansins expression is associated with grain size dynamics in wheat (Triticum aestivum L.). J Exp Bot 61:1147–1157
Lopes MA, Larkins BA (1993) Endosperm origin, development, and function. Plant Cell 5:1383
Lu PT, Kang M, Jiang XQ, Dai FW, Gao JP, Zhang CQ (2013a) RhEXPA4, a rose expansin gene, modulates leaf growth and confers drought and salt tolerance to Arabidopsis. Planta 237:1547–1559. doi:10.1007/s00425-013-1867-3
Lu X, Chen D, Shu D, Zhang Z, Wang W, Klukas C, Chen LL, Fan Y, Chen M, Zhang C (2013b) The differential transcription network between embryo and endosperm in the early developing maize seed (1 C W OA). Plant Physiol 162:440–455. doi:10.1104/pp.113.214874
Ma N, Wang Y, Qiu S, Kang Z, Che S, Wang G, Huang J (2013) Overexpression of OsEXPA8, improves rice growth and root system architecture by facilitating cell extension. PloS One 8:e75997
McQueen-Mason SJ, Cosgrove DJ (1995) Expansin mode of action on cell walls (analysis of wall hydrolysis, stress relaxation, and binding). Plant Physiol 107:87–100
McQueen-Mason S, Durachko DM, Cosgrove DJ (1992) Two endogenous proteins that induce cell wall extension in plants. Plant Cell 4:1425–1433
Nakaune M, Hanada A, Yin Y-G, Matsukura C, Yamaguchi S, Ezura H (2012) Molecular and physiological dissection of enhanced seed germination using short-term low-concentration salt seed priming in tomato. Plant Physiol Biochem 52:28–37
Oka M, Tasaka Y, Iwabuchi M, Mino M (2001) Elevated sensitivity to gibberellin by vernalization in the vegetative rosette plants of Eustoma grandiflorum and Arabidopsis thaliana. Plant Sci 160:1237–1245
Peng XJ, Zhao Y, Cao JG, Zhang W, Jiang HY, Li XY, Ma Q, Zhu SW, Cheng BJ (2012) CCCH-type zinc finger family in maize: genome-wide identification, classification and expression profiling under abscisic acid and drought treatments. PLoS ONE 7:e40120
Petersen TN, Brunak S, von Heijne G, Nielsen H (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786
Rombauts S, Déhais P, Van Montagu M, Rouzé P (1999) PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res 27:295–296
Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R (2003) DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496–2497. doi:10.1093/bioinformatics/btg359
Sampedro J, Cosgrove DJ (2005) The expansin superfamily. Genome Biol 6:242. doi:10.1186/Gb-2005-6-12-242
Schnable PS, Ware D, Fulton RS et al (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326:1112–1115
Schultz J, Copley RR, Doerks T, Ponting CP, Bork P (2000) SMART: a web-based tool for the study of genetically mobile domains. Nucleic Acids Res 28:231–234
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Valdivia ER, Stephenson AG, Durachko DM, Cosgrove D (2009) Class B β-expansins are needed for pollen separation and stigma penetration. Sex Plant Reprod 22:141–152
Vogler H, Caderas D, Mandel T, Kuhlemeier C (2003) Domains of expansin gene expression define growth regions in the shoot apex of tomato. Plant Mol Biol 53:267–272
Wei F, Coe E, Nelson W, Bharti AK, Engler F, Butler E, Kim H, Goicoechea JL, Chen M, Lee S, Fuks G, Sanchez-Villeda H, Schroeder S, Fang Z, McMullen M, Davis G, Bowers JE, Paterson AH, Schaeffer M, Gardiner J, Cone K, Messing J, Soderlund C, Wing RA (2007) Physical and genetic structure of the maize genome reflects its complex evolutionary history. PLoS Genet 3:e123
Wu Y, Meeley RB, Cosgrove DJ (2001) Analysis and expression of the α-expansin and β-expansin gene families in maize. Plant Physiol 126:222–232
Xin M, Yang R, Li G, Chen H, Laurie J, Ma C, Wang D, Yao Y, Larkins BA, Sun Q, Yadegari R, Wang X, Ni Z (2013) Dynamic expression of imprinted genes associates with maternally controlled nutrient allocation during maize endosperm development. Plant Cell 25:3212–3227. doi:10.1105/tpc.113.115592
Yan A, Wu M, Yan L, Hu R, Ali I, Gan Y (2014) AtEXP2 is involved in seed germination and abiotic stress response in Arabidopsis. PLoS ONE 9:e85208
Yennawar NH, Li LC, Dudzinski DM, Tabuchi A, Cosgrove DJ (2006) Crystal structure and activities of EXPB1 (Zea m 1), alpha, beta-expansin and group-1 pollen allergen from maize. Proc Natl Acad Sci USA 103:14664–14671. doi:10.1073/pnas.0605979103
Zhang X, Feng Y, Cheng H, Tian D, Yang S, Chen J-Q (2011) Relative evolutionary rates of NBS-encoding genes revealed by soybean segmental duplication. Mol Genet Genomics 285:79–90
Zhao Y, Li X, Chen W, Peng X, Cheng X, Zhu S, Cheng B (2011a) Whole-genome survey and characterization of MADS-box gene family in maize and sorghum. Plant Cell Tiss Org (PCTOC) 105:159–173
Zhao Y, Zhou YQ, Jiang HY, Li XY, Gan DF, Peng XJ, Zhu SW, Cheng BJ (2011b) Systematic analysis of sequences and expression patterns of drought-responsive members of the HD-Zip gene family in maize. PloS One 6:e28488
Zhao M-R, Han Y-Y, Feng Y-N, Li F, Wang W (2012) Expansins are involved in cell growth mediated by abscisic acid and indole-3-acetic acid under drought stress in wheat. Plant Cell Rep 31:671–685. doi:10.1007/s00299-011-1185-9
Acknowledgments
We are grateful to the members of the Key Laboratory of Crop Biology of Anhui Province for their assistance in this study. This work was supported by the Key Project of Chinese National Programs for Fundamental Research and Development (2014CB138200), the Ph.D. Programs Foundation of the Ministry of Education of China (20103418120001), and the National Key Technologies R&D Program of China (2012BAD20B02-2).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by S. Hohmann.
W. Zhang and H. Yan have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
438_2014_867_MOESM4_ESM.tif
Fig. S4 Distribution of 20 putative conserved motifs in ZmEXPs (as identified using the MEME web server). Box size is not proportional to actual motif size. Colored boxes were ordered manually according to MEME results. Conserved residues and motif sizes are shown in Table S1 (TIFF 3435 kb)
438_2014_867_MOESM5_ESM.jpg
Fig. S5 Phylogenetic tree of maize, rice, and Arabidopsis expansin genes. Green, α-expansins (EXPA); black, β-expansins (EXPB); red, expansin-like (EXPL) (JPEG 4171 kb)
Rights and permissions
About this article
Cite this article
Zhang, W., Yan, H., Chen, W. et al. Genome-wide identification and characterization of maize expansin genes expressed in endosperm. Mol Genet Genomics 289, 1061–1074 (2014). https://doi.org/10.1007/s00438-014-0867-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-014-0867-8